Abstract
FoF1-ATP synthase manufactures the energy “currency,” ATP, of living cells. The soluble F1 portion, called F1-ATPase, can act as a rotary motor, with ATP binding, hydrolysis, and product release, inducing a torque on the γ-subunit. A coarse-grained plastic network model is used to show at a residue level of detail how the conformational changes of the catalytic β-subunits act on the γ-subunit through repulsive van der Waals interactions to generate a torque that drives unidirectional rotation, as observed experimentally. The simulations suggest that the calculated 85° substep rotation is driven primarily by ATP binding and that the subsequent 35° substep rotation is produced by product release from one β-subunit and a concomitant binding pocket expansion of another β-subunit. The results of the simulation agree with single-molecule experiments [see, for example, Adachi K, et al. (2007) Cell 130:309–321] and support a tri-site rotary mechanism for F1-ATPase under physiological condition.
Keywords: ATP hydrolysis, coarse-grained, mechanical coupling, molecular motor, tri-site
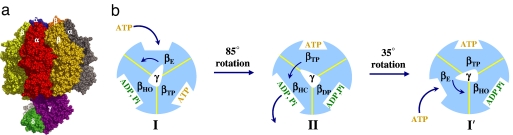
FoF1-ATP synthase, which is found in mitochondria, chloroplasts, and bacteria, uses the proton-motive force across membranes to synthesize adenosine triphosphate (ATP) from adenosine diphosphate (ADP) and inorganic phosphate H2PO4− (Pi) (1–4). This molecular machine consists of two moieties, a transmembrane portion (Fo), the rotation of which is induced by the proton gradient, and a globular catalytic moiety (F1) that synthesizes ATP. By itself, or as a part of FoF1-ATP synthase, the F1 moiety (F1-ATPase) can hydrolyze ATP to generate rotational motion of the γ-subunit. The mitochondrial F1-ATPase (MF1) is composed of nine subunits (Fig. 1a), six of which (α3β3) form an approximately spherical globular complex around a central stalk (5, 6), which consists of the subunits γδε. Under optimum conditions, the central shaft rotates (counterclockwise as seen from the membrane) at a rate of 7.6 rad/ms punctuated by dwell times of several milliseconds. When an external torque rotates the central shaft in the reverse direction (7), the F1 moiety by itself synthesizes ATP from ADP and Pi. During the rotational hydrolysis cycle of F1-ATPase, the three β catalytic subunits undergo consecutive conformational changes (1, 4, 5) that are believed to induce the central stalk rotation. The substrate binding, chemical steps, and product release are thought to take place in sites with different conformations (4, 8). The result is highly efficient catalysis with the observed ATP hydrolysis rate being accelerated by a factor of 5 × 108 to 8 × 109 in F1-ATPase compared with the reaction in solution (9).
Fig. 1.
Hydrolysis cycle of F1-ATPase. (a) Molecular representation of the mitochondrial F1-ATPase (α3β3γδε). (b) The proposed cycle of the tri-site ATP hydrolysis mechanism. The cycle starts from an ATP waiting state, with one β-subunit fully open (βE), the second β-subunit half-open (βHO), and the third β-subunit closed (βTP); βTP contains the ATP bound in the previous cycle. The calculated 85° rotation is caused by the ATP binding to βE and a cooperative opening of the βHO-subunit to adopt a “half-closed” (HC) conformation in preparation for product release. The subsequent calculated 35° rotation is related to hydrolysis product release from the βHC-subunit and a concomitant expansion of the βDP binding pocket to form βHO.
The overall mechanism for the rotary catalysis by F1-ATPase was originally suggested by Boyer (10), before the availability of a high-resolution crystal structure, based on the interpretation of thermodynamic and kinetic data. Boyer's “binding change mechanism” states that the three catalytic β-subunits at any given time have different nucleotide binding affinities and that they undergo sequential conformational changes during the catalytic cycle (11). Supporting evidence for the binding change mechanism came from the first high-resolution structure of bovine mitochondrial F1-ATPase (bMF1) (5) (referred to as Ab). Although now believed to be an azide-inhibited state (12), the Ab structure shows that the three β-subunits in one molecule have different conformations, apparently modulated by the asymmetric γ-stalk, a feature conserved in azide-free crystals (13, 14). In 1997, the unidirectional rotation of the central stalk in F1-ATPase was observed in single-molecule experiments (15). At low ATP concentrations, the hydrolysis drives the γ-stalk rotation in discrete steps of 120° (16), which can be further resolved into 90° and 30° substeps (16) [recently suggested to be 80°/40° (17)]. The duration of the dwell before the 90° substep is inversely proportional to ATP concentration and is therefore thought to be related to ATP binding, whereas the dwell time before the 30° substep is believed to be coupled to product release and/or chemical catalysis (8, 16–19).
A number of proposals have been put forward to develop semiquantitative models for the hydrolysis and synthesis cycle of F1-ATPase (see, for example, refs. 20 and 21). However, none of them included a full treatment of the binding change mechanism. In particular, they do not connect the different conformation of the β-subunits found in the crystal structures (e.g., ref. 5) with the different binding affinities measured in solution (22). The Ab structure has the β-subunits in three different conformations, called βTP (AMP-PNP bound), βDP (ADP bound), and βE (empty). Subunits βTP and βDP have very similar “closed” active-site conformations, whereas the empty βE-subunit is much more open. Given the ATP affinities of F1-ATPase measured in solution (<0.2 nM, ≈2 μM, and ≈25 μM) (9, 23, 24), identification of the lowest-affinity conformation as the open βE site is generally accepted, but it was not clear until recently which of the two, βTP and βDP, is the strong binding site for ATP. Free energy simulations, combined with solution measurements, led to identification of the βTP site in the crystal structures as the strong binding site for ATP and the βDP site as the weaker binding site (25). This simulation result has been confirmed recently by fluorescence resonance energy transfer measurements (26). Based on these results, an E1E2-type model was developed (4), to describe the role of the individual β conformations in ATP synthesis and hydrolysis by F1-ATPase (4). The model provides a quantitative reaction scheme and a chemical kinetic description, in agreement with the published experimental thermodynamic and kinetic data. It is in accord with the most complete single-molecule analysis published recently (8), although the latter does not consider the relation of the β-subunit conformations of F1-ATPase to the measurement.
What is still missing is a knowledge of the atomic mechanism by which the coupling between the structural changes of α3β3 globular moiety and the γ-shaft leads to energy transfer between them; i.e., how the γ-subunit rotation leads to the conformational changes in the β-subunits required for ATP synthesis, and conversely how the changes in the β-subunit conformations due to their occupations lead to γ-rotation in the opposite direction during the hydrolysis cycle. Molecular dynamics simulations have been done with an all-atom description of F1-ATPase in which a torque (27) or a biasing force (28) was applied to rotate the γ-subunit in the synthesis direction, starting with the Ab structure. In addition, a targeted molecular dynamics (TMD) was performed, in which a guiding force was applied to all heavy atoms (28); the full conformational transition of the α3β3-subunits that accompanies the 120° rotation of the γ-subunit was obtained from a trajectory of 500 ps. The change in the van der Waals repulsions and electrostatic attractions of the β-subunits with the γ-subunit, as a function of its rotation, provided an approximate description of the mechanism. One interesting observation was that the motion of the α3β3-subunits is guided by an ionic track on the globular part of the γ-subunit (28); the importance of this ionic track in thermophilic Bacillus PS3 F1 has been verified by mutation experiments (29).
In contrast to the simulation analysis of the subunit coupling in ATP synthesis by F1-ATPase, much less has been done for hydrolysis. A major problem is that, unlike synthesis, where applying a torque to the γ-subunit in the synthesis direction mimics what actually happens, albeit on a much shorter time scale, it is more difficult to develop a meaningful structural model to simulate hydrolysis. To follow the full rotational cycle of the γ-subunit induced by the conformational changes in the α3β3-crown, we here develop a coarse-grained model of F1-ATPase in which the α3β3-crown and the γ-stalk are represented by separate plastic network models (PNMs) (30), and they are coupled by a repulsive van der Waals-type interaction. The PNM represents each entity (α3β3-crown in a given conformation, γ-stalk) by an elastic network (EN), whose energy minimum corresponds to the known crystal structure. TMD (31) is applied to the α3β3-crown to gradually transform the conformation of the catalytic β-subunits (and their neighboring α-subunits) from the EN representing one structure to another, and the response of the γ-stalk is monitored. We were encouraged in this work by the publication of Koga and Takada (32) (KT) who used a simpler coarse-grained model that apparently supported a bi-site mechanism, in disagreement with experiment (3) (see Concluding Discussion).
The simulations with the PNM model produce essentially unidirectional rotation of the γ-stalk in the hydrolysis direction with 85°/35° rotational substeps. The structures of the α3β3-crown used in the model correspond to subunit occupations in agreement with a tri-site mechanism (see Fig. 1). A detailed analysis of the origin of the torque generation provides information concerning the essential residues involved in the 85° and 35° substep rotations; comparison with mutation studies are presented. Thus, the calculations reported here show not only how the in/out movement of the β-subunits causes the γ-subunit to rotate, but they also yield results in agreement with the most detailed single-molecule experiments (8) and the structure-based mechanism of Gao et al. (4).
Structures and Their Transitions on the Hydrolysis Pathway
The simulations use two high-resolution x-ray structures to construct the plastic network. The first (13) (referred to as Ka) has the β-subunits in the conformations βE (empty), βHO (AMP-PNP), and βTP (AMP-PNP), and the second (33) (referred to as Me) has them in the conformations βTP (ADP/AlF4−), βHC (ADP/SO42−), and βDP (ADP/AlF4−); we introduce the term βHO to indicate the somewhat more open conformation in the Ka structure, relative to the “closed” βDP conformation in Abrahams et al. (5). The internal structure of the γ-subunit is assumed not to change significantly during the transition, in accord with experiment [see supporting information (SI) Text (Internal Structural Difference of the γ-Subunit in the Ka and Me Structures) and SI Fig. 5], and is described by an elastic network based on the Ka structure. The simulations start with a PNM having the Ka α3β3-crown as the initial state and the Me α3β3-crown as the final state; i.e., during the transition, we apply the TMD method to the plastic networks of the α3β3-crown, so that βE goes to βTP (representing ATP binding), βHO goes to βHC, and βTP changes to βDP. The second step of the cycle involves the same PNM model, except that the initial and final states are interchanged. For each step, the initial orientation of the γ-subunit is essentially that observed in the crystal structure. A schematic representation of the nucleotide occupancy and β-subunit conformational transitions for the rotation cycle during the simulation is shown in Fig. 1b. No guiding force is applied to the γ-subunit, and the only interaction between the two moieties (α3β3-crown and γ-shaft) arises through a repulsive term between their Cα atoms. As the α3β3-crown goes through the conformational cycle, the γ-subunit responds to the repulsive term, which introduces a torque leading to the rotation. Details concerning the choice of structures and the simulation methodology are given in Materials and Methods.
Simulation Results: An 85°/35° Substep Rotation Cycle
To test the stability of the simulation systems, equilibrium simulations were done (≈1 ns in length) with the PNM network corresponding to the Ka α3β3-crown structure and the elastic model for the γ-subunit in the corresponding crystal orientation; the α3β3/γ repulsive interactions were present. The γ-stalk rotate angle is stabilized at −2° relative to the crystal structure. A series of 10 TMD simulations at constant temperature were then performed for the two parts of the rotation path with restraints on the system that mimic the single-molecule experiments (15) (see Materials and Methods). The TMD guiding force was chosen such that the conformational changes of the α3β3-crown between the end states took place in 130–220 ps. Test simulations showed that the relatively gradual conformational change modeled by TMD is necessary to achieve stable results; rapid switching of the conformations sometimes caused clashes and failed to generate unidirectional γ-rotation. After these TMD simulations (i.e., after the crown structure had made the transition), the system was simulated for ≈1 ns with the restricted perturbation targeted molecular dynamics (RP-TMD) method (34) applied to the α3β3-crown to restrain it to the end structure and allow the γ-stalk orientation to reach a stable value.
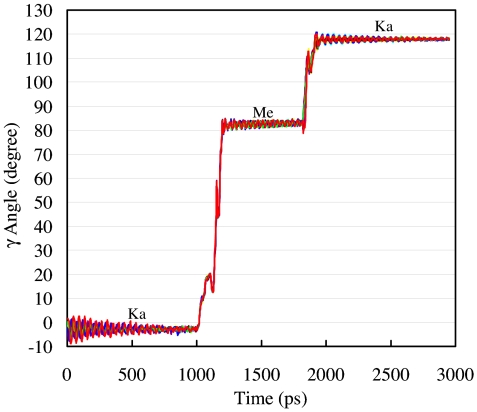
The rotation angles [see SI Text (Definition of the Rotation Angle) for how the rotation angle was defined] of the γ-subunit as a function of time during the Ka to Me and the Me to Ka simulations are shown in Fig. 2. As can be seen, the 10 independent trajectories for each transition show essentially the same behavior. Quantitatively, the substep rotation angles obtained from our simulations are 85°/35°; i.e., they are intermediate between the published experimental estimates of 90°/30° (16) and 80°/40° (17). The TMD simulation that transforms the (βE, βHO, βTP)-crown to the (βTP, βHC, βDP)-crown results in the 85° substep rotation of the γ-subunit. During this transition, βE closes to form βTP as a consequence of binding ATP, and βTP changes to βDP as a result of ATP hydrolysis. This rotation orients the “γ-bulge” (28) (see Fig. 4) to directly face the βHO-subunit and facilitates its opening to form βHC (the preproduct release conformation). The second TMD simulation transforms the (βHC, βDP, βTP)-crown to the (βE, βHO, βTP)-crown and yields a further 35° rotation of the γ-subunit. The two catalytic β-subunits with significant conformational changes in this step are βHC, which goes to βE, and βDP, which goes to βHO (a superposition of these subunits is shown in SI Fig. 6), whereas the βTP-subunit (with ATP bound) changes very little. The opening of the βHC-subunit to form βE is associated with the release of the hydrolysis product. The conformational change of the βDP-subunit from the closed to the half-open (βHO) conformation is likely to be due to the electrostatic repulsion between the newly formed ADP and Pi (35); i.e., a more open α/β interface allows ADP and Pi to separate from each other, thus lowering the Coulomb repulsion between the two negatively charged species. The synergistic effect of the conformation changes of the two β-subunits (βHC to βE and βDP to βHO) causes the F1-ATPase central cavity of the crown surrounding the γ-stalk to expand. This permits the torque on the γ-stalk to rotate it by 35°. The two-step TMD simulations result in an overall γ-stalk rotation of 120° in the hydrolysis direction. (See SI Movies 1 and 2 for a representative TMD trajectory.) This completes one cycle of ATP binding, hydrolysis, and product release with F1-ATPase having the same structure as it did initially, but the conformations of the crown subunits (see Fig. 2) interchanged, as if the crown had rotated by 120°. The results of using the Ab crown structure as the starting state for ATP binding are described in SI Fig. 7 and SI Text (Control Simulations Using Ab as the Starting Crown Structure).
Fig. 2.
Rotational angles of the γ-stalk during the molecular dynamics simulations. The system is first equilibrated for 1 ns with an elastic network representation of the Ka structure, then the crown conformation is gradually transformed to that of the Me structure by a TMD simulation (220 ps). The first transition yields an 85° γ-rotation. After a 600-ps further equilibration with the Me crown, the α3β3 subcomplex is gradually transformed back to the Ka structure over a period of 130 ps by a TMD simulation. The second transition introduces a 35° γ-rotation. The system was then equilibrated for another 1 ns until a plateau of the γ-rotational angle was obtained. Results for 10 independent simulations are plotted; they superpose so well that they cannot be distinguished. See also SI Text (Possible Additional Substeps).
Fig. 4.
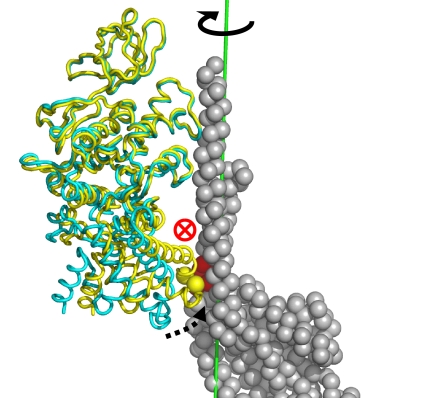
Representative snapshot of the torque generation configuration during the 85° substep rotation (see Fig. 3) in the neighborhood of 20°. Only the βE-subunit (yellow ribbon) and γ-subunit (gray spheres) are shown. The β/γ mechanical coupling interaction (βE:I390–L391 and γ:20–25) identified from the simulation is highlighted by spheres (β, yellow; γ, red). The initial conformation of the βE-subunit (fully open, before ATP binding) is shown as a cyan ribbon. During the ATP binding, the helix–turn–helix-motif domain of the βE-subunit swings up by ≈30° (represented by a dotted black arrow). This conformational change results in a close contact between βE:I390–L391 and the N-terminal helix of the γ-coiled-coil at γ:20–25 (most likely through hydrophobic interactions). The βE movement yields an off-axis force that pushes the N-terminal helix of the γ-stalk in a direction into the plane of the paper (indicated by the red ⊗). This results in a unidirectional γ-stalk rotation (indicated by the solid black arrow around the green rotation axis). The illustrations were made with the program PyMOL.
Simulation Analysis: A Tri-Site Mechanism
The simulation results shown in Fig. 2 yield a cooperative model in which the rotational substeps during the ATP hydrolysis cycle of F1-ATPase are reproduced. There has been considerable discussion in the literature concerning whether the hydrolysis corresponds to a bi-site mechanism (i.e., one or two β-subunits are occupied by nucleotide during the cycle) (11) or a tri-site mechanism (i.e., two or three β-subunits are occupied by nucleotide during the cycle) (3, 4). Because the nucleotides are included only implicitly in the model, we determine the occupations of each of the β-subunit conformations based on the crystal structures and the previously proposed model for ATP hydrolysis (4). As shown in Fig. 1b, the cycle begins with the (βE, βHO, βTP)-crown (state I), which is associated with the dwell intermediate before the 85° rotation. In state I, two β sites are occupied by nucleotides: βHO contains the hydrolysis product ADP+Pi, and βTP contains the ATP bound during the previous hydrolysis cycle; the third site, βE, is empty before binding of ATP. The 85° rotation of the γ-subunit is driven by binding an ATP to the βE-subunit of state I and ends up in state II (see above). The resulting conformation (state II) is an intermediate state, with a dwell time involving product release. In state II, all three β sites are occupied by nucleotides, with βHC containing ADP+Pi before product release, βTP being occupied by the newly bound ATP, and βDP containing the ADP+Pi produced by hydrolysis. The γ-rotation by an additional 35° is induced from state II as a consequence of product release from βHC, which restores the system to the ATP waiting mode (state I) (labeled as I′ in Fig. 1b to denote state I after the 120° rotation of the γ-subunit). In the transition, subunit βHC is fully opened to form βE after product release, and the βDP-subunit evolves to a “half-open” conformation (βHO) with ADP and Pi bound. The βTP-subunit, which binds ATP, is essentially unchanged.
The current model thus not only results in the experimentally observed 85°/35° substep rotation but also explains the ATP concentration dependence of the different dwell times. Single-molecule experiments have demonstrated that the duration of the dwell before the 85° rotation depends on ATP concentration, whereas the dwell before the 35° rotation does not (16). Analysis of kinetic data suggests that the latter dwell involves two kinetic processes, potentially chemical catalysis and/or ADP/Pi release (17, 19). The model presented here agrees with these experimental observations, because ATP binding drives the 85° γ-rotation and so the dwell time is expected to be concentration-dependent. The 35° γ-rotation is triggered by ADP and/or Pi release from the βHC-subunit, after hydrolysis, and so is independent of ATP concentration. Importantly, because the nucleotide occupation of F1-ATPase alternates between 2 (states I and I′) and 3 (state II), the proposed model is in accord with a tri-site mechanism (3).
It is useful to compare the present model with that proposed by Gao et al. (4); the latter is essentially the same as the model very recently proposed by Adachi et al. (8) in their figure 1c, without reference to Gao et al. Comparing Fig. 1b of the present article with figure 4 of Gao et al., we see that the former is essentially an abbreviated version of the latter. Fig. 1b includes only the rotation steps and does not show the ATP binding and ADP, Pi product release steps separately; in Gao et al., these were included explicitly because the model was used to develop a chemical kinetic description based on the rate constants for the individual steps. Thus, the cycle in Fig. 1b combines E1 and E1T (in state I and I′) and E2DP, E2P, and E2 (in state II) of figure 4 of Gao et al. The one difference between the two cycles is that the present article includes a new conformation for the β-subunits. This is the βHO conformation, which was observed in the Ka crystal structure (13) and replaces the βDP conformation of Ab crystal structure (5); the latter is now believed to be an azide-inhibited conformation (12). If the present model is correct, there are five conformations of the β-subunit, rather than four, that play a role in the F1-ATPase cycle. One consequence is that the kinetic analysis of the solution measurements is somewhat more complex than thought previously. However, given the structures of the conformations, the relative affinity for ATP, H2O vs. ADP, Pi of βHO should be in-between those of βDP and βHC used in the analysis of Gao et al.; i.e., it is expected to be strongly biased toward ADP, Pi, in contrast to βTP, which has nearly equal free energy for bound ATP, H2O and ADP, Pi (25).
Torque Generation
Although a simple functional form was used for the potential energy coupling of the α3β3-crown and the γ-stalk (see Materials and Methods), the unidirectional rotation with 85°/35° steps was produced. This suggests that an analysis of the torque generation mechanism is worthwhile. The torque τ⃗ acting on the stalk during the TMD trajectories describing the α3β3-crown conformational change during the 120° cycle is given by
where τ⃗i, the torque on the ith residue of the γ-stalk (for yMF1, i = 1 − 276), is the cross-product of the position vector r⃗i and the force vector F⃗i acting on residue i, coarse-grained to its Cα atom. Only the forces that couple the α3β3-crown and the γ-stalk were included in the torque calculation [for details, see Materials and Methods, SI Fig. 8, and SI Text (A Typical Torque Profile During the 85° Rotation and Absolute Torque Magnitude in the Simulations)].
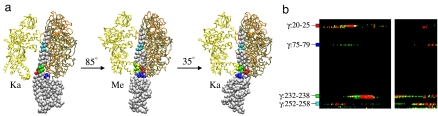
Fig. 3a shows the residues involved in the torque generation, whereas Fig. 3b shows the distribution of the torque over the γ-stalk residues as a function of time during the conformational transitions that produce the 85° and 35° substeps, respectively (see the legend of Fig. 3b). Four clusters of γ-residues are identified in Fig. 3b; they are the ones primarily responsible for the two stages of the 85°/35° rotation of the γ-stalk. The parts of the βE-subunit that generate the torque are indicated in Fig. 3a; the latter goes from βE to βTP during the 85° rotation and remains in the βTP conformation during the remaining 35° rotation, which completes the 120° rotation cycle. Structurally, the portion of the γ-subunit that inserts into the α3β3-subunits consists of a left-handed coiled-coil, formed by its N-terminal helix (short) and C-terminal helix (long) with the N and C helices antiparallel. Two of the torque-generating clusters are located in the “neck” region; i.e., the most convex curved part of the coiled-coil just above the globular base of the γ-subunit where close contacts with the surrounding β-subunits occur. They are γ:20–25 (red) on the N-terminal helix and γ:232–238 (green) on the C-terminal helix. The βE-subunit in the neighborhood of βE:I390–L391 (yellow) [these βE residues are “below” the DELSEED motif (residue 394–400)] interacts strongly with both clusters during its conformational change due to ATP binding. The third cluster, γ:252–258 (cyan), is located on the upper part of the C-terminal γ helix, making “catch” interactions with a βE-subunit loop via salt bridges (βE:D316/γ:R254 and βE:D319/γ:R254) or hydrogen bonds (βE:T318/γ:Q255 and βE:D316/γ:Q255) (5). Disruption of the β/γ interactions in the latter region has been shown to yield attenuated activity in both synthesis and hydrolysis (36). The last torque-generation cluster (dark blue) is located at γ:75–79. These residues form important β/γ interactions, as seen in crystal structures (5), and their role is confirmed by mutagenesis analysis [e.g., the γ:L77K mutant of a Thermophilic bacterial F1 (TF1) only displays 15% hydrolysis activity compared with the wild type (37); see also SI Text (Description of Mutation Studies)].
Fig. 3.
Torque generation. (a) Residues involved in the γ-subunit, shown as colored spheres. (b) Torque distribution over the residues (residue number on the y axis) of the γ-stalk. Torque profiles during the 85° (Left) and 35° (Right) substep rotations, respectively (see text). To focus on the mechanical coupling mechanism, only the internetwork repulsion (Eq. 2) is included in determining the torque; torques that act in the opposite direction are not shown. The productive torque is scaled to lie between 0 and 1 on each map; the maximum torque on each map is scaled to 1. The torque (τ) is then colored based on a relative scale, τ < 0.1 (black), 0.1 ≤ τ < 0.2 (green), 0.2 ≤ τ < 0.3 (yellow), and τ ≥ 0.3 (red). Residue numbers (bMF1 numbering is used, and the corresponding yMF1 number is given below in parenthesis) of four torque generation “hot spot” clusters on the γ-stalk are labeled by using the same color scheme for identifying residues in a: γ:20–25 (20–25) (red), γ:75–79 (81–85) (dark blue), γ:232–238 (237–243) (green), and γ:252–258 (257–263) (cyan). A region on the β-subunit that is tightly coupled to the torque generation, in particular βE:390–391 (390–391), is shown as yellow spheres in a. The torque generation for the 85° substep rotation displays a relaying pattern, where the first set of torques is generated primarily on the N-terminal helix of the coiled-coil (γ:20–25) and the second set of torques is generated on the C-terminal helix (γ:232–238). The illustrations were made with the program VMD.
The torque contributions from each set of γ-subunit residues are plotted in Fig. 3b as a function of the simulation time of the α3β3 conformational transition; relative units are used (see the legend of Fig. 3b). For the 85° rotation driven by the βE conformational change induced by ATP binding, the torque on the γ-stalk is generated in two successive steps. In the initial stage of the crown transition, there is essentially no torque on the γ-stalk because the βE- and γ-subunits are not in contact, and no rotation occurs. Once the two subunits come into van der Waals contact, a significant torque acts on γ:20–25, which induces the rotation to ≈40°. The initial βE closure brings the turn region (in particular β:I390–L391) of the helix–turn–helix motif region into contact with the N-terminal γ helix at γ:20–25. The inward motion of the βE helix–turn–helix motif pushes the N-terminal helix in a direction approximately perpendicular to the γ-rotational axis [see Fig. 4, SI Fig. 9, SI Text (Distances of βE:L391-γ:M25 and βE:L391-γ:K237 During the 85° Rotation) and SI Movie 3]. There is also a smaller contribution from βE interacting with γ:252–258 that complements the γ:20–25 contacts. The off-center arrangement of the coupling interface leads to the unidirectional rotation of the γ-stalk. After the γ-stalk rotates by ≈40° relative to the ATP waiting state (Ka-like structure), the torque on the γ:20–25 cluster is reduced because the original contact is no longer present. The second set of torques takes over, as indicated by torque signal on the C-terminal helix γ:232–238. The C-terminal helix has been rotated to a position that forms a new β/γ van der Waals interface between βE:I390, L391 and the cluster located at γ:232–238, particularly with residue γ:S236 and the bulky residue γ:K237. The further inward movement of βE results in a torque that activates the rest of the 85° rotation. This region contains residues M232, N234, and N238 that are conserved in the F1 family [see SI Text (Sequence Alignment of the β/γ Coupling Interface in the F1-ATPase Family) for a sequence alignment]. There appears to be little torque on the γ-stalk during the late stage of the 85° rotation (between 75° and 85°). This suggests that the completion of the 85° rotation is due to “release” of the γ-subunit when the βHO-subunit changes to the more open βHC conformation.
The analysis shows a significant torque acting on γ:232–238 due to βE:390–391 during much of the 35° rotation. In addition, γ:252–258, coupled to the catch loop at βE:316–319, and γ:75–79 are involved in the latter part of the 35° rotation. The notable torque generated on the cluster γ:75–79 arises from a close contact formed with the helix–turn–helix-motif region of the βHO-subunit after the 85° rotation (now βHC; see Fig. 1b).
Cross-linking (38) and deletion (39, 40) experiments have shown that the C-terminal helix of the γ tip [up to 12 residues (41); i.e., residues 276–287 of Escherichia coli F1:γ] is not required for torque generation. This leads to the question as to whether this tip is mobile, forming a “swivel” joint, or relatively fixed (40). The crystal structure suggests its role as a rotational pivot surrounded by a proline-rich hydrophobic bearing (5). Nonnegligible torque is found on the C-terminal γ tip in our simulations, which may be the cause of the partial unwinding suggested from the full γ-rotation observed for the F1 mutant with a crown cross-linked with the stalk at the γ top; i.e., between γ:A285C and α:P280C (38).
Concluding Discussion
We have used a coarse-grained model of F1-ATPase to show how the conformational changes of the catalytic β-subunits, particularly the in/out motions of the helix–turn–helix motif, induced by binding of ATP and product release, produce a torque that leads to the rotation of the γ-subunit. A plastic network model (PNM) was used in the simulations to produce the α3β3-crown conformational changes based on available crystal structures. Coupled to the γ-stalk by repulsive interactions, they generated experimentally observed 85°/35° rotational substeps of the γ-subunit in the 120° rotational cycle; three such cycles lead to the full 360° rotation. Interpretation of the results showed that the cycle corresponds to a tri-site mechanism in accord with experiments (3, 4, 8). This differs from KT (32), who concluded that their results supported a bi-site model. The coarse-grained models are significantly different (e.g., KT used a sudden “switching Gō” model, whereas we used the more gentle TMD-induced conformational changes based on a PNM), but the most important difference appears to be in the structures of the α3β3-crown. KT started with a conformation for βDP that is now known to correspond to an azide-inhibited state, which is likely to be off-pathway; and they constructed a hypothetical intermediate crown structure that contains both βE and βHC to obtain the 30° rotation. Our model replaced this starting crown structure with the Ka structure determined in the absence of azide (13), and the intermediate crown structure we used is based on the Me crystal structure (33) (see Comparison with KT “Tri-Site” Simulation in SI Text).
The present simulations demonstrated how a tri-site mechanism that is consistent with kinetics data (4) explains the generation of the 85°/35° rotational substeps. The torque analysis indicated that it is the hydrophobic side-chain (βE:I390–L391) region, rather than the DELSEED motif region, that is responsible for coupling the inward motion of βE to the rotation of the γ-stalk.
Introduction of a coarse-grained model has made possible simulations of the full rotation cycle of F1-ATPase. Although such a model is a simplified representation of the molecule, it is sufficiently detailed to provided new insights into how F1-ATPase works (particularly the nature of the induced rotation mechanism at a residue level), as a complement to the experimental data. The present approach is being used to guide simulations of this rotary motor with an all-atom force field (unpublished work) and can serve as a model for coarse-grained simulations of other molecular motors.
Materials and Methods
Simulation Methodology.
The coarse-grained model was constructed based on available crystal structures (see below), with each amino acid residue represented by its Cα atom; the actual residue mass used in the model was based on the Ka yMF1 sequence. Only the α3β3γ subcomplex of F1 was included in the model, in accord with many of the single-molecule experiments (8), and nucleotides were not explicitly represented. The positions of the Cα atoms in the crystal structures were used to define the nodes in the PNM connecting the Ka and Me conformations; i.e., two separate plastic network models (30) were constructed for the F1 crown (subunits α3β3) and the central stalk (γ-subunit). For a given protein conformation, atoms in the same network were connected to their neighboring atoms within a cutoff distance by a harmonic potential, and there was no connection between atoms that belong to different networks. Details of the model are given in SI Text (under Details of the Plastic Network Model (PNM) Used in the Simulations). The coupling term between the crown and stalk network was represented by a repulsive van der Waals-type interaction. We used the functional form (32)
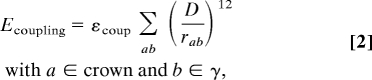 |
where εcoup = 0.36 kcal/mol and D = 6 Å. The total potential energy for the system corresponds to the sum of the PNM energies of the crown (Ecrown) and γ-stalk (Eγ) plus their interaction energy:
Crystal Structures Used to Define the PNMs.
Because the original 2.8-Å bovine mitochondrial F1-ATPase structure [Protein Data Bank (PDB) entry 1BMF] (5) (referred to as Ab), which was used in most previous simulations (27, 28, 32), has been shown to be an azide-inhibited state, we did not use it in the present study. In a newly reported structural study of yeast mitochondrial F1 (yMF1) (PDB entry 2HLD) (13), an ATP analog (AMP-PNP) rather than an ADP/Mg2+ is bound to the βDP-subunit, so that it cannot be ADP inhibited and is likely to be an intermediate state on the rotational pathway. The yMF1 structure was used and is denoted by Ka. It has an empty site and a novel αDP/βDP conformation with AMP-PNP bound, in which the nucleotide binding interface is more open than the βDP conformation in Ab [we refer to it as half-open (βHO)]; the third site has AMP-PNP and is in the βTP conformation. The other structure we used was that of Menz et al. (PDB entry 1H8E) (33) (referred to as Me). To complete the structures (i.e., account for missing portions of the γ-subunit or missing residues of the crown), modeling was done. For details, see SI Text (Missing Residues Modeling).
In all simulations based on the coarse-grained model (including equilibrations), harmonic restraints were applied to the center of mass (COM) of the N-terminal β-barrel domain (residues 9–84, bMF1 number) of three β-subunits with respect to their positions in the crystal structure, mimicking the effect of a stator on the periphery stalk (42, 43) present in the physical system and the use of His tags to immobilize the molecule on a glass surface in the single-molecule experiment (15); similar constraints were applied by Böckmann and Grubmüller (27). Details are given in SI Text (under Additional Details on the Restraints Used in the Simulations).
MD Simulations.
Constant-temperature molecular dynamics were performed with the CHARMM program (44). The Berendsen thermostat (45) was used to keep T at 50 K. The choice of a low simulation temperature was made to achieve stable simulations with the simplified potential. The equations of motion were integrated by using a 10-fs step. Nonbonded interaction pairs between the α3β3-crown residues and the γ-subunit residues were generated by using a cutoff distance of 14.0 Å along with a switching function in the region 12.0–13.0 Å to feather the interaction energy to zero; the list was updated heuristically during the dynamics. Additional details are given in SI Text (under MD Simulation Details).
Supplementary Material
ACKNOWLEDGMENTS.
We thank P. Maragakis for providing a version of the PNM code, which has been implemented in the CHARMM program, and P. Maragakis, A. van der Vaart, A. Golosov, K. Nam, Y. Kong, and W. Yang for discussions. The computations were supported through the Innovative and Novel Computational Impact on Theory and Experiment program at the Oak Ridge National Laboratory. Some of the calculations were made at the National Energy Research Scientific Computing Center and the National Cancer Institute. This work was supported in part by a grant from the National Institutes of Health and by contributions from the CHARMM Development Project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708746105/DC1.
References
- 1.Boyer PD. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 2.Junge W, Lill H, Engelbrecht S. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 3.Senior AE, Nadanaciva S, Weber J. Biochim Biophys Acta Bioenerg. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Yang W, Karplus M. Cell. 2005;123:195–205. doi: 10.1016/j.cell.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons C, Montgomery MG, Leslie AGW, Walker JE. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 7.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 8.Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K., Jr Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Gao YQ, Yang W, Marcus RA, Karplus M. Proc Natl Acad Sci USA. 2003;100:11339–11344. doi: 10.1073/pnas.1334188100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer PD. In: Membrane Bioenergetics. Lee CP, Schatz G, Ernster L, editors. Reading, MA: Addison–Wesley; 1979. pp. 461–479. [Google Scholar]
- 11.Boyer PD. Biochim Biophys Acta Bioenerg. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 12.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. Proc Natl Acad Sci USA. 2006;103:8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabaleeswaran V, Puri N, Walker JE, Leslie AGW, Mueller DM. EMBO J. 2006;25:5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. J Biol Chem. 2007;282:14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 15.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 16.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Nature. 2001;140:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 17.Shimabukuro K, Yasuda R, Muneyuki E, Hara KY, Kinosita K, Jr, Yoshida M. Proc Natl Acad Sci USA. 2003;100:14731–14736. doi: 10.1073/pnas.2434983100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishizaka T, Oiwa K, Noji H, Kimura S, Muneyuki E, Yushida M, Kinosita K., Jr Nat Struct Mol Biol. 2004;11:142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- 19.Ariga T, Muneyuki E, Yoshida M. Nat Struct Mol Biol. 2007;14:841–846. doi: 10.1038/nsmb1296. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Oster G. Nature. 1998;396:279–282. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 21.Liu MS, Todd BD, Sadus RJ. J Chem Phys. 2003;118:9890–9898. [Google Scholar]
- 22.Weber J, Wilke-Mounts S, Lee RSF, Grell E, Senior AE. J Biol Chem. 1993;268:20126–20133. [PubMed] [Google Scholar]
- 23.Weber J, Senior AE. Biochim Biophys Acta Bioenerg. 1997;1319:19–58. doi: 10.1016/s0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 24.Grüber G, Capaldi RA. Biochemistry. 1996;35:3875–3879. doi: 10.1021/bi952949h. [DOI] [PubMed] [Google Scholar]
- 25.Yang W, Gao YQ, Cui Q, Ma J, Karplus M. Proc Natl Acad Sci USA. 2003;100:874–879. doi: 10.1073/pnas.0337432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao HZ, Weber J. Proc Natl Acad Sci USA. 2007;104:18478–18483. doi: 10.1073/pnas.0709322104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Böckmann RA, Grubmüller H. Nat Struct Biol. 2002;9:198–202. doi: 10.1038/nsb760. [DOI] [PubMed] [Google Scholar]
- 28.Ma J, Flynn TC, Cui Q, Leslie AGW, Walker JE, Karplus M. Structure. 2002;10:921–931. doi: 10.1016/s0969-2126(02)00789-x. [DOI] [PubMed] [Google Scholar]
- 29.Bandyopadhyay S, Allison WS. Biochemistry. 2004;43:2533–2540. doi: 10.1021/bi036058i. [DOI] [PubMed] [Google Scholar]
- 30.Maragakis P, Karplus M. J Mol Biol. 2005;352:807–822. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 31.Schlitter J, Engels M, Krüger P. J Mol Graphics. 1994;12:84–89. doi: 10.1016/0263-7855(94)80072-3. [DOI] [PubMed] [Google Scholar]
- 32.Koga N, Takada S. Proc Natl Acad Sci USA. 2006;103:5367–5372. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menz RI, Walker JE, Leslie AGW. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 34.van der Vaart A, Karplus M. J Chem Phys. 2005;122:114903. doi: 10.1063/1.1861885. [DOI] [PubMed] [Google Scholar]
- 35.Ross J. J Phys Chem B. 2006;110:6987–6990. doi: 10.1021/jp0556862. [DOI] [PubMed] [Google Scholar]
- 36.Greene MD, Frasch WD. J Biol Chem. 2003;278:51594–51598. doi: 10.1074/jbc.M309948200. [DOI] [PubMed] [Google Scholar]
- 37.Bandyopadhyay S, Allison WS. Biochemistry. 2004;43:9495–9501. doi: 10.1021/bi0493012. [DOI] [PubMed] [Google Scholar]
- 38.Gumbiowski K, Cherepanov D, Müller M, Pänke O, Promto P, Winkler S, Junge W, Engelbrecht S. J Biol Chem. 2001;276:42287–42292. doi: 10.1074/jbc.M106884200. [DOI] [PubMed] [Google Scholar]
- 39.Hossain MD, Furuike S, Maki Y, Adachi K, Ali MY, Huq M, Itoh H, Yoshida M, Kinosita K., Jr Biophys J. 2006;90:4195–4203. doi: 10.1529/biophysj.105.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller M, Gumbiowski K, Cherepanov DA, Winkler S, Junge W, Engelbrecht S, Pänke O. Eur J Biochem. 2004;271:3914–3922. doi: 10.1111/j.1432-1033.2004.04328.x. [DOI] [PubMed] [Google Scholar]
- 41.Müller M, Pänke O, Junge W, Engelbrecht S. J Biol Chem. 2002;277:23308–23313. doi: 10.1074/jbc.M201998200. [DOI] [PubMed] [Google Scholar]
- 42.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker JE, Dickson VK. Biochim Biophys Acta Bioenerg. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 45.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.