Abstract
Myofibril elasticity, critical to muscle function, is dictated by the intrasarcomeric filament titin, which acts as a molecular spring. To date, the molecular events underlying the mechanics of the folded titin chain remain largely unknown. We have elucidated the crystal structure of the 6-Ig fragment I65–I70 from the elastic I-band fraction of titin and validated its conformation in solution using small angle x-ray scattering. The long-range properties of the chain have been visualized by electron microscopy on a 19-Ig fragment and modeled for the full skeletal tandem. Results show that conserved Ig–Ig transition motifs generate high-order in the structure of the filament, where conformationally stiff segments interspersed with pliant hinges form a regular pattern of dynamic super-motifs leading to segmental flexibility in the chain. Pliant hinges support molecular shape rearrangements that dominate chain behavior at moderate stretch, whereas stiffer segments predictably oppose high stretch forces upon full chain extension. There, librational entropy can be expected to act as an energy barrier to prevent Ig unfolding while, instead, triggering the unraveling of flanking springs formed by proline, glutamate, valine, and lysine (PEVK) sequences. We propose a mechanistic model based on freely jointed rigid segments that rationalizes the response to stretch of titin Ig-tandems according to molecular features.
Keywords: electron microscopy, poly-Ig tandem structure, small angle x-ray scattering, titin elasticity, x-ray crystallography
The striated muscle of vertebrate is characterized by a striking elasticity that allows it to store mechanical energy and stretch over twice its resting length without disrupting its structural integrity. At physiological amounts of stretch, most of the elastic response of the myofibril is generated by the intrasarcomeric titin filament (≈3.2 MDa, >1-μm length). This protein functions as a bidirectional spring that stretches and recoils during muscle function to return the myofibril to its resting length (1). The spring components of titin are located in its I-band fraction, which forms an elastic connection between the ends of the thick filaments and the Z-disk. Titin contains two main elastic components, a proline-rich PEVK (proline, glutamate, valine, and lysine) segment of up to 2,200 residues length and a poly-Ig array formed by up to 95 modules (2). Both segments straighten upon myofibril stretch developing a passive entropic tension in the sarcomere. Poly-Ig arrays extend at low force whereas PEVK-repeats unravel at higher load, with the combined action of both springs defining the mechanical stiffness of the sarcomere (1, 3). The importance of stretch–recoil control in muscle function is emphasized by the finely tuned composition of both poly-Ig and PEVK segments in titin, which through splicing undergo an extensive adaptation to the different physiological and pathological states of muscle.
The molecular basis of titin chain elasticity is currently unknown. The response to stretch of its I-band components, particularly poly-Ig arrays, has been extensively studied using nanotools and molecular simulations (refs. 1 and 4 and references within). These methodologies finger-print unfolding phenotypes determined by the secondary structure elements of the Ig fold (i.e., analyze events at the module level) but do not report on the behavior of the chain. Because domain unfolding is unlikely to drive titin elasticity at physiological loads (3, 5), elucidating the structure and dynamics of the titin chain (at a level higher than the Ig module) is crucial to establish its mechanistic principles. To date, atomic models of domain tandems from titin have been scarce and limited to the N-terminal Ig-doublet Z1Z2 (6, 7) part of the Z-disk, and A168–A170 (8) (subfragment in 9) located at the M-line near the C terminus of the filament. These tandems act as docking sites for sarcomeric binding partners and do not relate to I-band elasticity. In the current study, we report atomic models of poly-Ig tandems from the spring region of titin that can aid to elucidate the mechanistic principles of sarcomere elasticity.
Results
Atomic Structures of Poly-Ig Tandems from I-Band Titin.
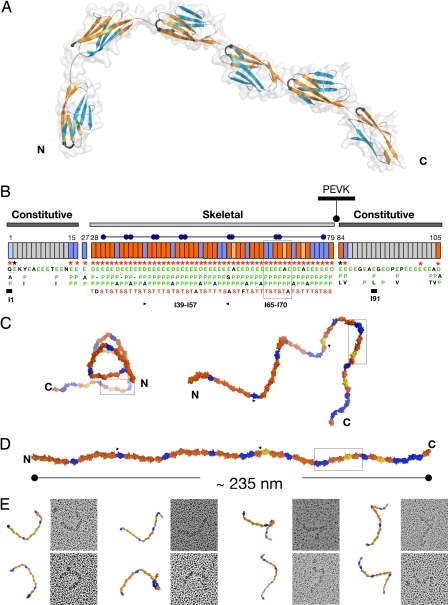
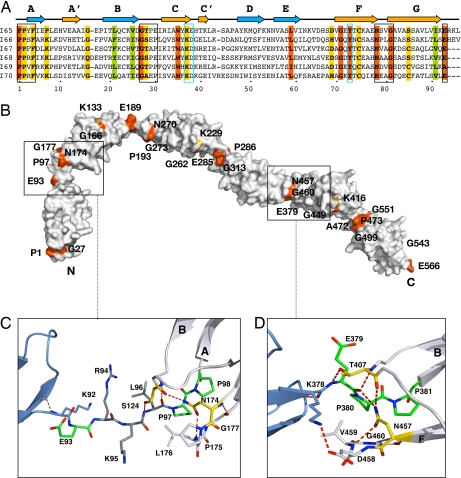
We have elucidated the crystal structure of a six-Ig fragment, I65–I70, from the skeletal I-band of soleus titin at 3.3-Å resolution [Fig. 1A and supporting information (SI) Movie 1]. X-ray data statistics and model parameters are given in Table 1. Its modules belong to the” N-conserved“ I-type of Ig folds (10), share a pairwise sequence identity of ≈35% and a high structural similarity [average rmsd 1.2 ± 0.1 Å for main chain atoms, calculated by using SPDBV (11)]. They are serially connected by linkers of diverse sequence composition and 0- to 3-residue length (Table 2). I65–I70 adopts a semiextended arrangement (≈21-nm end-to-end distance corresponding to ≈28-nm contour length) in L-shaped conformation, where the four C-terminal Ig lie almost perfectly coaxial, forming a straight section, and the N-terminal I65–I66 domains are bent away from the molecular axis, resulting in a frontal curvature of 114° (angle defined by the centers-of-mass of I65-I66-I67). Whereas domains in the linear fraction are joined tightly, the N-terminal Ig causing chain bending house longer three-residue linkers (Table 2). The long-range conformational definition of I65–I70 is unexpected because its contour length (≈28 nm) is about three times the persistence length estimated for this region of titin (≈9 nm) (12).
Fig. 1.
Structural order in the poly-Ig from I-band titin. (A) Crystal structure of I65–I70. β-sheets are color coded to emphasize domain torsions. The FG β-hairpin, which claps against the Ig–Ig transition motif EPP, is colored black. (B) Modular composition of the I-band of soleus titin from rabbit (N2A and N2B elements are omitted). Ig domains are represented as boxes, where orange indicates Ig tightly connected and blue represents Ig containing a C-terminal three-residue linker. Annotations refer to conserved features at the Ig–Ig interfaces, where (i) an FG β-hairpin containing an NxxG sequence is marked by red asterisks, (ii) interdomain EPP motifs in green are listed vertically under each domain (Ig exhibiting a natural E-to-A mutation in this motif are colored salmon), and (iii) the conserved S/T residue in the BC loop is shown in red. These features are characteristic of the skeletal but not of the constitutive Ig tandems. Super-repeats of 6 or 10 Ig are indicated by capped bars. Domains with previously known structure are marked with a thick bar. (C) Frontal (Left) and lateral (Right) views of a predicted model of the complete skeletal Ig-tandem in one of its putative slack conformations in solution as calculated from linker arrangements in I65–I70. (D) Model in extended conformation (C and D are color coded as in B). (E) EM images (70 × 70 nm2) of glycerol-sprayed/rotary shadowed I39–I57 accompanied by its corresponding model. The model (fragment indicated by arrows in C) has been oriented to match the micrographs, but no other manipulation has been applied (the 3D conformation of the I39–I57 model can be visualized in SI Movie 2).
Table 1.
Native x-ray data and model refinement statistics
| I65–I70 |
I67–I69 |
|||
|---|---|---|---|---|
| Spacegroup | P6522 | C2 | ||
| Unit cell dimensions | a = b = 141.43 Å, c = 166.01 Å | a = 86.68 Å, b = 86.29 Å, c = 44.70 Å | ||
| α = β = 90°, γ = 120° | α = 90°, β = 104.97°, γ = 90 | |||
| Solvent content, % | 67 | 52 | ||
| X-ray data | ||||
| X-ray source | ESRF ID-29–1 | ESRF ID-29–1 | ||
| Detector | ADSC Quantum Q210 | ADSC Quantum Q210 | ||
| Wavelength, Å | 0.9792 | 0.9792 | ||
| Resolution, Å | 17–3.3 | (3.35–3.30) | 19–1.6 | (1.62–1.60) |
| Unique reflections | 14,953 | (639) | 41,644 | (1,312) |
| Rsym (I) | 8.0 | (47.8) | 5.7 | (39.5) |
| Multiplicity | 6.84 | (6.83) | 4.57 | (3.75) |
| Completeness, % | 98.6 | (97.3) | 99.4 | (87.9) |
| I/σ (I) | 19.6 | (4.3) | 15.25 | (3.53) |
| Refinement | ||||
| No. of reflections in working/test set | 14,953/757 | 41,644/1,042 | ||
| No. of protein atoms/solvent molecules | 4,412/0 | 2,214/479 | ||
| R factor/Rfree, % | 22.0/26.7 | 17.1/21.6 | ||
| rmsd bond length, Å/bond angle, ° | 0.006/0.94 | 0.006/1.51 | ||
Values in parentheses correspond to the highest resolution shell.
Table 2.
Domain arrangements in the crystal structures of I65–I70 and I67–I69
*Linker is defined as those residues whose main chain is free from interactions. Residues within Ig domains are in bold. Domain boundaries are indicated by a hyphen. The conserved Ig-Ig transition pattern of skeletal I-band domains is boxed. The rabbit (but not human) sequence of I69–I70 hosts a natural E-to-A mutation at this point.
†Angle definition as in ref. 6.
‡Angle definition as in ref. 6.
§Distance between the centers of mass of individual domains.
¶Subscripts 6IG and 3IG refer to the crystal structure of I65–I70 and I67–I69, respectively.
‖Domain arrangement of Z1Z2 in solution as calculated from SAXS data and NMR residual dipolar couplings (6). It should be noted that Ig-doublets containing long linkers exhibit similar opening and torsion angles but differ in their swing angle.
The Ig domains in I65–I70 are finely ordered, showing regular intermodular spacings, hinge openings, and torsional angles (Table 2). The torsional pattern is particularly striking. The C-terminal modules I67–I70, joined by tight linkers, display relative torsions of ≈180° that result in an up-and-down domain alternation along the chain and give a straight appearance to this segment. In contrast, domains I65 and I66, connected by longer linkers to their successor Ig, consistently exhibit an ≈90° torsion that deviates the filament path from linearity. This regularity in modular parameters indicates the existence of an architectural order in the fine structure of the titin filament.
Conservation of Intermodular Transition Motifs.
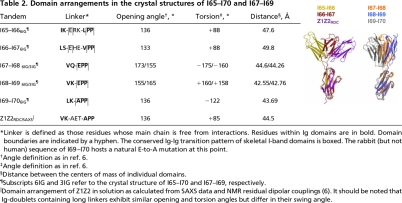
The I-band of skeletal-muscle titin (X90569) contains 52 specific Ig connected by either a tight linker or a three-residue hydrophilic sequence (Fig. 1B). The model of I65–I70 includes representatives of both types of domain interfaces, revealing the molecular principles of Ig arraying in the complete I-band tandem. The structure of I65–I70, however, was affected by partial crystallographic disorder in its I68–I69 subsection; thus, to analyze this region reliably, we elucidated independently the crystal structure of the three-Ig subfragment I67–I69 at 1.6-Å resolution (Table 1). Even though crystallization media and lattice parameters were unrelated, the conformation of I67–I69 agreed excellently with that of the equivalent fraction of I65–I70 (Table 2), confirming the well defined architecture of the tandem. In this array, tight modular transitions are mediated by a highly conserved EPP sequence motif N-terminal to each Ig domain (Figs. 1B and 2A). This motif packs against the groove formed by β-hairpin FG and the BC loop, which clap onto it via conserved N and S/T residues, respectively (Figs. 1A and 2D). In these tight connections, the regular torsional alternation of domains appears due to a combination of favorable steric orientation and interactions of the last residue of Igi, often a lysine, with the moderately variable positions of the NxxG motif of β-hairpin FG in Igi+1. The latter commonly contributes a small hydrophobic residue as well as an acidic group that leads to a salt bridge formation across domains (Fig. 2D). Tight Ig connections effectively result in a zero-length linker that can be predicted to sterically oppose the sharp bending and kinking of the titin chain.
Fig. 2.
Molecular features of I65–I70. (A) Structure-based sequence alignment where orange and yellow represent >90% and >70% sequence identity, respectively, across all Ig of the skeletal tandem. Green indicates conserved hydrophobic positions. The EPP motif, NxxG sequence in β-hairpin FG, and the BC loop are boxed in black. To ease comparison, the E group in tight linkers is given as the last residue of the preceding domain. A conserved set of residues (KD at the CC′ region and Y at β-strand F) responsible for the conformation of the CC′D loop characteristic of this Ig type is boxed in blue. (B) Molecular surface of I65–I70 colored according to sequence conservation as in A. (C) I65–I66 long linker interface. The three inserted residues are in dark gray. The conserved E is now an integral part of the linker, whereas L, the last of the inserted residues, has replaced it at the N terminus of the following Ig. (D) I68–I69 interface representative of tight connections. The transition motif EPP, an integral part of the N terminus of I69, is in green. The conserved N residue from β-hairpin FG and the T group from the BC loop are in yellow; their interactions are conserved in all Ig constituents of I65–I70. Hydrogen bonds are shown as dashed lines (experimental electron density for both linkers is shown in SI Fig 6).
Longer linker sequences, as those in I65-I66-I67, can be regarded as structural variants of the tight connections, where a three-residue insertion of variable sequence has occurred at E◇PP. The interactions of the PP motif with the FG and BC loops at Igi+1 are maintained, but the inserted residues as well as the conserved E group are now free from interactions (Fig. 2C). No direct Ig–Ig contacts are observed. Thus, long linkers could be expected to allow modular motions of large amplitude, probably acting as effective hinges in the chain. The Ig-doublet Z1Z2 from titin, which has been characterized recently through experiment (6) and simulation (13), contains a three-residue linker related to those of I65-I66-I67 (Table 2). At equilibrium in solution, the conformation of Z1Z2 is not random but it adopts a preferential, metastable semiextended state that resembles that of I65–I66 and I66–I67 in this study (Table 2). Nevertheless, its domain arrangement was shown to be adaptable, ranging from fully extended to compact “V”-shapes. It is conceivable that the conformational dynamics of long-linkers from I-band titin might approximate that of Z1Z2, thus introducing conformational freedom in the filament and permitting the pronounced bending of the chain at those points.
The conservation of EPP residues as Ig–Ig transition motifs is striking (Fig. 1B). To investigate the role of these residues in tandem conformation, the mutated variants I67–I69E93A and I67–I69P94A/P95I were studied using x-ray crystallography and EM, respectively (SI Text). This analysis revealed that, whereas the conserved glutamate is dispensable and does not influence significantly interdomain arrangement, the proline doublet is central to tandem architecture. The fact that variant I67–I69P94A/P95I had its solubility severely compromised and resulted in fibril formation (SI Fig. 5) indicated that the N-terminal PP motif acts as a gatekeeper of the Ig fold. It probably ensures the structural integrity of the tandem through stacking interactions that prevent fraying of β-strand A at domain interfaces.
Solution Structure of I65–I70 and Its Subfragments I67–I69 and I66–I69.
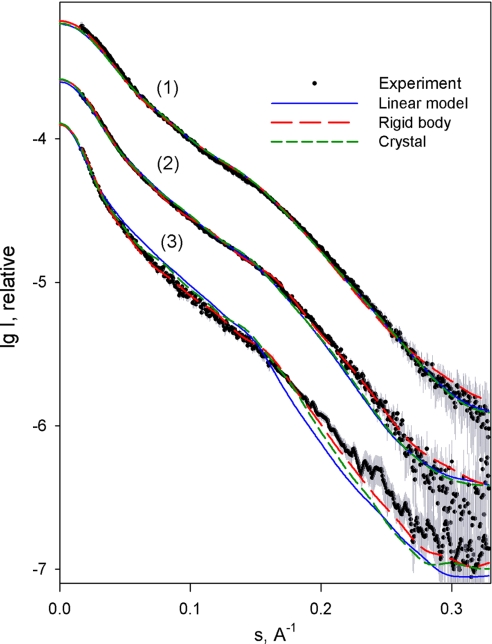
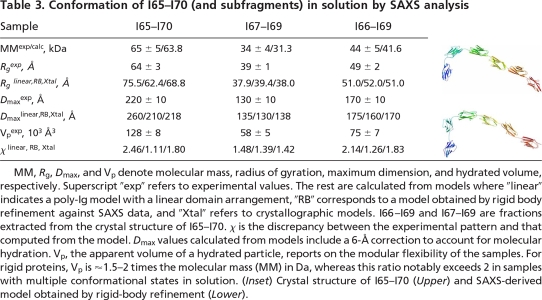
Given the potential flexibility of filamentous poly-Ig arrays and to ensure that crystalline conformations were not fortuitous, we investigated the structure of I65–I70 and its three- and four-Ig subfragments I67–I69 and I66–I69 in solution using small angle x-ray scattering (SAXS). Experimental scattering patterns are shown in Fig. 3, and calculated parameters are given in Table 3. A comparison of Rg and Dmax values with those calculated from theoretical linear models indicated that I67–I69 and I66–I69 are nearly linear in solution, whereas I65–I70 must be somewhat bent. Further, SAXS models of I65–I70 that were independently constructed ab initio using rigid-body refinement of six individual Ig by simultaneous fitting to scattering data from I65–I70, I67–I69, and I66–I69, were in remarkably good agreement with the crystal structure of I65–I70 (Table 3 Inset). Calculations on I67–I69 and I66–I69 yielded nearly linear models. Accordingly, the scattering patterns computed from the crystal structures agreed well with the experimental data (Fig. 3 and Table 3). These results suggest that the crystalline conformation of I65–I70 (and its subsections) is a good representative of this tandem in solution.
Fig. 3.
Scattering patterns of I65–I70, I67–I69, and I66–I69. Experimental scattering is displayed as dots with error bars. Patterns computed from linear, rigid-body, and crystallographic models are blue, red, and green, respectively. For better visualization, patterns are displaced along the logarithmic intensity scale. Patterns 1, 2, and 3 correspond to I67–I69, I66–I69, and I65–I70, respectively.
Table 3.
Conformation of I65–I70 (and subfragments) in solution by SAXS analysis
MM, Rg, Dmax, and Vp denote molecular mass, radius of gyration, maximum dimension, and hydrated volume, respectively. Superscript “exp” refers to experimental values. The rest are calculated from models where “linear” indicates a poly-Ig model with a linear domain arrangement, “RB” corresponds to a model obtained by rigid body refinement against SAXS data, and “Xtal” refers to crystallographic models. I66–I69 and I67–I69 are fractions extracted from the crystal structure of I65–I70. χ is the discrepancy between the experimental pattern and that computed from the model. Dmax values calculated from models include a 6-Å correction to account for molecular hydration. Vp, the apparent volume of a hydrated particle, reports on the modular flexibility of the samples. For rigid proteins, Vp is ≈1.5–2 times the molecular mass (MM) in Da, whereas this ratio notably exceeds 2 in samples with multiple conformational states in solution. (Inset) Crystal structure of I65–I70 (Upper) and SAXS-derived model obtained by rigid-body refinement (Lower).
In addition, we estimated the modular flexibility of these fragments by analyzing their apparent volume as a hydrated particle, Vp. For globular proteins, Vp in Å3 is ≈1.5–2 times the molecular mass (MM) in Da, but this ratio will noticeably exceed 2 if proteins adopt multiple conformations in solution (14). Vp values for all poly-Ig fragments in this study were 1.6–1.9 times their MM (Table 3), pointing to a restricted conformational variability in solution. This conclusion is in excellent agreement with a previous study of I65–I70 that, based on a distribution of end-to-end molecular distances measured in a sample population using EM, concluded that this tandem is a relatively stiff formation (10). Furthermore, the study estimated the average opening angle between any two consecutive Ig to be ≈143°, in close agreement with the average value of 146° in the current crystallographic model (Table 2). Taken together, SAXS and EM data suggest that the overall modular flexibility of I65–I70 is limited and that its conformation in solution closely resembles that of the crystal structure.
Model of the I-Band Tandem of Skeletal Titin.
The skeletal I-band of titin is arranged into domain super-repeats consisting of 6 or 10 Ig (15, 16) (Fig. 1B). The structure of I65–I70 (and its subfragment I67–I69) provide atomic data for representative domain and linker types of this section of titin and allow estimating the structural dynamics of the complete skeletal Ig-array. A constructed model suggests the presence of a supra-order in titin that results from a succession of modules joined through tight connections, interrupted at regular intervals by interspersed long linkers that alter the smooth “linearity” of the chain and lead to the formation of dynamic segmental units in the filament (Fig. 1C). Modules tightly connected are likely to be conformationally restrained, whereas Ig pairs joined by longer linkers can be expected to allow long-range intermodular motions and, thus, the bending of the chain. This conclusion is supported by EM images of a recombinant 19-Ig fragment, I39–I57, comprising three I-band super-repeats (Fig. 1E). Although all linkers might have a certain flexibility, images suggest that the shorter connections allow only moderate deformations whereas long linkers permit acute bending. This points to the existence of an architectural and dynamic supra-pattern in the filament caused by a regularity of molecular features. Speculatively, the inferred filament pattern might correspond to the observation of local pseudohelical substructure in EM (17) and atomic force microscopy (AFM) (18) studies of full-length titin molecules.
Segmental Flexibility as Mechanism of Titin Chain Elasticity.
These findings suggest that the elasticity of the skeletal I-band tandem is mediated by a regular pattern of domain pairs with two different bending stiffnesses, namely pairs that resist bending and have a tendency to retain an extended conformation and pairs that permit complex interdomain motions of large amplitude as observed in the doublet Z1Z2 (6). The stretch and recoil of the titin chain could then be expected to first induce conformational changes in the long permissive linkers, likely to act as weak “flexor” points. The deformation of these few points in the filament would be sufficient to achieve a complete extension of the chain (Fig. 1D). This implies that the entropic properties of titin poly-Ig are not homogeneous but that a regular distribution of variably entropic points exists along the filament resulting in a segmental distribution. We term this concept of dynamic super-repeats, where relatively rigid segments are intercalated with pliant connectors, the “carpenter ruler” model of titin elasticity.
Based on this concept, the elasticity of this poly-Ig tandem should be expressed in terms of a discrete formalism where the chain is represented by freely jointed rigid-segments (FJC) and not through continuous models such as the worm-like chain (WLC) form currently applied to this system (e.g., refs. 3 and 18–20) that assumes a smoothly flexible chain of random conformation and homogeneous composition. According to the segmental FJC model, the skeletal poly-Ig tandem of titin (comprising 52 Ig) can be described as containing 15 segments. On average, each segment consists of 3.5 Ig domains of 4.5-nm length each, resulting in an average segment length of ≈15.6 nm. For an FJC, the mean square end-to-end distance 〈R2〉FJC is defined as 〈R2〉FJC = n·l2, where n is the number of bonds (i.e., segments) and l is the average bond length (21). For this tandem (n = 15, l = 15.6 nm), 〈R2〉FJC = 3,650 nm2. This value allows now calculating the persistence length λ of a WLC with a contour length L of 52·4.5 = 234 nm. For a WLC in 3D equilibrium, 〈R2〉, L and λ are related by 〈R2〉WLC = 2λL[1 − (λ/L)(1 − e−L/λ)] (21). The persistence length so estimated is 8 nm, which is in excellent agreement with experimental values for this section of titin (≈9 nm) (12). Thus, our model, which now incorporates primary molecular information, explains overall statistical parameters currently available for the titin chain. It also shows that a chain can have fairly long stiff segments and be highly flexible as long as large freedom of motion is possible at discrete points. This conclusion emphasizes the fact that great care must be taken when attempting to interpret statistical concepts of polymer dynamics in terms of structural features of a chain.
Mechanistically, it could be speculated that the dominant role of pliant linkers is to permit the molecular shape variations that take place in the filament at moderate stretch (this being the regime where the end-to-end chain distance is much smaller than the contour length of the folded tandem). Thus, they primarily contribute conformational entropy to the system. Compared with the commonly used WLC model, which overestimates molecular shape contribution by assuming a continuous chain bending where entropic force arises from a narrowing in the distribution of end-to-end distances upon extension, our model has fewer accessible conformations and favors a lower conformational entropic component. In contrast, the stiffer segments seem to permit only minor molecular shape rearrangements. However, their mechanical contribution might be essential in resisting high forces that develop at a late stretch stage when the chain is fully extended. Considering that these segments must undergo thermal fluctuations of a certain amplitude and frequency (libration), additional stretch upon full chain extension would lead to a damping of the amplitude of the libration [as proposed for elastin (22)]. Here, librational entropy would oppose the over-stretch of the poly-Ig chain, acting as an energy barrier that protects it from domain unfolding by triggering, instead, the unraveling of the more compliant PEVK sequences in neighboring springs. As a result, the interplay of poly-Ig and PEVK serial springs causes a nonlinear response to stretch that defines the mechanical properties of titin (3, 5).
Discussion
The molecular events underlying titin mechanics remain largely unknown. Whereas techniques such as AFM have amply reported on the unfolding of secondary structure elements in its Ig domains, these data do not enlighten us on the primary mechanism of titin elasticity in vivo that resides on the properties of its folded chain. Given that the structure of this filament is poorly understood, its mechanics have remained described in terms of a first-approximation statistical model of polymer entropy, the WLC model, which considers titin as a continuous chain of random conformation and homogeneous composition. Although concerns about this model were echoed at an early stage (23), no alternative mechanistic principle has been proposed so far. Based on atomic structures, SAXS, and EM data of poly-Ig components from the I-band of titin, we now propose a model of poly-Ig elasticity based on a discrete organization of the chain into finely structured super-motifs displaying a segmental dynamics. Structural data indicate that the entropic properties of these tandems are not homogeneous, but that variably entropic points exist along the filament. We identify the location of flexible points in the chain and model them within the context of the full skeletal I-band tandem. This mechanical model, here termed “carpenter-ruler,” is based on freely jointed rigid segments of variable segment length. The model allows calculating the physical properties of this spring and offers great promise for the future study, rationalization, and modeling of the stretch-response phenotype of skeletal myofibrils.
Experimental Procedures
Cloning.
Domains I65–I70 (amino acids 7946–8511), I67–I69 (amino acids 8137–8417), and I66–I69 (amino acids 8137–8511) from rabbit soleus titin, and I39–I57 (amino acids 5498–7287) from the human variant (X90569) were cloned independently into pET-M11 (EMBL collection), containing a His6-tag and a TEV protease cleavage site before the target gene. The mutated variant I67–I69P94A/P95I was generated using the QuikChange method (Stratagene).
Protein Production.
Expression was in Escherichia coli Rosetta (DE3) grown in LB medium supplemented with 34 μg/ml chloramphenicol and 25 μg/ml kanamycin up to an OD600 of 0.6 at 30°C. Induction used 1 mM isopropyl β-d-thiogalactoside (IPTG), and growth was continued at 20°C overnight. Bacterial cells were harvested by centrifugation, resuspended in 50 mM Tris·HCl (pH 7.5), 50 mM NaCl and sonicated in the presence of lysozyme, DNase, and a protease inhibitor mixture (Roche Diagnostics). The homogenate was applied to a HiTrap column (GE Healthcare) equilibrated in lysis buffer. Tag removal was by incubation with TEV protease overnight at 4°C in dialysis against 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT. Subtractive purification of the protease (His6-tagged) was carried out on a further chelating step. The eluted protein was purified to homogeneity by ion exchange chromatography using a Resource Q column and gel filtration on a Superdex 200 HiLoad 16/60 column (both from GE Healthcare) in 50 mM Tris·HCl (pH 7.5). Samples were then concentrated and stored at 4°C. Tags were removed in samples used in crystallization. Samples used in SAXS and I39–I57 retained the tag and were not incubated with protease.
Crystal Structure Elucidation of I65–I70.
Crystals grew as large hexagonal obelisks from 2 M NaCl, 100 mM sodium acetate (pH 4.9–5.1), and 3–5% (vol/vol) glycerol or NDSB-201 using a hanging drop setting at protein concentrations of 10–20 mg/ml. Cryoprotection for data collection used 20% (vol/vol) glycerol as supplement. Data processing used XDS/XSCALE (24) (statistics are listed in Table 1). Phasing by molecular replacement failed. Experimental phasing used MIRAS on CH3HgCl, Ta6Br12, and Se-Met derivatized crystals (SI Table 5). Heavy atom sites were identified with CNS (25), and phases were calculated in SHARP (26). The positions of Se-Met and Hg scatterers (the latter predictably vicinal to Cys residues) were used as sequence markers for the recognition and positioning of individual Ig during model building. Manual model building was in O (27). Initial refinement used CNS (25) and applied bulk solvent correction, overall anisotropic B-factor, grouped isotropic B-factor, and energy minimization. At a later stage, once the high-resolution structure of I67–I69 became available, its individual domains were used to replace equivalent Ig in I65–I70 on a final series of refinement that used TLS and B-group refinement in PHENIX (28). Model parameters are given in Table 1.
Crystal Structure Elucidation of I67–I69.
Crystals of I67–I69 grew as thin plates (≈20 μm in thickness) from 0.2 M ammonium acetate, 0.1 M sodium acetate (pH 4.6), and 30% (wt/vol) PEG-4000 in hanging-drops. For data collection, crystals were shock-frozen at 100 K in solutions supplemented with 20% (vol/vol) glycerol. Data processing used XDS (24). Phasing was by molecular replacement in Phaser (29), using a model derived from telokin (PDB code 1FHG) as search model, accounting for 1/3 of the asymmetric unit and with 28% average sequence identity to the target domains. Phase improvement used ARP/wARP (30), and model building was in O (27). Model refinement used Refmac5 (31). Data and model statistics are given in Table 1.
Small Angle X-Ray Scattering.
Synchrotron SAXS data from I65–I70, I67–I69, and I66–I69 were collected at X33 (DESY, Hamburg, Germany) using a linear gas detector and a MAR Image plate. All samples were measured at least at three concentrations (1.0–20.0 mg/ml) in the range of momentum transfer 0.01 < s < 0.35 Å−1 (s = 4π sinθ/λ, where the wavelength λ is 1.5 Å and 2θ is the scattering angle). Radiation damage, monitored by repetitive 1- or 2-min exposures of the sample, was not detected. Data were processed using PRIMUS (32) and GNOM (33). Standard procedures were applied to the computation of the forward scattering I(0), radius of gyration Rg, maximum dimension Dmax, and the hydrated volume Vp. The molecular mass of the solutes was evaluated by comparison with reference solutions of BSA.
The scattering amplitudes of individual Ig domains and the intensities of I65–I70, I67–I69, and I66–I69 were calculated from atomic coordinates using CRYSOL (34). SASREF (35) was used to determine the conformation of I65–I70 in solution through rigid body refinement, which assembled individual Ig while maintaining chain connectivity and avoiding steric clashes. Starting from random arrangements, SASREF used simulated annealing to simultaneously fit the three experimental patterns by minimizing the overall discrepancy χ2tot = χ2I65–I70 + χ2I66–I69 + χ2I67–I69 (definition of χ2 as in ref. 35).
Electron Microscopy.
Aliquots (5 μl) of protein samples were adsorbed (1 min) onto glow-discharged carbon-coated copper grids and negatively stained with 2% (wt/vol) uranyl acetate. Alternatively, aliquots (20 μl) of protein samples at a concentration of 0.1 mg/ml were glycerol-sprayed and rotary shadowed according to standard procedures (36). Visualization used a Hitachi H-7000 transmission electron microscope (Hitachi, Tokyo, Japan).
Supplementary Material
ACKNOWLEDGMENTS.
We thank the staff at ID29-1 (European Synchrotron Radiation Facility) and X06SA (Swiss Light Source) for excellent support during data collection and Pavel Afonine and Garib Murshudov for helpful discussions. Special thanks go to Prof. Henk Granzier for critical reading of this manuscript. Z.U.-F. was supported by SNF Grant 3100A0-112595. A.U. was supported by Pole Intelligence Logicielle CPER Lorraine. D.S. and P.K. were supported by the European Union Design Study SAXIER (011934). D.L and S.L. were supported by Deutsche Forschungsgemeinschaft Grants La668/9-1 and La1619/1-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Model coordinates and experimental x-ray amplitudes for I65–I70, I67–I69, and I67–I69E93A have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3B43, 2RIK, and 2RJM, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707163105/DC1.
References
- 1.Granzier HL, Labeit S. Circ Res. 2004;94:284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- 2.Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 3.Trombitás K, Greaser M, Labeit S, Jin JP, Kellermayer M, Helmes M, Granzier H. J Cell Biol. 1998;140:853–859. doi: 10.1083/jcb.140.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellermayer MS, Grama LJ. J Muscle Res Cell Motil. 2002;23:499–511. doi: 10.1023/a:1023414624092. [DOI] [PubMed] [Google Scholar]
- 5.Linke WA, Ivemeyer M, Olivieri N, Kolmerer B, Rüegg JC, Labeit S. J Mol Biol. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- 6.Marino M, Zou P, Svergun D, Garcia P, Edlich C, Simon B, Wilmanns M, Muhle-Goll C, Mayans O. Structure. 2006;14:1437–1447. doi: 10.1016/j.str.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Zou P, Pinotsis N, Lange S, Song YH, Popov A, Mavridis I, Mayans O, Gautel M, Wilmanns M. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
- 8.Mrosek M, Labeit D, Witt S, Heerklotz H, von Castelmur E, Labeit S, Mayans O. FASEB J. 2007;7:1383–1392. doi: 10.1096/fj.06-7644com. [DOI] [PubMed] [Google Scholar]
- 9.Müller S, Lange S, Gautel M, Wilmanns M. J Mol Biol. 2007;371:469–480. doi: 10.1016/j.jmb.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 10.Marino M, Svergun DI, Kreplak L, Konarev PV, Maco B, Labeit D, Mayans O. J Muscle Res Cell Motil. 2005;26:355–365. doi: 10.1007/s10974-005-9017-6. [DOI] [PubMed] [Google Scholar]
- 11.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 12.Di Cola E, Waigh T, Trinick J, Tskhovrebova L, Houmeida A, Pyckhout-Hintzen W, Dewhurst C. Biophys J. 2005;88:4095–4106. doi: 10.1529/biophysj.104.054908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee EH, Hsin J, Mayans O, Schulten K. Biophys J. 2007;93:1719–1735. doi: 10.1529/biophysj.107.105528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller WT. Acta Crystallogr D. 2005;61:33–44. doi: 10.1107/S0907444904025855. [DOI] [PubMed] [Google Scholar]
- 15.Gautel M. Adv Biophys. 1996;33:27–37. doi: 10.1016/s0065-227x(96)90020-9. [DOI] [PubMed] [Google Scholar]
- 16.Kenny PA, Liston EM, Higgins DG. Gene. 1999;232:11–23. doi: 10.1016/s0378-1119(99)00122-5. [DOI] [PubMed] [Google Scholar]
- 17.Tskhovrebova L, Trinick J. J Mol Biol. 2001;310:755–771. doi: 10.1006/jmbi.2001.4700. [DOI] [PubMed] [Google Scholar]
- 18.Kellermayer MS, Bustamante C, Granzier HL. Biochim Biophys Acta. 2003;1604:105–114. doi: 10.1016/s0005-2728(03)00029-x. [DOI] [PubMed] [Google Scholar]
- 19.Leake MC, Wilson D, Gautel M, Simmons RM. Biophys J. 2004;87:1112–1135. doi: 10.1529/biophysj.103.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivetti C, Guthold M, Bustamante C. J Mol Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 21.Flory PJ. Statistical Mechanics of Chain Molecules. New York: Interscience; 1969. [Google Scholar]
- 22.Urry DW. J Protein Chem. 1988;7:1–34. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]
- 23.Politou AS, Thomas DJ, Pastore A. Biophys J. 1995;69:2601–2610. doi: 10.1016/S0006-3495(95)80131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 25.Brunger AT, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 30.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. Acta Crystallogr D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 33.Svergun DI. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 34.Svergun DI, Barberato C, Koch MHJ. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 35.Petoukhov MV, Svergun DI. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fowler WE, Aebi U. J Ultrastruct Res. 1983;83:319–334. doi: 10.1016/s0022-5320(83)90139-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.