Abstract
Aquaporins and aquaglyceroporins form a family of pore proteins that facilitate the efficient and selective flux of small solutes across biological membranes. We studied the selectivity of aquaporin-1 (AQP1) and the bacterial glycerol facilitator, GlpF, for O2, CO2, NH3, glycerol, urea, and water. Using molecular dynamics simulations, we calculated potentials of mean force for solute permeation along the aquaporin channels and compared them with the alternative pathway across the lipid bilayer. For small solutes permeating through AQP1, a remarkable anticorrelation between permeability and solute hydrophobicity was observed, whereas the opposite trend was observed for permeation through the membrane. This finding renders AQP1 a selective filter for small polar solutes, whereas GlpF was found to be highly permeable for small solutes and permeable for larger solutes. Surprisingly, not solute-channel but water-channel interactions were found to be the key determinant underlying the selectivity mechanism of aquaporins. Hence, a hydrophobic effect, together with steric restraints, determines the selectivity of aquaporins.
Keywords: free-energy profile, potential of mean force, molecular dynamics simulations, umbrella sampling, membrane permeation
Aquaporins are a ubiquitous family of intrinsic membrane proteins present throughout nature. They have been discovered in organisms as diverse as archae, bacteria, plants, insects, and mammals (1, 2). The aquaporin family is divided into two subfamilies. Ordinary aquaporins were initially seen as pure water channels, whereas aquaglyceroporins are additionally permeated by small organic compounds such as glycerol or urea (3).
The elucidation of the structure of aquaporins (4, 5) and aquaglyceroporins (6) gave the first insights into their selectivity mechanisms and formed the basis for the analysis of dynamics and energetics of water and glycerol conduction (7–9). Numerous experiments revealed diverse permeation characteristics of aquaporins. Some aquaglyceroporins are permeable to both glycerol and urea or even arsenite (10). Others such as the glycerol facilitator, GlpF, are permeable for various polyols, but hardly for urea (11). The permeability with respect to water differs significantly within the aquaporin family. Aquaporin-6 can act as an anion channel (12), and some aquaporins were reported to be permeated by ammonia (13). CO2 permeation through aquaporins has been a matter of debate since AQP1 was reported to enhance CO2 flux (14). Recently, permeation of O2 and NO across membranes has been reported to correlate with the inhibition of AQP1 (15, 16).
Two main constriction sites have been identified in the aquaporin channels (4–6). In the center of the pore, two conserved Asn-Pro-Ala motifs (NPA; compare Fig. 1A) with their two asparagine side chains pointing into the pore are located at the end of two half helices. The dipoles of the half helices generate an electrostatic barrier in the NPA region, which is, together with desolvation effects, essential for proton exclusion (17–20). Close to the extracellular exit of the channel, the aromatic/arginine (ar/R) constriction region forms the narrowest part of the pore and is therefore generally assumed to be important for the channel selectivity. This interpretation has been supported by theoretical studies (21, 22) and mutation experiments (23). The residues in the ar/R region differ in the aquaporin family, rendering the constriction site diverse in size and hydrophobicity. The highly conserved arginine, together with a nearby histidine in water-specific aquaporins, was considered to be essential for the isolation of water molecules from their solvation shell in the bulk (5). This interpretation has been questioned by recent experiments on oocyte permeability (23).
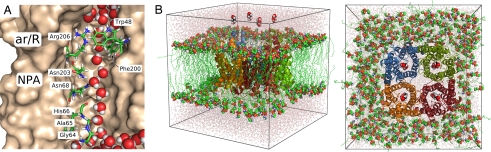
Fig. 1.
Glycerol facilitator GlpF. (A) Snapshot of an MD simulation showing a single file of water inside the pore of GlpF. Some water-interacting residues are shown in stick representation. (B) Simulation box of a GlpF tetramer solvated in a membrane of POPE and water in side view (Left) and top view (Right). Glycerol molecules (displayed in sphere representation) are placed along the channel axes as starting configurations for umbrella sampling simulations.
So far, however, a unifying picture that explains the diverse permeation properties of both aquaporins and aquaglyceroporins has not evolved. Therefore, we applied molecular dynamics simulations to determine permeation barriers of a wide range of solutes permeating through human AQP1 (hAQP1) and GlpF as typical members of the two aquaporin subfamilies. Potentials of mean force (PMFs) for O2, CO2, NH3, glycerol, urea, and water were derived by using the technique of umbrella sampling simulations. Because these solutes differ strongly in hydrophobicity and size, the permeation barriers allow one to derive a unifying picture of the selectivity of aquaporins and aquaglyceroporins. The physiological relevance of permeation through aquaporins is investigated by comparing the pathway through the aquaporin pores with the pathway across two distinct model membranes. This approach also allows one to rationalize permeation experiments on aquaporins and aquaglyceroporins embedded in oocyte membranes or liposomes. The direct evaluation of interactions that govern the selectivity enables one to identify the molecular determinants of selectivity. The results reveal a comprehensive mechanism underlying the permeation characteristics of aquaporins and aquaglyceroporins.
Results
Solute Permeability.
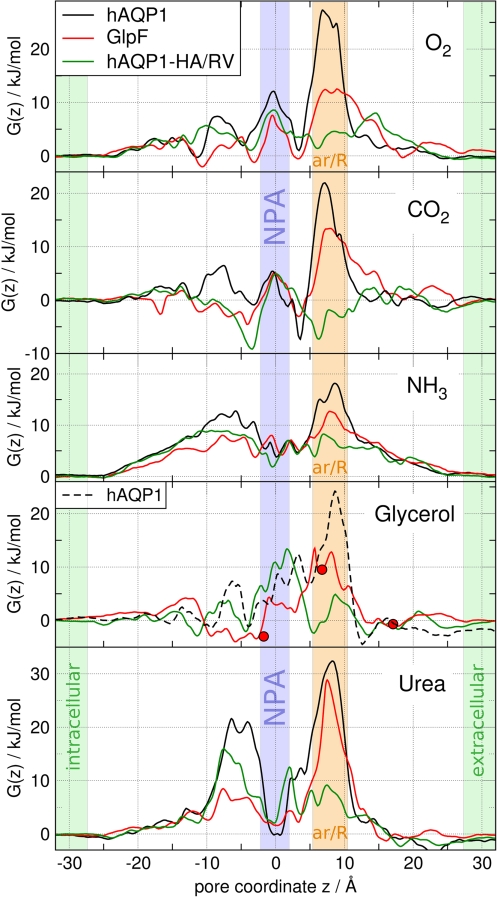
Fig. 2 presents the PMFs for permeation of O2, CO2, NH3, glycerol, and urea permeating through hAQP1 (black curves) and GlpF (red curves). The PMFs G(z) are shown as a function of the pore coordinate z, where z = 0 corresponds to the center of mass of the conserved NPA motifs.
Fig. 2.
Potentials of mean force G(z) for O2, CO2, NH3, glycerol, and urea permeating through the monomeric water pores of human AQP1 (black curves), GlpF (red curves), and the H180A/R195V mutant of AQP1 (green curves). z = 0 corresponds to the center of the NPA motifs. The NPA region is highlighted by a blue bar, the aromatic/arginine constriction region by an orange bar. The red circles correspond to the glycerol positions in the GlpF crystal structure (6).
Fig. 2 shows that the main barrier for each solute permeating through hAQP1 or GlpF is located in the ar/R region, demonstrating its role as the selectivity filter for uncharged solutes. For all solutes, the barrier in GlpF at the ar/R site is lower than the barrier in hAQP1. Hence, GlpF is more permeable to the solutes and is less selective than hAQP1. In contrast to the ar/R region, the highly conserved NPA motifs are not involved in the selectivity for uncharged solutes because they form only a minor barrier against permeation of the rather apolar O2 and CO2 molecules. The heights of the main barriers are summarized in Table 1.
Table 1.
Summary of free energy barriers ΔGmax in kJ/mol
| Solute | hAQP1 | GlpF | hAQP1-HA/RV | POPE | POPC |
|---|---|---|---|---|---|
| O2 | 27 | 12.5 | 8.5 | 6 | 4 |
| CO2 | 22 | 13.5 | 5 | 4 | 1.5 |
| NH3 | 18 | 12.5 | 9 | 19 | 15 |
| H2O | 14† | 13.5† | 13† | 31 | 27 |
| Glycerol | (24) | 13.5 | 13.5 | 30 | 27 |
| Urea | 32.5 | 29 | 16 | 32 | 34 |
†Determined from water density. All other barriers are from umbrella sampling simulations.
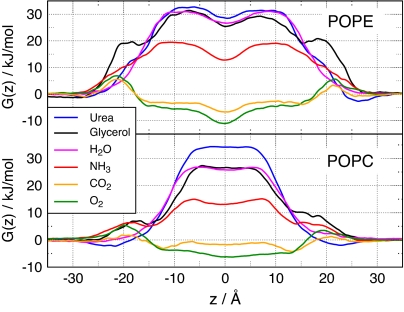
To address the physiological relevance of permeation through aquaporins, the barriers were compared with barriers against permeation across the lipid bilayer. Fig. 3 shows the PMFs for permeation of urea, glycerol, H2O, NH3, CO2, and O2 through two phospholipid membranes, one composed of pure 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and the other of pure 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). The highest barrier for the apolar O2 and CO2 molecules permeating through either POPE or POPC is ≤6 kJ/mol, indicating that these membranes are highly permeable to small apolar molecules such as O2 and CO2 (Fig. 3, green and yellow curves). The barriers against O2 and CO2 permeation through hAQP1 and GlpF are significantly higher than the corresponding membrane barriers. Hence, these aquaporin channels embedded in membranes similar to POPE or POPC are not expected to increase O2 or CO2 flux across the membrane.
Fig. 3.
PMFs for permeation of O2, CO2, NH3, H2O, glycerol, and urea through membranes of POPE (Upper) and POPC (Lower). z = 0 corresponds to the membrane center.
The barriers against ammonia permeation through POPE (19 kJ/mol) or POPC (15 kJ/mol) are considerably higher than the barriers for apolar gas molecules, reflecting its more hydrophilic nature. Compared with O2 or CO2, these barriers refer to a permeability reduced by a factor of 80–400. Here we assumed the permeability to be proportional to exp(−ΔGmax/kBT), where ΔGmax denotes the maximum barrier height, kB denotes the Boltzmann constant, and T denotes the temperature. When permeating through hAQP1 and GlpF, ammonia encounters barriers of 18 and 12.5 kJ/mol, respectively, indicating that hAQP1 is unlikely to increase NH3 permeation, whereas GlpF can significantly enhance the NH3 flux across the membrane. This observation is in line with an experimental study on ammonia permeation across the oocyte membrane (13). In that study, no influence of AQP1 on NH3 flux was measured, whereas aquaglyceroporins such as AQP3 and AQP9 were found to increase NH3 permeation.
Both glycerol and urea encounter substantial barriers between 27 and 34 kJ/mol against permeation through POPE or POPC, underlining the need for membrane channels if a substantial glycerol or urea flux is required by the metabolism. As expected, GlpF is permeated efficiently by glycerol with a maximum barrier of 13.5 kJ/mol. This value is significantly lower than the activation energy of 30.5 kJ/mol reported in a previous molecular dynamics (MD) study (9). Compared with membranes of pure POPE or POPC, the barrier of 13.5 kJ/mol results in an increase in the permeability by a factor of 750 or 230, respectively, when assuming dense expression of GlpF. These values compare favorably with the reported increase by a factor of 400 for proteoliposomes reconstituted with GlpF (24). In contrast, urea encounters a large barrier of 29 kJ/mol against permeation through GlpF, confirming that GlpF is not an efficient urea channel. Solvating GlpF in a membrane of POPE or POPC is expected to increase the permeability for urea only 3- or 7-fold, respectively, in agreement with an only 3-fold increase measured for liposomes (24). As expected, hAQP1 does not enhance urea flux across the membrane because its ar/R region forms a barrier against urea permeation of more than 30 kJ/mol.
After inserting glycerol into the ar/R region of hAQP1, we observed a widening of the pore during the energy minimization (EM) of the structure. The widening was visible from the RMSD drift of the protein atoms during the EM and did not occur in any other simulation (data not shown). These findings confirm that the ar/R site of hAQP1 is too narrow to allow passage of glycerol. The unphysical widening of the pore led to an underestimation of the barrier of 24 kJ/mol for glycerol permeation through hAQP1 (Fig. 2, dashed line). The true barrier is likely to be substantially higher, which is in line with experiments that did not observe glycerol permeation through AQP1 (25).
The role of the ar/R region as the selectivity filter for uncharged solutes can be further investigated by point mutations in the ar/R region. Here we studied the H180A/R195V double mutant of hAQP1 (hAQP1-HA/RV) because H180 and R195 are generally considered key residues for the selectivity of AQP1. A recent experimental study investigated permeation properties of several hAQP1 mutants, including hAQP1-HA/RV (23). The authors found that some of the mutations enhanced NH3 permeability or allowed urea or glycerol permeation through hAQP1. To further rationalize these findings, Fig. 2 shows the PMFs for O2, CO2, NH3, glycerol, and urea permeating through hAQP1-HA/RV as green curves. For all solutes, the barrier in the ar/R region is drastically reduced by the mutation, emphasizing the importance of the ar/R residues for channel selectivity. In contrast to the wild type, the main barriers are not located in the ar/R region anymore. O2 and CO2 can almost freely diffuse through hAQP1-HA/RV. In agreement with ref. 23, the mutation renders hAQP1 an efficient NH3 channel and allows passage of urea, whereas water permeability is hardly affected [Table 1 and supporting information (SI) Fig. 6]. In addition, our results indicate that glycerol is likely to permeate through hAQP1-HA/RV, although ref. 23 reported glycerol permeation through the hAQP1-F56A/H180A mutant only.
Solute Hydrophobicity.
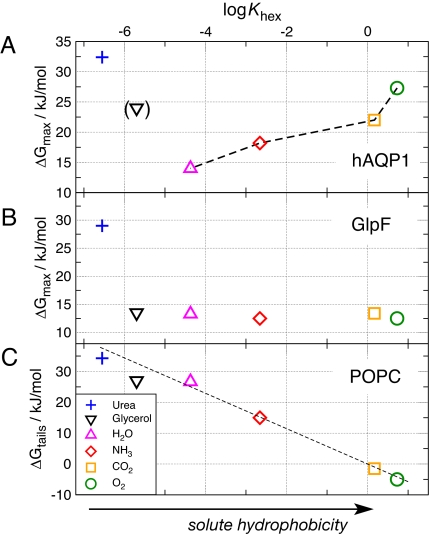
Fig. 4A displays the height of the main barrier for urea, glycerol, H2O, NH3, CO2, and O2 against permeation through hAQP1 as a function of solute hydrophobicity. The solute hydrophobicity is measured as the logarithm of the hexadecane/water partition coefficient log Khex of the solute at 298 K. Khex values were taken from refs. 26–29. For the small solutes H2O, NH3, CO2, and O2, the maximum barrier clearly increases with the solute hydrophobicity. The larger and hydrophilic glycerol and urea molecules are not excluded by hydrophobicity, but rather sterically. Hence, the ar/R region of hAQP1 can be considered as both a hydrophobicity filter and a size filter.
Fig. 4.
Permeability as a function of hydrophobicity of the solute. (A and B) Free-energy barrier ΔGmax for urea (+), glycerol (inverted triangle), H2O (triangle), NH3 (diamond), CO2 (square), and O2 (circle) permeating along the pores of AQP1 (A) and GlpF (B). ΔGmax is plotted versus the logarithm of the hexadecane–water partition coefficient log Khex of the solute, which is a common measure for hydrophobicity. (C) Solvation free-energy difference ΔGtails between the solute in water and the solute in the hydrophobic tail region of the POPC membrane as determined by umbrella sampling simulations. The dashed line indicates the energetic cost ΔGhex = −kBT ln Khex for moving the solute from bulk water into hexadecane.
Fig. 4B shows the corresponding plot for GlpF. The larger and more hydrophobic ar/R region of GlpF is less selective than the ar/R region of hAQP1, forming a significant barrier only against permeation of urea, but efficiently allowing the passage of glycerol. The difference between Fig. 4 A and B demonstrates that the differing residues in the ar/R regions of hAQP1 and GlpF account for highly different permeation characteristics.
Fig. 4C shows the energetic cost ΔGtails for moving the solute from bulk water to the hydrophobic lipid tail region of a POPC membrane. As expected, the bilayer forms a filter permeated by hydrophobic molecules. The linear dependence between ΔGtails and log Khex favorably compares to Overton's rule, stating that the permeability of a membrane with respect to different solutes is approximately proportional to their oil/water partition coefficients. For comparison, the dashed line indicates the energetic cost ΔGhex = −kBT ln Khex for moving the solute from bulk water into hexadecane.
Water–Protein Interactions as Selectivity Mechanism.
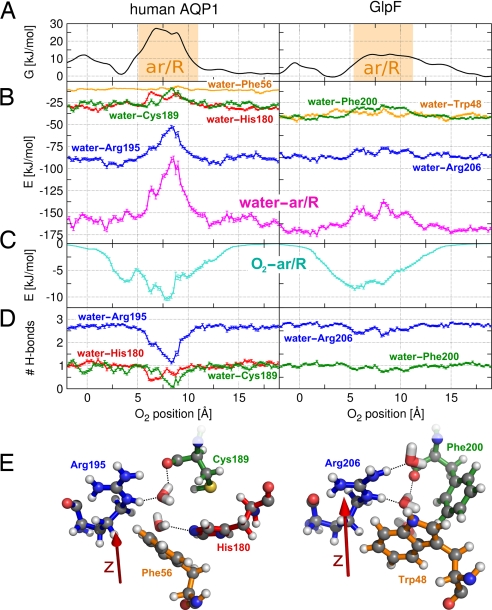
As demonstrated in the previous section, the ar/R region of hAQP1 is a filter permeated by small polar molecules. A previous MD study showed that the barrier that CO2 encounters in the ar/R region is not because of unfavorable CO2–protein interactions (22). Instead, water–Arg-195 hydrogen bonds were observed to open upon CO2 passage. Here we investigate the selectivity mechanism of hAQP1 in detail. As an example, we consider a permeation event of the apolar O2 molecule (Fig. 5). To guide the eye, Fig. 5A shows the PMFs for O2 permeating through hAQP1 and GlpF, respectively. Fig. 5B presents the interaction between water and key residues of the ar/R region versus the position of an O2 molecule inside the channel. In Fig. 5B Left, water–protein interactions in hAQP1 are presented, whereas in Fig. 5B Right, the interactions in GlpF are presented. Likewise, Fig. 5C displays the interaction between the O2 molecule and the ar/R residues as a function of O2 position, and Fig. 5D shows the average number of water–protein hydrogen bonds. In Fig. 5E, MD snapshots of the ar/R regions of hAQP1 and GlpF are shown in ball-and-stick representation. The residues are colored according to the curves in the graph, and possible water–protein hydrogen bonds are indicated by dashed lines.
Fig. 5.
Water–protein interaction as selectivity mechanism for aquaporins. Comparison between hAQP1 (Left) and GlpF (Right). (A) PMFs for O2 permeating through the ar/R regions of hAQP1 and GlpF versus the position of O2. (B) Interactions between water and the ar/R residues as a function of the position of a permeating O2 molecule (magenta curves). In AQP1, water–ar/R interactions are reduced by ≈60 kJ/mol when an O2 molecule is present in the ar/R region, and the loss of water–protein interaction cannot be compensated by O2–protein interactions, which are displayed in C. In GlpF, water–ar/R interactions are hardly affected by the O2. The water–ar/R interaction can be decomposed into interactions between water and single residues (blue, orange, green, and red curves). (D) Number of hydrogen bonds between water and protein residues versus the position of a permeating O2 molecule. (E) MD snapshots of the ar/R regions of hAQP1 and GlpF, including several water molecules. The residues are colored according to the curves in B and D. Possible water–protein hydrogen bonds are denoted by dashed lines. The red arrow indicates the pore coordinate.
The key finding is that, upon O2 passage through hAQP1, favorable interactions between water and the ar/R residues (magenta curve) are reduced by ≈60 kJ/mol. The loss of water–protein interaction cannot be compensated by O2–protein interaction because the apolar O2 interacts with the ar/R residues on the order of 10 kJ/mol. The reduction in water–protein interaction occurs at the barrier in the PMF. Decomposing the ar/R in the single residues (blue, orange, green, and red curves for Arg-195, Phe-56, Cys-189, and His-180, respectively) shows that the water–protein interaction is mainly affected by reduced interaction to Arg-195 (≈35 kJ/mol). In addition, interaction to Cys-189 and His-180 are reduced by ≈15 kJ/mol each. In the wider and more hydrophobic ar/R region of GlpF, water–ar/R interactions are hardly affected by the position of a passing O2 molecule (Fig. 5B Right). Hence, small solutes pass the ar/R region of GlpF without reducing the water–protein interaction. The same trend can be observed in Fig. 5D, which presents the average number of water–protein hydrogen bonds as a function of O2 position. Upon O2 passage through hAQP1, more than one hydrogen bond between water and Arg-195 is lost (blue line). In addition, the probability for a hydrogen bond between water and both His-180 and Cys-189 is significantly reduced (red and green lines). In contrast, water–Arg-206 hydrogen bonds in GlpF are hardly affected by a permeating O2 molecule.
Discussion
From the extensive set of MD simulations presented here, a detailed understanding of the selectivity mechanism of aquaporins can be derived. We showed that, in both aquaporins and aquaglyceroporins, the ar/R region is the selectivity site for uncharged solutes. In hAQP1, the narrow and hydrophilic ar/R site forms a filter permeated by small polar solutes. Upon passage of a solute, favorable interactions between water and protein (mainly to Arg-195 but also to Cys-189 and His-180) are reduced and need to be replaced by solute–protein interactions. The more polar the solute, the stronger it can interact with hydrophilic ar/R residues, rendering a lower energetic cost to replace a water molecule. Larger solutes such as urea or glycerol are sterically excluded in hAQP1.
The hAQP1-HA/RV double mutant was found to be highly permeable to water, with an even lower barrier against water permeation compared with the hAQP1 wild type (SI Fig. 6). Hence, Arg-195 and His-180 are not necessary to isolate single water molecules from the bulk, which is in agreement with experiments that measured high water flux across hAQP1-HA/RV (23). However, we showed that these residues are important for the selectivity of uncharged solutes because mutations in the ar/R region have drastic effects on the channel characteristics. The mutations result in a larger and more hydrophobic pore, thus eliminating the selectivity mechanism for small polar solutes, which is present in the hAQP1 wild type. Without the selectivity mechanism and because of the larger pore size, the hAQP1-HA/RV mutant is highly permeable to apolar solutes, as well as to NH3 and urea.
In aquaglyceroporins, the ar/R region is wider than in AQP1 and has a more hydrophobic pocket opposite to the arginine. In GlpF, this environment allows passage of polyols, whose hydroxyl groups can interact with Arg-206 replacing water–Arg-206 hydrogen bonds, whereas its apolar backbone matches into the hydrophobic pocket (6). Our results demonstrate that, because of the wider and more hydrophobic ar/R site, the selectivity mechanism present in AQP1 does not apply in aquaglyceroporins similar to GlpF. Therefore, GlpF-like aquaglyceroporins are efficiently permeated by small solutes such as NH3, CO2, or O2.
We stress that, from the permeabilities of the wide range of solutes that were probed in the present study, the permeability of AQP1 and GlpF for other small solutes may be estimated. The results suggest that small hydrophobic solutes such as NO, CO, or N2O are unlikely to permeate through AQP1 with barriers >20 kJ/mol. In contrast, aquaglyceroporins similar to GlpF are expected to be permeable to such solutes.
To address the possible flux of molecules such as CO2 or NO through AQP1, we suggest permeation experiments on GlpF or the hAQP1-HA/RV mutant. If permeation occurs along the AQP1 water channels and not through the lipid bilayer, the different ar/R region should drastically increase the permeability. For GlpF, for example, we would expect a 30-fold increase of the CO2 flux, compared with hAQP1.
Conclusions
A simple filter mechanism governs the permeability of AQP1. For small solutes permeating through AQP1, a hydrophobic effect leads to a remarkable anticorrelation between permeability and hydrophobicity. Large solutes are sterically excluded. Hence, the ar/R region of AQP1 is a filter against both hydrophobicity and size. This filter mechanism does not apply in GlpF, which efficiently permeates all investigated solutes except for urea. Surprisingly, not solute–pore, but water–pore interactions complemented by steric effects emerged as the key determinants underlying the selectivity mechanism and the barrier height in aquaporins and aquaglyceroporins.
Methods
Simulation Setup.
The simulation boxes of hAQP1 or GlpF contained the protein tetramer, 271 POPE lipids, and 19,769 (18,443 for GlpF) TIP4P (30) water molecules. The GlpF structure was taken from the Protein Data Bank (PDB ID code 1FX8) (6). The starting structure of hAQP1 was modeled based on the x-ray structure of bovine AQP1 (PDB ID code 1J4N) (5) by mutating differing residues by using the WHAT IF modeling software (31). Crystal water molecules were kept in the structures, and chloride ions were added to neutralize the simulation systems. The OPLS all-atom force field (32, 33) was used for the protein, and lipid parameters were taken from Berger et al. (34). All simulations were equilibrated for 2 ns before production. A typical simulation box is shown in Fig. 1B.
The simulations were carried out by using the GROMACS simulation software (35, 36). Electrostatic interactions were calculated at every step with the particle-mesh Ewald method (37, 38). Short-range repulsive and attractive dispersion interactions were described together by a Lennard–Jones potential, which was cut off at 1.0 nm. The Settle (39) algorithm was used to constrain bond lengths and angles of water molecules, and Lincs (40) was used to constrain all other bond lengths, allowing a time step of 2 fs. The simulation temperature was kept constant by weakly (τ = 0.1 ps) coupling the protein, lipids, and solvent separately to a temperature bath (41) of 300 K. Likewise, the pressure was kept constant by weakly coupling the system to a pressure bath of 1 bar with a coupling constant τ of 1 ps. During umbrella sampling simulations only, the box length in the z direction (perpendicular to the membrane) was kept fixed.
The starting frames for the umbrella simulations were taken from 20-ns equilibrium simulations of hAQP1 and GlpF. The aquaporin channels were divided into 0.25-Å-wide equidistant sections parallel to the membrane, with the center of each section representing an umbrella center. Subsequently, the solute was placed into the channel at the umbrella center. Water molecules that overlapped with the solute were removed. To enhance sampling, two to four solute molecules were placed in each pore at different positions. A distance between the solutes along the pore of at least 25 Å for water, ammonia, CO2, and O2 and at least 30 Å for glycerol and urea was imposed to ensure that interactions between the solute molecules (if any) were negligibly small. During the subsequent EM of the structure, the protein RMSD was observed to ensure that the insertion of a larger solute into the pore did not lead to an unphysical widening of the pore.
Umbrella sampling calculations were carried out by applying a harmonic restraint force along the pore coordinate with force constants between 400 and 4,000 kJ·mol−1·nm−2. In ammonia, the nitrogen atom was restrained; in water, the oxygen was restrained; in CO2, urea, and glycerol, the central carbon was restrained; and in O2, a dummy atom centered between the two oxygen atoms was restrained. Additionally, the solutes were restrained to a cylinder of radius rc = 5 Å whose axis was centered along the pore by applying an additional harmonic force F(r) = −kc(r − rc) H(r − rc) pointing toward the cylinder axis. Here r denotes the distance from the cylinder axis, kc = 400 kJ·mol−1·nm−2 the force constant, and H is the Heaviside step function. The umbrella simulations of O2, CO2, NH3, and water were carried out for 400 ps, and the simulations of urea and glycerol were carried out for 600 ps and 1 ns, respectively.
Umbrella simulations for the lipid bilayers were performed by inserting the solute molecules at various positions into random snapshots taken from a 20-ns equilibrium run of a bilayer patch. The POPE and POPC patches contained 128 lipid molecules each and 4,777 or 5,788 TIP4P water molecules, respectively. Up to 12 solute molecules were inserted into one simulation, keeping a minimum distance of 25 Å to each other parallel and perpendicular to the bilayer. After EM, the simulations ran for ≥500 ps.
Ammonia parameters were taken from ref. 33, and urea parameters were from refs. 42 and 43. Lennard–Jones parameters for O2 were taken from the CHARM22 force field (44). The O2 quadrupole was calculated from wave functions obtained at the MP2/6–311G* level to Qzz = −0.82 DÅ and modeled by a positively charged dummy atom centered between two negatively charged oxygen atoms. CO2 parameters were taken from ref. 22, and glycerol was modeled from OPLS atom types for alcohols. The parameters were validated by comparing the experimental Khex to Khex calculated by umbrella simulations (see SI Fig. 7).
Construction of PMFs.
After removing the first 100 ps for equilibration, umbrella histograms were extracted from the z coordinate of the restrained atom. Subsequently, the umbrella positions were corrected with respect to the center of mass of the two NPA motifs of the corresponding channel. This procedure avoids a possible unphysical flattening of the PMF because of fluctuations of the monomers within the tetramer. It was ensured that all positions along the channel were well sampled. In case of poor sampling at maxima in the PMF, additional umbrella simulations with higher force constants were performed. In total, 34,432 histograms were collected from 1.6 μs of simulation of the aquaporin systems. The PMFs for permeation through the lipid bilayer were constructed from 15,232 histograms that were extracted from 695 ns of simulation.
The construction of the PMFs is described in more detail in SI Methods and is illustrated in SI Fig. 8. PMFs were calculated separately for each monomeric channel by using the weighted histogram analysis method (45). The single channels were considered as independent pathways. Therefore, the effective PMF Geff(z) was calculated by exp(−Geff(z)/kBT) = 4−1Σj=14 exp(−Gj(z)/kBT). Because of the cylindric restraint, the umbrella samplings yield a PMF that refers to channel density of one channel per cross-section of the cylinder (46). We corrected the PMFs by a trapezial correction in the entrance and exit regions of the pore, such that the final PMFs refer to a density of one channel per membrane area occupied by an AQP monomer (see also SI Methods). Statistical errors were calculated by using bootstrap analysis. The procedure is illustrated in SI Fig. 9. The uncertainty at the main barriers was ≤2.5 kJ/mol in the aquaporin channels and ≤2 kJ/mol in the lipid bilayer systems.
To validate the umbrella sampling calculations, the PMFs for water permeating through hAQP1 and GlpF were independently calculated from the water density in the equilibrium trajectories. SI Fig. 10 shows the PMFs for hAQP1 and GlpF determined by umbrella sampling and calculated from the water density. The two methods agree within 3 kJ/mol.
Water–protein and solute–protein interactions (Fig. 5) were calculated as the sum of Lennard–Jones and short-range coulomb interactions. Only configurations that contribute to permeation events were taken into account. Therefore, snapshots were evaluated only when the distance between Arg-195 (Arg-206 in GlpF) and the opposite His-180 (Phe-200 in GlpF) was not reduced by >1 Å, compared with the crystal structures (5, 6). The analysis of arginine interactions was restricted to arginine atoms along the pore.
Supplementary Material
ACKNOWLEDGMENTS.
We thank I. Tremmel, G. Vriend, E. Beitz, and H. Grubmüller for carefully reading the manuscript and G. Groenhof for assistance in the calculation of partial charges. This work was supported by European Union Grant LSHP-CT-2004-012189.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707662104/DC1.
References
- 1.Preston GM, Carroll TP, Guggino WB, Agre P. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 2.Zeidel ML, Ambudkar SV, Smith BL, Agre P. Biochemistry. 1992;31:7436–7440. doi: 10.1021/bi00148a002. [DOI] [PubMed] [Google Scholar]
- 3.Maurel C, Reizer J, Schroeder JI, Chrispeels MJ, Saier MH. J Biol Chem. 1994;269:11869–11872. [PubMed] [Google Scholar]
- 4.Murata K, Mitsuoka K, Walz T, Agre P, Heymann J, Engel A, Fujiyoshi Y. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 5.Sui H, Han B.-G., Lee JK, Walian P, Jap BK. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 6.Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, Stroud RM. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 7.de Groot BL, Grubmüller H. Science. 2001;294:2353–2357. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 8.Tajkhorshid E, Nollert P, Jensen MØ, Miercke LJW, O'Connell J, Stroud RM, Schulten K. Science. 2002;296:525–530. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- 9.Jensen MØ, Park S, Tajkhorshid E, Schulten K. Proc Natl Acad Sci USA. 2002;99:6731–6736. doi: 10.1073/pnas.102649299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King LS, Kozono D, Agre P. Nat Rev Mol Cell Biol. 2004;5:687–698. doi: 10.1038/nrm1469. [DOI] [PubMed] [Google Scholar]
- 11.Heller KB, Lin EC, Wilson TH. J Bacteriol. 1980;144:274–278. doi: 10.1128/jb.144.1.274-278.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasui M, Hazama A, Kwon TH, Nielsen S, Guggino WB, Agre P. Nature. 1999;402:184–187. doi: 10.1038/46045. [DOI] [PubMed] [Google Scholar]
- 13.Holm LM, Jahn TP, Møller ALB, Schjoerring JK, Ferri D, Klaerke DA, Zeuthen T. Pflügers Arch. 2005;450:415–428. doi: 10.1007/s00424-005-1399-1. [DOI] [PubMed] [Google Scholar]
- 14.Nakhoul NL, Davis BA, Romero MF, Boron WF. Am J Physiol. 1998;274:C543–C548. doi: 10.1152/ajpcell.1998.274.2.C543. [DOI] [PubMed] [Google Scholar]
- 15.Herrera M, Hong NJ, Garvin JL. Hypertension. 2006;48:157–164. doi: 10.1161/01.HYP.0000223652.29338.77. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov II, Loktyushkin AV, Guskova RA, Vasilev NS, Fedorov GE, Rubin AB. Dokl Biochem Biophys. 2007;414:137–140. doi: 10.1134/s160767290703012x. [DOI] [PubMed] [Google Scholar]
- 17.de Groot BL, Frigato T, Helms V, Grubmüller H. J Mol Biol. 2003;333:279–293. doi: 10.1016/j.jmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti N, Roux B, Pomes R. J Mol Biol. 2004;343:493–510. doi: 10.1016/j.jmb.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 19.Ilan B, Tajkhorshid E, Schulten K, Voth GA. Proteins. 2004;55:223–228. doi: 10.1002/prot.20038. [DOI] [PubMed] [Google Scholar]
- 20.Kato M, Pisliakov AV, Warshel A. Proteins. 2006;64:829–844. doi: 10.1002/prot.21012. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Schulten K, Tajkhorshid E. Structure (London) 2005;13:1107–1118. doi: 10.1016/j.str.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Hub JS, de Groot BL. Biophys J. 2006;91:842–848. doi: 10.1529/biophysj.106.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. Proc Natl Acad Sci USA. 2006;103:269–274. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borgnia MJ, Agre P. Proc Natl Acad Sci USA. 2001;98:2888–2893. doi: 10.1073/pnas.051628098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Verkman AS. J Biol Chem. 1997;272:16140–16146. doi: 10.1074/jbc.272.26.16140. [DOI] [PubMed] [Google Scholar]
- 26.Walter A, Gutknecht J. J Membr Biol. 1986;90:207–217. doi: 10.1007/BF01870127. [DOI] [PubMed] [Google Scholar]
- 27.Finkelstein A. J Gen Physiol. 1976;68:127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham MH, Buist GJ, Grellier PL, McGill RA, Doherty RM, Kamlet MJ, Taft RW, Maroldo SG. J Chromatogr. 1987;409:15–27. doi: 10.1016/s0021-9673(01)86779-0. [DOI] [PubMed] [Google Scholar]
- 29.Simon SA, Gutknecht J. Biochim Biophys Acta. 1980;596:352–358. doi: 10.1016/0005-2736(80)90122-4. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 31.Vriend G. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen WL, Maxwell DS, Tirado-Rives J. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 33.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 34.Berger O, Edholm O, Jähnig F. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindahl E, Hess B, Van der Spoel D. J Mol Model. 2001;7:306–317. [Google Scholar]
- 36.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. J Comput Chem. 2005;26:701–1719. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 37.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8592. [Google Scholar]
- 39.Miyamoto S, Kollman PA. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 40.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 41.Berendsen HJC, Postma JPM, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 42.Duffy EM, Severance DL, Jorgensen WL. Isr J Chem. 1993;33:323–330. [Google Scholar]
- 43.Smith LJ, Berendsen HJC, van Gunsteren WF. J Phys Chem B. 2004;108:1065–1071. [Google Scholar]
- 44.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 46.Allen TW, Andersen OS, Roux B. Biophys Chem. 2006;124:251–267. doi: 10.1016/j.bpc.2006.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.