Abstract
Monotremes have left a poor fossil record, and paleontology has been virtually mute during two decades of discussion about molecular clock estimates of the timing of divergence between the platypus and echidna clades. We describe evidence from high-resolution x-ray computed tomography indicating that Teinolophos, an Early Cretaceous fossil from Australia's Flat Rocks locality (121–112.5 Ma), lies within the crown clade Monotremata, as a basal platypus. Strict molecular clock estimates of the divergence between platypus and echidnas range from 17 to 80 Ma, but Teinolophos suggests that the two monotreme clades were already distinct in the Early Cretaceous, and that their divergence may predate even the oldest strict molecular estimates by at least 50%. We generated relaxed molecular clock models using three different data sets, but only one yielded a date overlapping with the age of Teinolophos. Morphology suggests that Teinolophos is a platypus in both phylogenetic and ecological aspects, and tends to contradict the popular view of rapid Cenozoic monotreme diversification. Whereas the monotreme fossil record is still sparse and open to interpretation, the new data are consistent with much slower ecological, morphological, and taxonomic diversification rates for monotremes than in their sister taxon, the therian mammals. This alternative view of a deep geological history for monotremes suggests that rate heterogeneities may have affected mammalian evolution in such a way as to defeat strict molecular clock models and to challenge even relaxed molecular clock models when applied to mammalian history at a deep temporal scale.
Keywords: Mammalia, Monotremata, phylogeny, molecular clock
The timing of divergence of the two living monotreme clades is of general interest because it bears on basal events in mammalian history and provides independent calibration for understanding temporal aspects of the great radiation of therian mammals. Strict molecular clock estimates of the platypus-echidna divergence time range from 17 Ma to 80 Ma (1–11), with most opinions favoring the younger end of the spectrum (Table 1). Recent discoveries of Mesozoic and earliest Cenozoic fossils hint at a far deeper geological history for monotremes (12, 13); however, these fossils consist mostly of isolated teeth and jaws and their precise relationships are controversial. They are generally relegated to positions at the base of the monotreme stem, and viewed as consistent with a relatively recent divergence between the living platypus and echidna clades.
Table 1.
Published estimates of echidna–platypus divergence times, including our relaxed molecular clock estimates (bottom three rows)
| Date | Data/method |
|---|---|
| 17–25 Ma (10) | Immunoglobulin IgM |
| 18–27 Ma (4) | Molecular clock estimates based on Protamine P1 genes (290 bp for platypus, 311 bp for echidna) |
| 20–45 Ma (6) | Molecular estimates based on mitochondrial ND1 protein sequences and assuming that the echidna-platypus split is 20–30% as old as the monotreme-marsupial split |
| 25 Ma (7) | Molecular clock estimate based on single copy DNA–DNA hybridization data |
| 25–30 Ma (5) | Molecular clock estimates based on partial mitochondrial 12S rRNA gene sequences |
| 28–73 Ma (2) | Molecular clock estimates based on myoglobin, α-globin, and β-globin protein sequences |
| 34 Ma (9) | Molecular clock estimate based on amino acid sequences for 12 mitochondrial proteins |
| 54 Ma (1) | Molecular clock estimate based on α-and β-haemoglobin and myoglobin protein sequences |
| 50–57 Ma (8) | Molecular clock estimate based on α-lactalbumin protein sequences |
| 63.6 Ma (11) | Molecular clock estimate based on 66 nDNA, mDNA, tRNA genes |
| >63.2 - 61.8 Ma (13) | Fossil (Monotrematum), paleomagnetic date |
| 64–80 Ma (3) | Molecular clock estimates based on single copy DNA–DNA hybridization data |
| 63.7 (95.0–39.7) Ma | Relaxed molecular clock estimate based on Woodburne et al. IGF2 data (41) |
| 79.5 (110.4–51.6) Ma | Relaxed molecular clock estimate based on van Rheede et al. Dataset I DNA data (29) |
| 88.9 (130.8–55.6) Ma | Relaxed molecular clock estimate based on van Rheede et al. Dataset I amino acid data (29) |
High-resolution x-ray computed tomography (HRXCT) is enabling a systematic reappraisal of these important and controversial fossils by generating new information on the comparative internal structure of the mandible and dentition (14, 15). In this report, we focus on one such fossil, Teinolophos trusleri, and describe new information on derived features of its jaw morphology. We also re-analyzed a large data set of morphological characters relevant to basal mammalian phylogeny, and our results shifted the phylogenetic position of Teinolophos, from stem to crown Monotremata. The implications of this seemingly minor adjustment are magnified and amplified by the great antiquity of this fossil. As we report below, this adjustment may broadly affect our understanding of early mammalian history, with special implications for molecular clock estimates of basal divergence times.
Monotremata today comprises five species that form two distinct clades (16). The echidna clade includes one short-beaked species (Tachyglossus aculeatus; Australia and surrounding islands) and three long-beaked species (Zaglossus bruijni, Z. bartoni, and Z. attenboroughi, all from New Guinea). The platypus clade includes only Ornithorhynchus anatinus (Australia, Tasmania). At first glance, the platypus and echidnas may seem as different from each other as they are from therian mammals, yet monotreme monophyly is supported by skeletal morphology (17–22), brain architecture (23, 24), facial electroreceptor arrays of unique structure (25), karyotype (26), and mitochondrial (27, 28) and nuclear gene sequences (29). Monotremes are conventionally recognized as the sister clade to therian mammals, and to retain many plesiomorphic mammalian features that were transformed during therian evolution (30, 31).
Challenges to this conventional view were raised recently, when either an echidna or the platypus was found to be phylogenetically nested within, or as sister taxon to, marsupials. Evidence came from sequence analyses of 18s rRNA (32), and both mitochondrial (9, 32–37) and nuclear genes (7, 38, 39). These findings resurrected the obscure “Marsupionta” hypothesis, which contended that monotremes are derived marsupials who secondarily reacquired such ancient characters as ovipary. Formulated in 1947 by the great morphologist W. K. Gregory (17), this hypothesis has been largely disregarded ever since (29, 31). If true, it would profoundly alter the framework in which mammalian history is understood today.
The Marsupionta hypothesis has been unanimously rejected in favor of the conventional view of monotreme-therian relationships in all recent computed phylogenetic analyses that incorporated fossils and large samples of skeletal characters (18–22, 40–43), including our analysis. The molecular arguments favoring Marsupionta have also been challenged. Some analyses did not simultaneously sample both monotreme clades (9, 32), whereas those that sampled both platypus and echidna clades supported the conventional view of monotreme relationships (11, 27–29, 44). Analyses of large concatenated sets of nuclear gene sequences suggest that ribosomal and mitrochondrial sequences are most successful in modeling and resolving branching events in relatively shallow time whereas long nuclear sequences preserve stronger signal on the timing of Mesozoic and more ancient events (6, 26, 29, 41, 45). The underlying clock models and calibration rationale for molecular estimates are also matters of debate (5–8, 41).
Owing to the sparseness of crown monotreme fossils, molecular clock models have been the method of choice for most estimates of echidna–platypus divergence timing, and various events in therian history were used as calibration. Fossils still provide decisive evidence on minimum divergence times and are required for molecular clock calibration (46, 47). However, the oldest verifiable echidna fossil is from the middle Miocene (48, 49) and until now, the oldest unequivocal platypus (Obdurodon, Fig. 1) dated to the late Oligocene (49–51). These taxa are based on fairly complete skulls whose identities are unmistakable because they possess nearly all of the apomorphies that are so distinctive of their living relatives. None has offered much insight into more distant monotreme history, into what the last common ancestor of the platypus and echidnas looked like, or when it lived. Different events in therian history were used to calibrate the molecular clock models applied to monotreme history, and they produced conflicting timing estimates of the echidna–platypus split (Table 1).
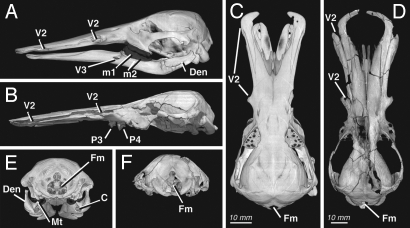
Fig. 1.
Ornithorhynchus anatinus, in lateral (A), dorsal (C), and posterior (E) views, compared with its extinct relative Obdurodon dicksoni (lacking mandible) in lateral (B), dorsal (D), and posterior (F) views. Volumetric reconstructions based on HRXCT, illustrated at same lengths (not to scale). Abbreviations: Den (dentary), Fm (foramen magnum); m1 (sclerified first lower molar); m2 (sclerified second lower molar); Mt (mandibular tubercle); P3 (upper third premolar); P4 (upper fourth premolar); V2 (foramina for maxillary nerve), and V3 (foramen for mandibular nerve).
The monotreme stem was long thought to be represented by the extinct Mesozoic taxa Morganucodontidae (Late Triassic–Middle Jurassic) and Multituberculata (Late Jurassic–Paleogene) (52–53). Under that interpretation, the fossil record testified that monotremes and therians had separated by the Late Triassic. However, those relationships were unanimously overturned by computed phylogenetic analyses that incorporated large samples of skeletal characters. Morganucodotids are now thought to be the sister taxon to crown Mammalia, whereas multituberculates are stem therians (18–21, 40–43). The revised phylogeny also altered the evidentiary standing of the Triassic fossils, placing them on the mammalian stem rather than within the crown, and leaving us without any Triassic mammals or any stem monotremes.
Recent discoveries of Mesozoic fossils from the Southern Hemisphere have renewed hope of acquiring genuine clues to early monotreme history. One record is an isolated humerus from the Early Cretaceous (≈106 Ma) Dinosaur Cove locality of Victoria, Australia, that resembles an echidna, but is sufficiently incomplete as to deny unequivocal attribution (54). All of the other fossils consist of isolated teeth and jaws, and the phylogentic placement of each has been challenged on different grounds (40, 41, 55, 56, 60). The Early Cretaceous Australian fossil Teinolophos is among these.
A number of researchers have noted that dental morphology allies Teinolophos with monotremes (40, 41, 58–60), and specifically with the platypus (31, 56, 61). However, the complete absence of teeth in all known echidnas has left equivocal the nature of dental similarities that Teinolophos shares with Obdurodon and Ornithorhynchus. They could either be indications of true platypus affinities, or plesiomorphic characters present in monotremes ancestrally. Other researchers argued that Teinolophos retains a Meckelian canal that held postdentary elements, another plesiomorphic feature placing Teinolophos outside of crown monotremes (22, 61). Previous phylogenetic analyses have all favored the hypothesis that Teinolophos branched from the monotreme stem (11, 21, 57, 59, 61, 62), in a relationship consistent with a comparatively recent divergence of the platypus and echidna clades.
Also assigned to the monotreme stem by several analyses are Steropodon, Ausktribosphenos, Bishops (Early Cretaceous, Australia), Ambondro (Middle Jurassic, Madagascar), and Asfaltomylos (Middle–Late Jurassic, Argentina) (57, 58, 62). Believed to represent a clade that includes living monotremes, this assemblage was named “Australosphenida,” and this hypothesis of relationships formed the basis for controversial arguments that the tribosphenic molar (57) and middle ear (61, 63) evolved independently in australosphenidans and therians.
For our study, three specimens of Teinolophos (specimen numbers: NMV P216750, NMV P216575, and NMV P212933) were scanned at the University of Texas High Resolution x-ray Computed Tomography facility (Figs. 2 and 3). Two were edentulous jaws, and the third was a dentary with three teeth, all from the Flat Rocks locality, Wonthaggi Formation, Bunarong Marine Park, Victoria, Australia, where the type specimen of Teinolophos was collected (64). These data were compared with scans of living monotremes including an adult Ornithorhynchus and a juvenile still in possession of its deciduous teeth, the fossil platypus Obdurodon, the fossil mammaliaform Morganucodon, several eutriconodont fossils, several multituberculates, and a large collection of extant therian mammals scanned with HRXCT over the last decade (Fig. 4). The new morphological data were used to modify a large morphological data set published by Luo and Wible (21), which we analyzed using parsimony. Insofar as our results were inconsistent with all strict molecular clock estimates for the echidna–platypus split, we applied three different relaxed clock modes using different sequences, to evaluate whether any molecular clock models is consistent with the interpretation of a deep geological age for the echidna–platypus divergence. Our matrix, modified character scores, sequence analyses, and methodology are described in supporting information (SI) Text.
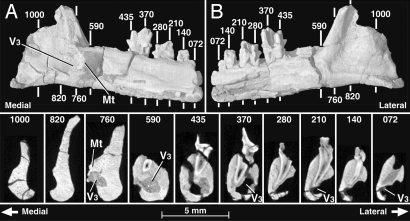
Fig. 2.
Teinolophos trusleri (NMV P216575). Volumetric reconstruction of left dentary, from HRXCT, in medial (A) and lateral (B) views, and in selected coronal cross sections (slice thickness = 0.019 mm). Medial tubercle (Mt) and mandibular canal (V3) are labeled. Complete CT serial section stacks and volumetric animations are available at http://digimorph.org/specimens/Teinolophos_trusleri/216575/.
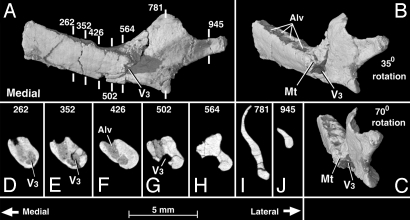
Fig. 3.
Teinolophos trusleri (NMV P216750). Volumetric reconstruction of right dentary, built from HRXCT, in medial (A) and increasingly oblique views (B and C). (D–J) Selected coronal cross sections (slice thickness = 0.012 mm) of this jaw, with slice sequence positions indicated by numbers and vertical white lines in A to show position of the slice plane. Abbreviations: Alv, tooth alveoli; V3, mandibular canal; Mt, Medial tubercle. Complete CT serial section stacks and volumetric animations are available at http://digimorph.org/specimens/Teinolophos_trusleri/216750/.
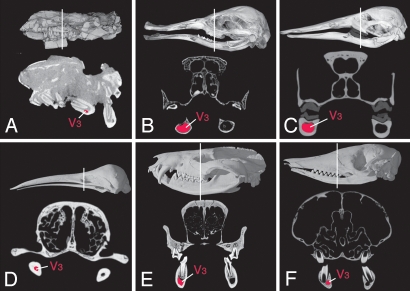
Fig. 4.
Comparative diameters of the mandibular canal (in red) shown in HRXCT coronal slice planes, with the location of the slice marked by a white line on three-dimensional reconstructions of complete skulls in lateral view. (A) Morganucodon sp. (IVPP 8685), Early Jurassic fossil from China. (B) Ornithorhynchus anatinus (AMNH 252512), juvenile specimen still in possession of its deciduous dentition. (C) Ornithorhynchus anatinus (AMNH 200255) adult specimen with keratinous adult dentition. (D) Zaglossus bruijni (AMNH 157072) adult specimen. (E) Didelphis virginiana (TMM M-2517) adult specimen. (F) Dasypus novemcinctus (TMM M-7417), adult specimen. The mandibular canal is labeled (V3). Images not to scale; scaled imagery and complete CT data sets for each are available at www.DigiMorph.org.
Results
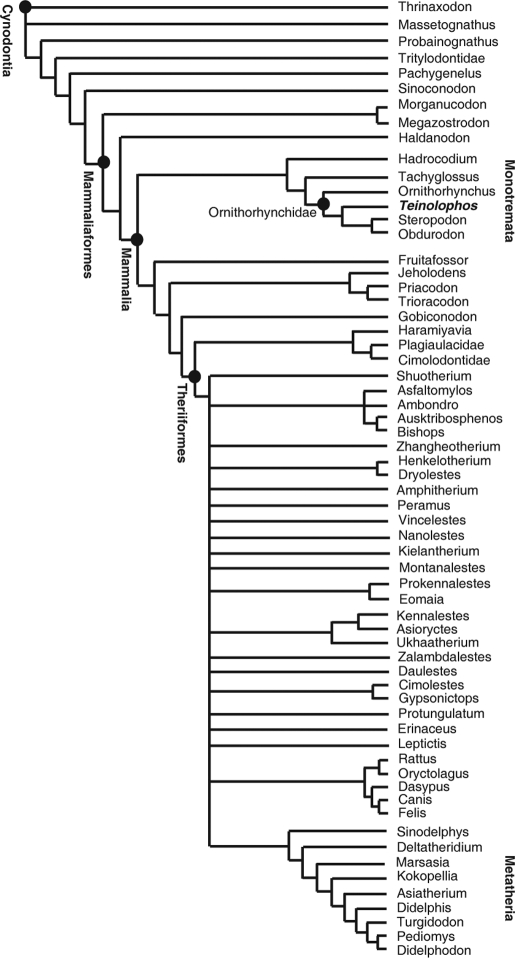
Our analysis found that Teinolophos lies within the monotreme crown, as the most ancient member of the platypus clade, Ornithorhynchidae (Fig. 5). Its precise position among ornithorhynchids was sensitive to different taxon samples, but invariably Teinolophos clustered with other ornithorhynchids (see SI Text). The most compelling evidence to us of its platypus affinities was provided by the HRXCT scans, which revealed the presence of a hypertrophied mandibular canal coursing along the entire length of dentary in a position lateral to the molariform tooth roots, and which exits the ramus medially beneath a large medial tubercle (Figs. 1–3). Among extant mammals, only the platypus has such a hypertrophied canal and medial tubercle (Fig. 4). Whereas diprotodont marsupials have an enlarged canal near the back of the jaw for insertion of the pterygoideus musculature (65), our comparative analysis indicates that only in ornithorhynchids is the mandibular canal hypertrophied along its entire length.
Fig. 5.
Cladogram showing relationships of Teinolophos to other mammals and their extinct relatives among Cynodontia (see SI Text for matrix and details of methodology).
The ornithorhynchid mandibular canal transmits the mandibular artery and hypertrophied mandibular branch of the trigeminal nerve, in support of the electroreceptive bill that gives the duckbilled platypus its common name (66). The bill deploys an array of 60,000 mechanoreceptors along with 40,000 mucous gland electroreceptors. Electroreceptive nerve terminals lie in the ducts of glands that secrete mucous when immersed in water, and they measure electrical profiles of aquatic prey items (25, 67–72). Stimului received by receptors in the bill are transmitted via comparatively huge mandibular and maxillary branches of the trigeminal nerve to an expansive population of neurons in the S1 somatosensory cortex that are bimodally responsive to both mechanical and electrical signals (25, 69, 70). Echidnas also have electroreceptive capabilities in their beaks (72), but their sensitivity is far less than in the platypus bill. The long-beaked echidnas have only ≈2,000 electroreceptors, whereas the short-beaked echidna has only ≈400 (25), which they use in probing moist substrate for food. Both have comparatively narrow mandibular canals, reflecting the plesiomorphic condition that is found in Morganucodon and all therians sampled (Fig. 4). Electroreception therefore appears to be an apomorphic characteristic of Monotremata, whereas the evolution of a specialized duckbill for high-resolution aquatic electroreception is unique to the platypus clade. Teinolophos preserves the oldest evidence of a duckbill in its hypertrophied mandibular canal.
Our analysis also affirms that the slightly younger (110 Ma) Australian fossil Steropodon (12, 73), known from a single broken jaw, is also a platypus. This unique specimen was preserved as an opal infilling of a natural mold, left after the actual bone and teeth dissolved. However, the dental resemblances to Teinolophos and Obdurodon are thoroughly documented (40, 41, 59, 60), and it preserves the edge of a large mandibular canal. The Paleocene fossil Monotrematum (74), based on three teeth from Argentina, is probably also a member of the platypus clade (Ornithorhynchidae) (31).
Optical data (61) has been interpreted as evidence for the presence of postdentary bones in Teinolophos. However, HRXCT data show no evidence of a postdentary trough or postdentary bones, suggesting that Teinolophos had a “standard” mammalian middle ear in which the auditory ossicles were separate from the lower jaw and hung suspended beneath the otic capsule, as in the platypus today (75, 76). This finding adds to the mounting evidence (60) that “Australosphenida” is a polyphyletic assemblage, with several of its members (Teinolophos, Steropodon) belonging to crown monotremes, whereas others (Ausktribosphenos, Bishops, Ambondro, Asfaltomylos) clustered consistently with therians. The precise positions of the latter taxa were also sensitive to taxon sampling in our analyses, but invariably they clustered with therian mammals (see SI Text). Should the Australosphenida hypothesis fail, then so too would assertions based thereupon that the mammalian middle ear and tribosphenic molars evolved convergently (57, 58, 61, 62, 77).
The Flat Rocks locality was dated by using the fission track method at 121–112.5 Ma (64). The finding that Teinolophos is a platypus indicates that the platypus and echidna clades diverged during or before the Early Cretaceous. This date is more ancient by a factor of 7 than the youngest, and 50% older than the oldest strict molecular clock estimates (Table 1). The recent characterization of monotreme history as a “long-fuse” clade, whose diversification into platypus and echidna clades postdated the Cretaceous–Tertiary boundary (11), is difficult to reconcile with our more ancient divergence estimate, nor is there evidence of a diversity “explosion” at any time in monotreme history.
Although far older than any previous estimate, the accumulation of anatomical novelty in even the oldest monotreme fossils suggests that our estimate may underrepresent the actual timing of the split between platypus and echidna. Viewing Teinolophos and Steropodon as platypuses in their ecological aspect suggests that, since the Early Cretaceous, rates of ecological exploitation and morphological diversification among monotremes may have been far slower than is the case in most or all therian clades. The 1,000-fold difference in species diversity found today (5 monotremes vs. 5,362 therian species; ref. 16) may be another indication that monotremes evolved at far slower rates than therians. Several molecular clock studies (6–9) have also suggested the possibility of a molecular evolution rate slowdown in monotremes. A rate slowdown in monotremes will result in estimates for the platypus–echidna divergence that are too young if calibrations are derived from therian taxa with faster rates of molecular evolution.
Given differences in rates of molecular evolution, we applied a relaxed molecular clock method (refs. 78–80; see SI Text) to reanalyze both the DNA and amino acid sequence versions of van Rheede et al.'s (29) data set I for 21 mammalian taxa. Each analysis gave a different estimate. The amino acid data of van Rheede et al.'s (29) data set I yielded a point estimate of 88.9 Ma for the platypus–echidna split, with 95% credibility intervals ranging from 130.8 to 55.6 Ma. This result overlaps broadly with the 121–112.5 Ma date for Teinolophos. However, the DNA data set of van Rheede et al. yielded a point estimate of 79.5 Ma (credibility interval 110.4 to 51.6 Ma). Previously, Woodburne et al. (41) reported a relaxed clock point estimate of 63.7 Ma (credibility interval 95.0–39.7 Ma) for the platypus–echidna split based on IGF2 amino acid sequences.
Discussion
Even considering Teinolophos as a crown montreme, the monotreme fossil record remains dismally sparse and open to interpretation. Our results are consistent with estimates of a Triassic date for the monotreme–therian split (41) (SI Text), although as yet we have no Triassic crown-mammalian fossils that would offer direct corroboration. If the new position of Teinolophos is upheld, crown monotremes had originated and the platypus and echidna clades were established by the Early Cretaceous.
Monotremes have left only a meager fossil record, but what is known at present is consistent with the view that soon after their divergence, in or before the Early Cretaceous, monotremes settled into rates of molecular and morphological evolution and speciation far slower than in the living clades of therian mammals. Even the monotreme metabolic rates and ventilation rates are much slower than in therian mammals of similar body mass, and their body temperature is lower as well (81). In what measure and to what degree these various rates are coupled or were independently evolving phenomena remains to be determined. It is also difficult to discern in which respects monontremes are simply expressing plesiomorphic mammalian rates, whether there have been apomorphic slowdowns in monotremes that evolved following their divergence from therians, or to what degree therian history can be characterized by rate accelerations over the ancestral states for mammals. In any case, our results suggest that different mammalian clades were subject to evolutionary rate heterogeneities that are incompatible with strict molecular clocks and difficult to accommodate even when relaxed molecular clock models are applied to mammalian history on a deep temporal scale.
Supplementary Material
ACKNOWLEDGMENTS.
This study was funded by National Science Foundation Grants IIS-0208675 and AToL 0531767, and the Geology Foundation of the Jackson School of Geosciences, The University of Texas at Austin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Digital Morphology data have been deposited online at www.digimorph.org (http://digimorph.org/specimens/Teinolophos_trusleri/216575, http://digimorph.org/specimens/Teinolophos_trusleri/216750, and http://digimorph.org/specimens/Teinolophos_trusleri/212933)
This article contains supporting information online at www.pnas.org/cgi/content/full/0706385105/DC1.
References
- 1.Clemens WA, Richardson BJ, Baverstock PR. In: Faunas of Australia: Biogeography and Phylogeny of the Metatheria. Walton DW, Richardson BJ, editors. Canberra: Austr Gov Publ Serv; 1989. pp. 527–548. [Google Scholar]
- 2.Hope RM, Cooper S, Wainwright B. Austral J Zool. 1989;37:289–313. [Google Scholar]
- 3.Westerman M, Edwards D. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 28–34. [Google Scholar]
- 4.Retief JD, Winkfein RJ, Dixon GH. Eur J Biochem. 1993;218:457–461. doi: 10.1111/j.1432-1033.1993.tb18396.x. [DOI] [PubMed] [Google Scholar]
- 5.Gemmell NJ, Westerman MJ. Mamm Evol. 1994;2:3–23. [Google Scholar]
- 6.Cao Y, Janke A, Waddell PJ, Westerman M, Takenaka O, Murata S, Okada N, Pääbo S, Hasegawa M. J Mol Evol. 1998;47:307–322. doi: 10.1007/pl00006389. [DOI] [PubMed] [Google Scholar]
- 7.Kirsch JAW, Mayer GC. Philos Trans R Soc B. 1998;353:1221–1237. doi: 10.1098/rstb.1998.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messer M, Weiss A, Shaw D, Westerman M. J Mamm Evol. 1998;5:95–105. [Google Scholar]
- 9.Janke A, Magnell O, Wieczorek G, Westerman M, Arnason U. J Mol Evol. 2002;54:71–80. doi: 10.1007/s00239-001-0019-8. [DOI] [PubMed] [Google Scholar]
- 10.Belov K, Hellman L. Austral Mamm. 2003;25:87–94. [Google Scholar]
- 11.Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Rutger AV, Gittleman JL, Purvis A. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 12.Archer M, Flannery TF, Ritchie A, Molnar RE. Nature. 1985;318:363–366. [Google Scholar]
- 13.Pascual R, Archer M, Jaureguizar EO, Prado JL, Godthelp H, Hand SJ. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 1–14. [Google Scholar]
- 14.Rowe T, Kappelman J, Carlson WD, Ketcham RA, Denison C. Geotimes. 1997;42:23–27. [Google Scholar]
- 15.Carlson WD, Rowe T, Ketcham RA, Colbert MW. In: Applications of X-ray Computed Tomography in the Geosciences. Mees F, Swennen R, Van Geet M, Jacobs P, editors. London: Geol Soc; 2003. pp. 7–22. [Google Scholar]
- 16.Wilson DE, Reeder DM, editors. Mammal Species of the World. Baltimore: Johns Hopkins Univ Press; 2005. [Google Scholar]
- 17.Gregory WK. Bull Amer Mus Nat Hist. 1947;88:1–52. [Google Scholar]
- 18.Rowe T. J Vert Paleo. 1988;8:241–264. [Google Scholar]
- 19.Rowe T. In: Mammalian Phylogeny. Szalay FS, Novacek MJ, McKenna MC, editors. New York: Springer; 1993. pp. 129–145. [Google Scholar]
- 20.Rougier GW, Wible JR, Hopson JA. Am Mus Novitates. 1996;3138:1–38. [Google Scholar]
- 21.Luo Z-X, Wible J. Science. 2005;308:103–107. doi: 10.1126/science.1108875. [DOI] [PubMed] [Google Scholar]
- 22.Meng J, Hu Y, Wang Y, Wang X, Li C. Nature. 2006;444:889–893. doi: 10.1038/nature05234. [DOI] [PubMed] [Google Scholar]
- 23.Macrini TE, Rowe T, Archer M. J Morph. 2006;267:1000–1015. doi: 10.1002/jmor.10452. [DOI] [PubMed] [Google Scholar]
- 24.Rowe MJ, Bohringer RC. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 177–193. [Google Scholar]
- 25.Pettigrew JD. J Exp Biol. 1999;202:1447–1454. doi: 10.1242/jeb.202.10.1447. [DOI] [PubMed] [Google Scholar]
- 26.Grützner F, Deakin J, Rens W, El-Mogharbel N, Graves JAM. Comp Biochem Physiol A. 2003;136:867–881. doi: 10.1016/j.cbpb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Phillips MJ, Penny D. Mol Phylogenet Evol. 2003;28:171–185. doi: 10.1016/s1055-7903(03)00057-5. [DOI] [PubMed] [Google Scholar]
- 28.Reyes A, Gissi C, Catzeflis F, Nevo E, Pesole G, Sacconell C. Mol Biol Evol. 2004;21:397–403. doi: 10.1093/molbev/msh033. [DOI] [PubMed] [Google Scholar]
- 29.van Rheede T, Bastiaans T, Boone DN, Hedges SB, de Jong WW, Madsen O. Mol Biol Evol. 2006;23:587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- 30.Burrell H. The Platypus. Sydney: Angus & Robertson; 1927. [Google Scholar]
- 31.McKenna MC, Bell SO. Classification of Mammals Above the Species Level. New York: Columbia Univ Press; 1997. [Google Scholar]
- 32.Janke A, Xu X, Arnason U. Proc Natl Acad Sci USA. 1997;94:1276–1281. doi: 10.1073/pnas.94.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zardoya R, Meyer A. Proc Natl Acad Sci USA. 1998;95:14226–14231. doi: 10.1073/pnas.95.24.14226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumanzawa Y, Nishida M. Mol Biol Evol. 1999;16:784–792. doi: 10.1093/oxfordjournals.molbev.a026163. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson MA, Arnason U, Spencer PB, Janke A. Gene. 2004;340:189–196. doi: 10.1016/j.gene.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 36.Penny D, Hasegawa M. Nature. 1997;387:549–550. doi: 10.1038/42352. [DOI] [PubMed] [Google Scholar]
- 37.Penny D, Hasegawa M, Wadell PJ, Hendy MD. Syst Biol. 1999;48:76–93. doi: 10.1080/106351599260454. [DOI] [PubMed] [Google Scholar]
- 38.Vernesson M, Aveskogh M, Munday B, Hellman L. Eur J Immunol. 2002;32:2145–2155. doi: 10.1002/1521-4141(200208)32:8<2145::AID-IMMU2145>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.Nowak MA, Parra ZE, Hellman L, Miller RD. Immunogenetics. 2004;56:555–563. doi: 10.1007/s00251-004-0720-z. [DOI] [PubMed] [Google Scholar]
- 40.Woodburne MO. J Mamm Evol. 2003;10:195–248. doi: 10.1007/s10914-010-9144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodburne MO, Rich TH, Springer MS. Mol Phylo Evol. 2003;28:360–385. doi: 10.1016/s1055-7903(03)00113-1. [DOI] [PubMed] [Google Scholar]
- 42.Ji Q, Luo Z-X, Yuan C-X, Wible JR, Zhang JP, Georgi JA. Nature. 2002;416:816–822. doi: 10.1038/416816a. [DOI] [PubMed] [Google Scholar]
- 43.Ji Q, Luo Z-X, Yuan C-X, Tabrum AR. Science. 2006;311:1123–1127. doi: 10.1126/science.1123026. [DOI] [PubMed] [Google Scholar]
- 44.Toyosawa S, O'hUigin C, Figueroa F, Tichy H, Klein J. Proc Natl Acad Sci USA. 1998;95:13056–13061. doi: 10.1073/pnas.95.22.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy WJ, et al. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 46.Reisz RR, Müller J. Trends Genet. 2004;20:237–241. doi: 10.1016/j.tig.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Müller J, Reisz RR. BioEssays. 2005;27:1069–1075. doi: 10.1002/bies.20286. [DOI] [PubMed] [Google Scholar]
- 48.Woodburne MO, Tedford RH, Archer A, Turnbull WD, Plane MD, Lundelius EL. Spec Pub S Austral Dept Mines Energy. 1985;5:347–363. [Google Scholar]
- 49.Musser AM. Comp Biochem Physiol. 2003;136:927–942. doi: 10.1016/s1095-6433(03)00275-7. [DOI] [PubMed] [Google Scholar]
- 50.Woodburne MO, Tedford RH. Am Mus Novitates. 1975;2588:1–11. [Google Scholar]
- 51.Woodburne MO, Macfadden BJ, Case JA, Springer MS, Pledge NS, Power JD. J Vert Paleo. 1993;13:483–515. [Google Scholar]
- 52.Hopson JA, Crompton AW. Evol Biol. 1969;3:15–72. [Google Scholar]
- 53.Kermack KA, Kielan-Jaworowska Z. Zool J Linn Soc. 1971;50(Suppl 1):103–115. [Google Scholar]
- 54.Pridmore PA, Rich TH, Vickers-Rich P, Gambaryan P. J Mamm Evol. 2005;12:359–378. [Google Scholar]
- 55.Bever GS, Rowe T, Ekcale EG, Macrini TE, Colbert MW, Balanoff AM. Science. 2005;309:1492a. doi: 10.1126/science.1112248. [DOI] [PubMed] [Google Scholar]
- 56.Rougier GW, Forasiepi AM, Martinelli AG. Science. 2005;309:1496. doi: 10.1126/science.1111294. [DOI] [PubMed] [Google Scholar]
- 57.Luo ZX, Cifelli RL, Kielan-Jaworowska Z. Nature. 2001;409:53–56. doi: 10.1038/35051023. [DOI] [PubMed] [Google Scholar]
- 58.Kielan-Jaworowska Z, Cifelli RL, Luo Z-X. Mammals from the Age of Dinosaurs. New York: Columbia Univ Press; 2004. [Google Scholar]
- 59.Rich TH, Vickers-Rich P, Trusler P, Flannery TF, Cifelli R, Constantine A, Kool L, van Klaveren N. Acta Palaeont Polon. 2001;46:113–118. [Google Scholar]
- 60.Rich TH, Flannery TF, Trusler P, Kool L, van Klaveren NA, Vickers-Rich J Vert Paleo. 2002;22:466–469. [Google Scholar]
- 61.Rich TH, Hopson JA, Musser AM, Flannery TF, Vickers-Rich P. Science. 2005;307:910–914. doi: 10.1126/science.1105717. [DOI] [PubMed] [Google Scholar]
- 62.Luo Z-X, Cifelli RL, Kielan-Jaworowska Z. Acta Palaeont Polon. 2002;47:1–78. [Google Scholar]
- 63.Martin T, Luo ZX. Science. 2005;307:861–862. doi: 10.1126/science.1107202. [DOI] [PubMed] [Google Scholar]
- 64.Rich TH, Vickers-Rich P, Constantine A, Flannery TF, Kool L, van Klaveren N. Rec Queen Victoria Mus. 1999;106:1–29. [Google Scholar]
- 65.Gregory WK. Bull Amer Mus Nat Hist. 1910;27:1–524. [Google Scholar]
- 66.Zeller U. U Abh Senckenberg Naturforsch Gesell. 1989;545:1–188. [Google Scholar]
- 67.Bohringer RC. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 194–203. [Google Scholar]
- 68.Proske U, Gregory JE, Iggo A. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 204–210. [Google Scholar]
- 69.Taylor NG, Manger PR, Pettigrew JD, Hall LS. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 216–224. [Google Scholar]
- 70.Iggo A, Gregory JE, Proske U. J Physiol. 1992;447:449–465. doi: 10.1113/jphysiol.1992.sp019011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manger PR, Pewttigrew JD. Brain Behav Evol. 1996;48:27–54. doi: 10.1159/000113185. [DOI] [PubMed] [Google Scholar]
- 72.Augee ML, Gooden BA. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 211–215. [Google Scholar]
- 73.Flannery T, Archer M, Rich TH, Jones R. Nature. 1995;377:418–420. [Google Scholar]
- 74.Pascual R, Archer M, Jaureguizar EO. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 1–14. [Google Scholar]
- 75.Rowe T. Science. 1996;273:651–654. doi: 10.1126/science.273.5275.651. [DOI] [PubMed] [Google Scholar]
- 76.Rowe T. In: New Perspectives on the History of Life. Ghiselin M, Pinna G, editors. San Francisco: California Acad Sci; 1996. pp. 71–96. Memoir 20. [Google Scholar]
- 77.Rauhut OWM, Martin T, Ortiz-Jaureguizar E, Puerta P. Nature. 2002;416:165–168. doi: 10.1038/416165a. [DOI] [PubMed] [Google Scholar]
- 78.Thorne JL, Kishino H, Painter IS. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 79.Kishino H, Thorne JL, Bruno WJ. Mol Biol Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- 80.Thorne JL, Kishino H. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 81.Bech C, Nicol S, Andersen NA. In: Platypus and Echidna. Augee ML, editor. Sydney: Roy Zool Soc New South Wales; 1992. pp. 134–139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.