Abstract
Targeted gene expression is a powerful approach to study the function of genes and cells in vivo. In Drosophila, the P element-mediated Gal4-UAS method has been successfully used for this purpose. However, similar methods have not been established in vertebrates. Here we report the development of a targeted gene expression methodology in zebrafish based on the Tol2 transposable element and its application to the functional study of neural circuits. First, we developed gene trap and enhancer trap constructs carrying an engineered yeast Gal4 transcription activator (Gal4FF) and transgenic reporter fish carrying the GFP or the RFP gene downstream of the Gal4 recognition sequence (UAS) and showed that the Gal4FF can activate transcription through UAS in zebrafish. Second, by using this Gal4FF-UAS system, we performed large-scale screens and generated a large collection of fish lines that expressed Gal4FF in specific tissues, cells, and organs. Finally, we developed transgenic effector fish carrying the tetanus toxin light chain (TeTxLC) gene downstream of UAS, which is known to block synaptic transmission. We crossed the Gal4FF fish with the UAS:TeTxLC fish and analyzed double transgenic embryos for defects in touch response. From this analysis, we discovered that targeted expression of TeTxLC in distinct populations of neurons in the brain and the spinal cord caused distinct abnormalities in the touch response behavior. These studies illustrate that our Gal4FF gene trap and enhancer trap methods should be an important resource for genetic analysis of neuronal functions and behavior in vertebrates.
Keywords: targeted gene expression, Gal4-UAS, tetanus toxin, touch response, interneuron
Targeted expression of a desired gene in desired cells, tissues, and organs is a powerful approach to study the function of genes and cells in model animals. In Drosophila, the GAL4 enhancer trap method was developed by using the P transposable element for this purpose (1). When the P enhancer trap construct that contains a minimal promoter and the yeast GAL4 transcription activator was integrated in the genome and the minimal promoter was activated by a chromosomal enhancer, Gal4 was expressed in a temporally and spatially regulated fashion. Gal4 can activate transcription through its recognition sequence, UAS, and therefore theoretically any genes of interest placed downstream of UAS can be expressed in the Gal4-expressing cells. One important application of this system has been the study of neural circuits and behavior. The tetanus toxin light chain (TeTxLC) cleaves a vesicle membrane protein, synaptobrevin-2, and thereby blocks neurotransmitter release from synaptic vesicles (2). When transgenic flies carrying the TeTxLC gene downstream of UAS were crossed with enhancer trap fly lines expressing Gal4 in the embryonic nervous system, TeTxLC was expressed in the Gal4-expressing neurons and the embryos displayed uncoordinated muscle movements (3). Thus, the Gal4-UAS system has been powerful to study neuronal functions in vivo in Drosophila.
Zebrafish is a promising vertebrate model for genetic analysis of development and behavior. In an attempt to develop a targeted gene expression method in this model animal, it has been shown that Gal4 can activate expression of a gene placed downstream of UAS (4, 5). Despite these promising results, the usefulness of the Gal4-UAS approach in zebrafish has been limited mainly because the availability of specific promoters that can target Gal4 expression to specific cells has been limited. To circumvent this problem, development of an efficient gene trap or enhancer trap method that generates fish expressing Gal4 in specific cells has been highly desired. Implementation of such methods, however, has been hampered by the lack of a transgenesis method that can generate random insertions efficiently throughout the genome. In addition, whereas the Gal4-VP16 fusion gene was used in zebrafish to overcome the weak gene expression activity of the full-length Gal4 (6), it has been known that expression of Gal4-VP16, a transcription factor with strong activator domains, causes nonspecific toxic effects in vertebrate cells (7, 8) and in zebrafish (6, 9). Hence, construction of a less toxic transcription activator would be advantageous for large-scale screens in zebrafish.
We have been developing transposon technologies in zebrafish by employing the medaka fish Tol2 transposable element. Tol2 is an autonomous transposon that encodes a fully functional transposase capable of catalyzing transposition in the zebrafish germ lineage (10–12). Recently, we reported a highly efficient transgenesis method and gene trap and enhancer trap methods by using the Tol2 transposon system (13, 14). More recently, we and others have taken advantage of these methods to isolate fish lines expressing Gal4-VP16 in specific tissues (9, 15). In the present study we aim to further develop methodologies in zebrafish that enable targeted expression of a desired gene in desired cells. First, we used an improved version of Gal4 and developed gene trap and enhancer trap constructs and UAS reporter systems. Second, we performed large-scale screens for fish expressing the modified Gal4 in specific patterns and demonstrated that our method can indeed create a large number of such fish efficiently. Finally, we illustrated that our method is applicable to functional studies of neural circuits. Our present study provides a methodology that will facilitate functional studies of genes and cells in zebrafish and therefore increase our understanding of vertebrate development and behavior.
Results
Development of the Gal4FF-UAS System.
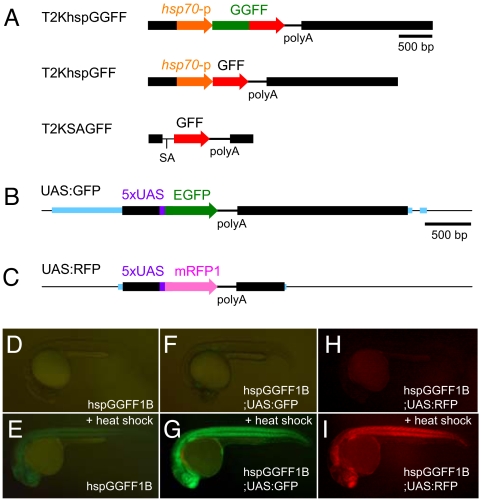
To avoid the possible toxicity of Gal4-VP16, we created a transcription activator, Gal4FF, that consists of the DNA binding domain from Gal4 and two transcription activation modules from VP16, which consists of 13 aa containing a critical phenylalanine (16, 17). Injection of ≈1 nl of 25 ng/μl mRNA encoding Gal4FF into fertilized eggs did not cause any obvious defects whereas injection of the same amount of mRNA encoding Gal4-VP16 caused severe developmental defects (data not shown), indicating that Gal4FF is less toxic than Gal4-VP16. Also, we created the GGFF fusion gene, in which the GFP gene was fused to the Gal4FF gene, to visualize synthesis of the activator protein. We constructed T2KhspGGFF (Fig. 1A), which contained the 0.65-kb DNA from the zebrafish hsp70 promoter and the GGFF gene, injected a transposon-donor plasmid containing T2KhspGGFF and the transposase mRNA to fertilized eggs, and created a transgenic fish line, hspGGFF1B, that carried a single-copy insertion of T2KhspGGFF in the genome and showed GFP fluorescence ubiquitously upon heat shock.
Fig. 1.
The gene trap and enhancer trap constructs and the UAS reporter system. The Tol2 vector sequences are shown as thick black lines. (A) The structures of T2KhspGGFF, T2KhspGGFF, and T2KSAGFF. (B) The UAS:GFP reporter fish carries a single-copy insertion of T2KUASGFP within a gene encoding a homolog of Nedd4-binding protein 1. Blue boxes indicate exons. (C) The UAS:RFP reporter fish carries a single-copy insertion of T2ZUASRFP within a gene encoding a solute carrier protein homolog. Blue boxes indicate exons. (D and E) The hspGGFF1B embryos before (D) and after (E) heat shock. (F and G) The hspGGFF1B;UAS:GFP embryos before (F) and after (G) heat shock. (H and I) The hspGGFF1B;UAS:RFP embryos before (H) and after (I) heat shock.
In parallel, we constructed T2KUASGFP and T2KUASRFP, transposon constructs containing the GFP and RFP gene downstream of UAS, respectively, and created the UAS:GFP and UAS:RFP transgenic fish that carried single-copy insertions of these constructs (Fig. 1 B and C). We crossed the hspGGFF1B fish with the UAS:GFP and UAS:RFP fish and analyzed the double transgenic embryos. The hspGGFF1B embryo showed GFP expression upon heat shock (Fig. 1 D and E). Both of the hspGGFF1B;UAS:GFP and hspGGFF1B;UAS:RFP embryos showed much stronger GFP and RFP expression in the whole body (Fig. 1 F–I). These results indicated that GGFF can activate a gene placed downstream of UAS and that the UAS:GFP and UAS:RFP transgenic fish can be used as reporters.
Tissue-Specific Gal4FF Expression in Enhancer Trap Lines.
We injected the T2KhspGGFF construct and the transposase mRNA to fertilized eggs and crossed 60 injected fish at first with wild-type fish. The F1 embryos that showed GFP expression at normal temperatures (≈26°C) were raised. Because in most cases GFP fluorescence generated from GGFF was rather weak (Fig. 1D), we further outcrossed these fish with the UAS:GFP fish to confirm the GFP expression patterns. During this process, some new patterns were found. In total, we established 29 hspGGFF fish lines that harbored single T2KhspGGFF insertions [supporting information (SI) Fig. 5] and caused specific GFP expression patterns in their offspring when crossed with the UAS:GFP fish. We cloned and sequenced genomic DNA flanking the transposon insertions and found that the integration loci were mapped throughout the genome (SI Table 1). These analyses clearly identified the insertions that are responsible for the specific GFP expression patterns.
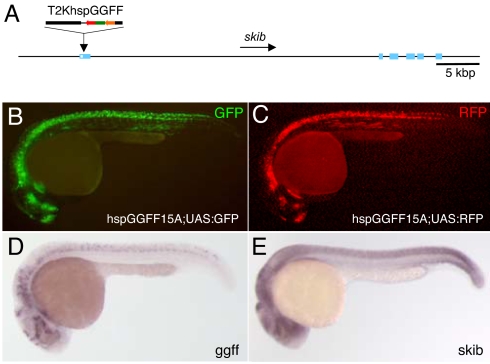
One of them, the hspGGFF15A fish line, harbored a single-copy insertion of T2KhspGGFF in the first exon of the skib gene (Fig. 2A). In the hspGGFF15A embryo, GFP expression was hardly detected (data not shown). In the hspGGFF15A;UAS:GFP double transgenic embryo, GFP was restrictedly expressed in the spinal cord, hindbrain, diencephalons, hypothalamus, and dorsal telencephalon (Fig. 2B). Whole-mount in situ hybridization revealed that the ggff mRNA was expressed in a pattern similar to the GFP expression pattern (Fig. 2D), indicating that expression of the UAS:GFP gene was activated by GGFF. The skib mRNA was expressed broadly in the central nervous system (Fig. 2E), which included the regions where GGFF was expressed, suggesting that a putative skib enhancer activated transcription from the hsp70 promoter. When the hspGGFF15A fish was crossed with UAS:RFP fish, RFP was expressed in the hspGGFF15A;UAS:RFP embryos in almost the same pattern as the GFP expression pattern in the hspGGFF15A;UAS:GFP embryos (Fig. 2C), indicating that the RFP expression was also activated by GGFF. These studies revealed that T2KhspGGFF can indeed be used as an enhancer trap construct and that our Gal4FF-UAS system can express appropriately any gene placed downstream of UAS.
Fig. 2.
GFP and RFP expression in the hspGGFF15A enhancer trap line. (A) The structure of the hspGGFF15A insertion. T2KhspGGFF is integrated in the first exon of the skib gene. (B) GFP expression in the hspGGFF15A;UAS:GFP embryo at 24 hpf. (C) RFP expression in the hspGGFF15A;UAS:RFP embryo at 24 hpf. (D) Whole-mount in situ hybridization of the hspGGFF15A embryo at 24 hpf using the GGFF probe. (E) Whole-mount in situ hybridization of a wild-type embryo at 24 hpf and the skib probe.
Large-Scale Screens for Gal4FF Enhancer Trap and Gene Trap Lines.
Because we established the UAS:GFP fish line and found that this line indeed can be used as a reporter, the GFP part of the GGFF protein was not needed anymore. Therefore, we constructed enhancer trap and gene trap constructs, T2KhspGFF and T2KSAGFF, both of which contained the Gal4FF gene (Fig. 1A). First, we performed an enhancer trap screen. We crossed 69 fish injected with a plasmid containing T2KhspGFF and the transposase mRNA with homozygous UAS:GFP reporter fish and identified embryos exhibiting unique GFP expression patterns in their offspring. These fish were raised, further outcrossed, and analyzed by Southern blot hybridization. From this screen, we established 27 fish lines that expressed Gal4FF in specific cells and tissues and revealed that the T2KhspGFF construct can be used for enhancer trapping. It should be noted that GFP expression in the heart at day 1 and in the heart and skeletal muscle at day 5 was observed in these lines in common (data not shown and Fig. 2B). We think that this common expression should be due to basal transcription activity of the hsp70 promoter.
Next, we performed a gene trap screen. We crossed ≈250 fish injected with a plasmid containing T2KSAGFF and the transposase mRNA with homozygous UAS:GFP reporter fish, and we identified embryos exhibiting unique GFP expression patterns in their offspring. These fish were raised, further outcrossed, and analyzed by Southern blot hybridization. From this screen, we established 129 fish lines expressing Gal4FF in specific cells and tissues and revealed that the T2KSAGFF construct can be used for gene trapping. We performed the gene trap screen more extensively than the enhancer trap screen because in the SAGFF;UAS:GFP embryos the basal GFP expression observed in the hspGFF;UAS:GFP embryos was not detected and the GFP expression patterns were found to be more restricted. This confirmed that the basal GFP expression was caused by the enhancer trap construct itself. Approximately 20% of the SAGFF insertions, however, caused UAS:GFP expression in nonneuronal cells in the spinal cord at days 3–5 (data not shown). We infer that this is probably due to a weak or cryptic enhancer activity contained in the sequence of T2KSAGFF itself.
By performing the gene trap and enhancer trap screens using T2KhspGGFF, T2KhspGFF, and T2KSAGFF, we established a total of 185 fish lines that expressed Gal4FF in spatially and temporally regulated fashions and identified insertions responsible for the expression patterns by Southern blot hybridization. These are a useful resource for zebrafish researchers.
Gal4FF-Mediated TeTxLC Expression Can Inhibit Neuronal Function.
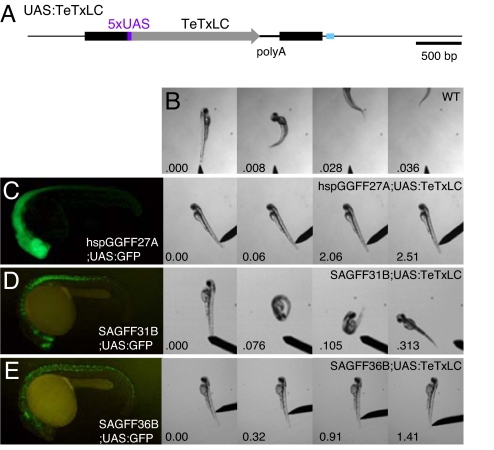
We aimed to perform functional studies by employing the collection of the Gal4FF transgenic fish lines. The tetanus toxin light chain (TeTxLC) gene has been shown to inhibit the neuronal function in transgenic flies and mice (3, 18). To examine whether TeTxLC can inhibit neuronal function in zebrafish, we constructed a transgenic fish line that harbored a single-copy insertion of the TeTxLC gene downstream of 5xUAS (UAS:TeTxLC) (Fig. 3A). At 48 h postfertilization (hpf), wild-type embryos respond to a gentle touch in the tail and escape rapidly from the stimulus (Fig. 3B). We crossed the UAS:TeTxLC fish with the hspGGFF27A;UASGFP heterozygous fish, which expressed GFP in a large population of neurons in the brain (Fig. 3C), and performed the touch response assay. Of 54 GFP-positive embryos that carried both hspGGFF27A and UAS:GFP, 30 embryos responded to the touch as well as wild-type embryos, but 24 embryos did not (Fig. 3C). We analyzed these embryos by PCR. All of the 24 embryos defective in the touch response were hspGGFF27A;UAS:GFP;UAS:TeTxLC triple transgenic embryos, and 27 of 30 embryos that responded to the touch did not carry the UAS:TeTxLC transgene. Thus, 89% (24/27) of the hspGGFF27A;UAS:GFP;UAS:TeTxLC embryos showed the behavioral defect, and none (0/27) of the hspGGFF27A;UAS:GFP embryos showed the defect. Furthermore, at 96 hpf, 100% (27/27) of the hspGGFF27A;UAS:GFP embryos showed spontaneous swimming whereas 100% (27/27) of the hspGGFF27A;UAS:GFP;UAS:TeTxLC embryos did not. From these results we concluded that, in the hspGGFF27A line, Gal4FF is expressed in neurons essential for motility and Gal4FF-mediated TeTxLC expression inhibited their activities.
Fig. 3.
Abnormal touch response phenotypes in the UAS:TeTxLC double transgenic embryos. (A) The UAS:TeTxLC effector fish carries a single-copy insertion of T2MUASTeTxLC in the myosin heavy chain gene. A blue box indicates an exon. (B) The touch response behavior of a wild-type embryo at 48 hpf. (C) GFP expression in the hspGGFF27A;UAS:GFP embryo at 24 hpf and the touch response behavior of the hspGGFF27A;UAS:TeTxLC embryo at 48 hpf. (D) GFP expression in the SAGFF31B;UAS:GFP embryo at 24 hpf and the touch response behavior of the SAGFF31B;UAS:TeTxLC embryo at 48 hpf. (E) GFP expression in the SAGFF36B;UAS:GFP embryo at 24 hpf and the touch response behavior of the SAGFF36B;UAS:TeTxLC embryo at 48 hpf.
Then we crossed 29 hspGGFF, 23 hspGFF, and 69 SAGFF fish lines, a total of 121 lines, with the UAS:TeTxLC heterozygous fish and analyzed their progeny by the touch response assay. We found that the progeny from nine lines showed abnormal behavioral phenotypes (SI Fig. 6). We analyzed these embryos by PCR to detect the UAS:TeTxLC transgene and found that 37% (15/41), 89% (24/27), 74% (46/62), 78% (18/23), 100% (20/20), 21% (5/24), 90% (9/10), 92% (11/12), or 47% (8/17) of double transgenic embryos carrying both UAS:TeTxLC and Gal4FF (hspGGFF15A, hspGGFF27A, SAGFF31B, SAGFF36B, SAGFF73A, SAGFF128A, SAGFF158A, SAGFF193A, or SAGFF206A, respectively) showed abnormal touch response behaviors, and none of embryos that did not carry UAS:TeTxLC showed the phenotypes. These results indicated that the abnormal behavioral phenotypes were caused by Gal4FF-mediated TeTxLC expression.
We further found that the nine Gal4FF lines could be divided into three groups based on the differences in the phenotypes. The hspGGFF27A and SAGFF73A embryos neither responded to the touch nor showed spontaneous movements (class I) (Fig. 3C). The SAGFF31B and SAGFF193A embryos responded to the touch and bend but could not swim away from the stimulus (class II) (Fig. 3D). The hspGGFF15A, SAGFF36B, SAGFF128A, SAGFF158A, and SAGFF206A embryos did not respond to the touch (Fig. 3E) but could move spontaneously (class III). Among them, the SAGFF31B;UAS:GFP (class II) and SAGFF36B;UAS:GFP (class III) embryos showed restricted GFP expression (Fig. 3 D and E), suggesting that the specific populations of neurons may be responsible for the distinct behavioral phenotypes. Because these fish carried multiple insertions in the F1 or F2 generation, fish lines with single insertions of hspGGFF27A, SAGFF31B, and SAGFF36B were established by performing several outcrosses for generations. The hspGGFF27A insertion is located within a predicted gene encoding a homolog of human ADAMTSL-3 (SwissProt accession no. P82987). The SAGFF31B insertion is located within a gene encoding a homolog of Xenopus X-Myt1 that is involved in neuronal differentiation (19), and trapped its 5′ noncoding exon. The SAGFF36B insertion was located ≈7 kb upstream of a gene encoding β-tubulin.
Visualization of TeTxLC Expression via a CFP Fusion Protein in the Gal4FF Lines.
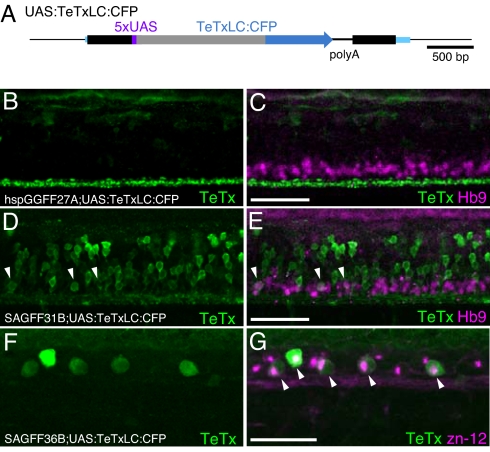
To visualize the candidate neural circuits whose functions were inhibited by the TeTxLC expression, we constructed a transgenic line that carried a single-copy insertion of the TeTxLC:CFP fusion gene downstream of UAS (Fig. 4A). We crossed the UAS:TeTxLC:CFP fish with the hspGGFF27A (class I), SAGFF31B (class II), and SAGFF36B (class III) fish and analyzed TeTxLC:CFP expression in the spinal cord. These double transgenic embryos showed essentially the same behavioral phenotypes as the double transgenic embryos carrying the untagged TeTxLC gene, indicating that the TeTxLC:CFP fusion protein can function properly in zebrafish as well as the TeTxLC protein.
Fig. 4.
Expression of TeTxLC:CFP in double transgenic embryos. (A) The UAS:TeTxLC:CFP fish carries a single-copy insertion of T2SUASTeTxLCCFP within the CSPP1 gene. Blue boxes indicate exons. (B–G) Lateral views of the trunk of double transgenic embryos immunostained with the anti-GFP antibody (green) and the anti-Hb9 (B–E) or the zn-12 (F and G) antibody (red). Arrowheads indicate costaining with anti-GFP and anti-Hb9 (E) or anti-GFP and zn-12 (G). Anterior is to the left, and dorsal is to the top. (Scale bars: 50 μm.) (B and C) The hspGGFF27A;UAS:TeTxLC:CFP embryo at 48 hpf. The anti-GFP antibody detects the TeTxLC:CFP fusion protein but does not detect the GGFF protein in this condition (data not shown). Axons of descending hindbrain interneurons were strongly stained (green). The anti-Hb9 antibody detected spinal motor neurons (red). (D and E) The SAGFF31B;UAS:TeTxLC:CFP embryo at 48 hpf. The anti-GFP antibody detected spinal interneurons and motor neurons (green). (F and G) The SAGFF36B;UAS:TeTxLC:CFP embryo at 30 hpf. Both the anti-GFP and zn-12 antibodies detected Rohon-Beard neurons (green and red).
We analyzed the hspGGFF27A;UAS:TeTxLC:CFP double transgenic embryo by immunostaining using the anti-GFP antibody, which can bind to the CFP portion of TeTxLC:CFP, and we found that descending hindbrain interneurons were strongly stained by the antibody (Fig. 4B). Interneurons in the spinal cord were also stained weakly (Fig. 4B and SI Fig. 7). Hb9 is a marker for motor neurons (20). Coimmunostaining using the anti-Hb9 and anti-GFP antibodies showed that TeTxLC:CFP was not detected in motor neurons (Fig. 4C). These results suggest that the immotility observed in the hspGGFF27A;UAS:TeTxLC:CFP embryo may be caused by inhibition of the TeTxLC:CFP-expressing interneurons.
In the SAGFF31B;UAS:TeTxLC:CFP embryos, the TeTxLC:CFP expression was detected in numbers of neurons located in the ventral part of the spinal cord, which were thought to include motor neurons and interneurons (Fig. 4D). Coimmunostaining using the anti-Hb9 and anti-GFP antibodies revealed that only a small portion of Hb9-positive cells were detected by the anti-GFP staining (Fig. 4E), suggesting that TeTxLC:CFP was expressed in a small population of motor neurons and more largely in spinal interneurons. This finding is consistent with the observation that the SAGFF31B;UAS:TeTxLC embryos retained motility and may imply that the abnormal escape movement was caused mainly by inhibition of the spinal interneurons.
In the SAGFF36B;UAS:TeTxLC:CFP embryos, the TeTxLC:CFP expression was detected in the somata of dorsal spinal neurons (Fig. 4F). Those TeTxLC:CFP-positive somata were also stained with the zn-12 antibody, which specifically stained Rohon-Beard sensory neurons (Fig. 4G) (21). Therefore, the touch response defect observed in the SAGFF36B;UAS:TeTxLC:CFP embryo is likely to be caused by inhibition of the Rohon-Beard sensory neurons.
Discussion
Gene and Enhancer Trapping Using the Gal4FF-UAS System.
In this study we constructed a transcription activator, Gal4FF, and demonstrated that it can drive UAS-dependent expression of reporter and effector genes in zebrafish. In the previous studies, the Gal4-UAS system in zebrafish used the full-length Gal4 (4) or Gal4-VP16 (6). Although Gal4-VP16, which contains a strong transcription activator domain, was used to enhance weak expression observed with the full-length Gal4, developmental defects were observed in 8% of embryos injected with Gal4-VP16 mRNA (6) or 4% of transgenic embryos expressing Gal4-VP16 (9). On the other hand, we did not observe such toxic effects by using Gal4FF in the following cases: in embryos injected with Gal4FF mRNA, in Gal4FF;UAS:GFP embryos that showed fairly high levels of GFP expression, and in hspGGFF and hspGFF transgenic embryos treated by heat shock. Therefore, we think that Gal4FF is less toxic and more tolerable for zebrafish cells than Gal4-VP16.
Mosaic (or variegated) expression of a UAS:GFP reporter has been reported; i.e., when Gal4-VP16 was expressed under the control of the islet1 enhancer, GFP was not expressed in all of the islet1-positive neurons (22). In contrast, we did not observe such mosaic expression in the Gal4FF;UAS:GFP embryos; i.e., expression of the UAS:GFP (and UAS:RFP) reporter was consistent with the Gal4FF expression as revealed by whole-mount in situ hybridization, and the GFP expression pattern was reproducibly the same in individual fish carrying the same single Gal4FF insertion. Although the mechanism that causes mosaic expression is not known, there may be two possibilities. One is a phenomenon termed “squelching,” in which overproduction of a strong transcription activator causes inhibition of gene expression (7, 8). The other is “gene silencing.” Because the islet1:Gal4-VP16 and the UAS:GFP fish were created by the plasmid injection method, these constructs may be integrated as concatemers (22). Expression from such transgenes may suffer from gene silencing. If these were the cases, our present system suffered from neither squelching nor gene silencing because Gal4FF was weaker than Gal4-VP16 and our transgenic fish insertions were created by transposon-mediated transgenesis that should create single-copy insertions.
In this study we demonstrated that both gene trap and enhancer trap approaches can create fish expressing Gal4FF in specific cells, tissues, and organs. Also, we observed GFP expression in various regions of the embryo, indicating that Gal4FF can activate transcription through UAS in nearly all cell types. Although specific GFP expression patterns were observed, we also detected basal expression associated with the trap constructs. In a screen carried out in our laboratory by using an enhancer trap construct containing the 1.5-kb zebrafish hsp70 promoter, we observed basal expression of a reporter in the heart, skeletal muscle, and lens (14). The lens expression was not associated with T2KhspGFF carrying the 0.65-kb hsp70 promoter, suggesting that the shorter hsp70 promoter did not contain the putative lens enhancer. The gene trap construct T2KSAGFF also showed weak basal expression in nonneural cells in the spinal cord at day 5. Our preliminary result suggests that this basal expression may be caused by the sequence around the splice acceptor (data not shown). At present, we think that T2KSAGFF is superior to T2KhspGFF from the aspect of basal expression.
Inhibition of Neuronal Activities by Using the UAS:TeTxLC Effector Fish.
In this study we discovered that expression of TeTxLC:CFP in distinct populations of neurons caused distinct abnormal behavioral phenotypes. Thus, the UAS:TeTxLC and UAS:TeTxLC:CFP effector fish lines we described here are useful to inhibit neuronal activities in zebrafish. Although we observed basal (background) expression in the gene trap and enhancer trap lines, we isolated only nine lines that showed selectable behavioral abnormalities by screening 121 Gal4FF lines, indicating that the basal expression is not an obstacle for such a behavioral screen. We also noticed that, when the same Gal4FF driver line was used, the CFP expression pattern, which reports the UAS:TeTxLC:CFP expression, always appeared more restricted than the GFP expression pattern from UAS:GFP, likely correlated with the long stability and strong fluorescent intensity of the GFP protein.
The hspGGFF27A;UAS:TeTxLC:CFP embryos showed immotility. These embryos expressed TeTxLC:CFP strongly in the descending hindbrain interneurons and weakly in the spinal interneurons. The previous studies used spinalized embryos created by surgical ablation of the brain to analyze roles of the hindbrain in the touch response behavior (23, 24). It was shown that even surgically severed embryos could perform a touch response after a recovery time (24). In the hspGGFF27A;UAS:TeTxLC:CFP embryos, the TeTxLC:CFP-expressing interneurons should include neurons essential for the motility. Thus, the genetic system developed in the present study will allow investigation of such neuronal activities using intact embryos.
The TeTxLC:CFP expression pattern in the SAGFF31B;UAS:TeTxLC:CFP embryo suggested that the abnormal escape movement was caused by inhibition of the interneurons. In this embryo, the CFP expression was also observed in the brain and a small population of motor neurons (Fig. 4E and data not shown); therefore, a possibility that inhibition of these neurons is involved in the phenotype, at least partly, cannot be ruled out. Different subsets of spinal interneurons can be marked by expression of different transcription factors (25). For instance, in zebrafish, Engrailed-1 and alx (Chox10) are expressed in distinct spinal interneurons, circumferential ascending interneurons, and ipsilateral descending interneurons, respectively. The activities of these neurons during swimming have been studied by electrophysiological analysis of transgenic fish expressing GFP under the control of the En-1 and the alx promoter (26, 27). To probe the function of interneurons in the spinal cord, inhibiting the activity of each subset in vivo by isolating lines expressing Gal4FF more specifically will be necessary.
The analysis of the SAGFF36B;UAS:TeTxLC:CFP embryo suggested that expression of TeTxLC:CFP in Rohon-Beard sensory neurons inhibited their activity and abolished the touch response behavior. It has been shown by electrophysiological analyses using immobilized Xenopus embryos that Rohon-Beard neurons are responsible for detecting a light touch on the skin and for initiating swimming (28). Our result is consistent with this notion and provides in vivo evidence showing that the activity of Rohon-Beard neurons is essential for generating escape movements in response to a touch.
In the present study we developed the Gal4FF gene and enhancer trap methods along with UAS reporter and effector systems, demonstrating that targeted expression of Gal4FF resulted in selective expression of the reporter and effector genes in zebrafish. Although we focused on the application of these tools to the analysis of touch response behavior and spinal neural circuits, our method is widely applicable to the study of other neural systems in the intact zebrafish. In addition, our methods offer obvious advantages for developmental biology. In the future, generation and screening of new Gal4FF lines should be performed according to the interests of individual researchers. The current frequency of obtaining Gal4FF gene trap lines is reasonably high (129 of 215 injected fish screened) to permit a small laboratory to efficiently collect hundreds of different patterns. Thus, our present methods will be important for studies of vertebrate neurobiology and developmental biology using zebrafish.
Materials and Methods
Tol2 Constructs.
T2KhspGGFF, T2KhspGFF, T2KSAGFF, T2KUASGFP, T2ZUASRFP (Fig. 1 A–C), T2MUASTeTxLC, and T2SUASTeTxLCCFP (Figs. 3A and 4A) were constructed based on Tol2 vectors, T2KXIG (13), T2AL200R150G, T2KXIGΔin (29), and T2MUASMCS, which contains a synthetic multicloning site downstream of five tandem repeats of the Gal4 binding sequence (5xUAS) and a TATA sequence. The Gal4FF gene (or referred to as GFF) is a fusion of the DNA binding domain from the Gal4 protein and two transcription activation modules (2xPADALDDFDLDML) from VP16. GGFF is a fusion of EGFP and Gal4FF. The zebrafish hsp70 promoter is a kind gift from J. Kuwada (University of Michigan, Ann Arbor, MI) (30). T2KSAGFF uses a splice acceptor from the rabbit β-globin gene (13). T2KUASGFP, T2ZUASRFP, T2MUASTeTxLC, and T2SUASTeTxLCCFP contain the EGFP gene, the monomeric RFP gene (a kind gift from R. Tsien, University of California, San Diego, CA) (31), the tetanus toxin light chain (TeTxLC) gene (a kind gift from S. Sweeney, Univeristy of York, York, U.K.) (3), and the TeTxLC:CFP (cyan fluorescent protein) fusion gene (a kind gift from A. Craig, Washington University School of Medicine, St. Louis, MO) (32) downstream of 5xUAS and TATA.
Microinjection of mRNA.
Microinjection of mRNA was carried out by using PV830 (World Precision Instruments). An approximate volume was determined by measuring the diameter of a sphere of the injected solution.
Construction of Gal4FF Gene Trap and Enhancer Trap Lines.
A plasmid DNA containing T2KhspGGFF, T2KhspGFF, or T2KSAGFF was coinjected with the transposase mRNA into fertilized eggs as described previously (13, 33). The T2KhspGGFF-injected fish were crossed with wild-type fish. The progeny were analyzed for GFP expression upon heat shock or at normal temperatures. The T2KhspGFF- or T2KSAGFF-injected fish were crossed with homozygous UAS:GFP reporter fish. Double transgenic embryos were analyzed for GFP expression at day 1 and day 5 by using a fluorescence stereoscope (MZ16 FA; Leica) and a CCD camera (DFC300 FX; Leica). For each cross, at least 40 F1 embryos were analyzed. The integration sites of the transposon insertions were analyzed by inverse PCR and linker-mediated PCR (29, 33, 34). These transgenic fish are designated as hspGGFF, hspGFF, and SAGFF, respectively.
Construction of UAS Reporter and Effector Fish.
Fish injected with the T2KUASGFP plasmid and the transposase mRNA were crossed with wild-type fish. A Gal4FF expression plasmid DNA was injected to their offspring, and F1 embryos that showed GFP fluorescence were raised. The F1 fish were further crossed with the hspGGFF1B fish to establish UAS:GFP reporter fish. Fish injected with the T2ZUASRFP plasmid and the transposase mRNA were crossed with the hspGGFF15A fish. F1 embryos that showed the brightest RFP expression were raised to establish UAS:RFP reporter fish. These reporter fish lines carry a single-copy insertion of the transposon construct and show no detectable background GFP or RFP expression. Fish homozygous for respective insertions are viable and fertile. Fish injected with the T2MUASTeTxLC plasmid and the transposase mRNA were crossed with wild-type fish. Three F1 fish that carried single-copy insertions at different loci were identified and crossed with the hspGGFF27A fish. The progeny were analyzed by the touch response assay. One of them, which caused the severest motility defects in the offspring, was established as the UAS:TeTxLC effector fish. Fish injected with the T2SUASTeTxLCCFP plasmid and the transposase mRNA were crossed with the SAGFF73A fish that expressed Gal4FF in the whole body. F1 embryos that showed strong CFP expression were selected and raised to establish the UAS:TeTxLC:CFP effector fish. These effector fish lines carry single-copy insertions of the transposon construct and do not show any behavioral defects in the absence of Gal4FF. The UAS:GFP, UAS:RFP, UAS:TeTxLC, and UAS:TeTxLC:CFP insertions were mapped within genes encoding a homolog of human Nedd4-binding protein 1 (Fig. 1B), a solute carrier protein (Slc12a8) (Fig. 1C), myosin heavy chain (Fig. 3A), and the centrosome and spindle pole-associated protein 1 (CSPP1) (Fig. 4A), respectively.
Heat Shock Treatment.
Approximately 20–30 embryos at 24 hpf were placed into 500 μl of E3 buffer preheated at 38°C on Block Incubator BI-516S (Astec) and incubated for 15 min. After the heat shock treatment, the embryos are transferred to the E3 buffer at 28.5°C. The heat shock treatment causes neither an anomaly nor a decrease in viability.
The Touch Response Assay.
The hspGGFF, hspGFF;UASGFP, and SAGFF;UASGFP fish were crossed with the UAS:TeTxLC fish, and double or triple transgenic embryos were analyzed by the touch response assay (35). Throughout the screen, more than six embryos from each cross were analyzed, and a gentle tactile stimulus at the tail was applied to each embryo at least three times. The images of escape movements were taken by a high-speed digital video camera (FASTCAM-512PCI; Photoron).
Immunohistochemistry and Confocal Microscopy.
Primary antibodies used are rabbit polyclonal anti-GFP antibody (1:500 dilution; Invitrogen A6455), mouse monoclonal anti-Hb9 antibody (1:25 dilution; Developmental Studies Hybridoma Bank) (20), and mouse monoclonal zn-12 antibody (1:250 dilution; obtained from th Zebrafish International Resource Center) (21). The anti-GFP antibody can recognize CFP. Secondary antibodies used are goat anti-rabbit Alexa Fluor 488 (1:1,000) and goat anti-mouse Alexa Fluor 633 (1:1,000) (Molecular Probes). Embryos were fixed for 2–3 h in PBS containing 4% paraformaldehyde. The samples were soaked in PBS-TX (PBS/0.5% Triton X-100)/10% BSA for 1 h, then incubated in the primary antibody diluted in PBS-TX/1% BSA overnight at 4°C, washed with PBS-TX, and then incubated in the secondary antibody diluted in PBS-TX/1% BSA for 2 h. After a final wash with PBS-TX, the samples were subjected to confocal microscopy using a Zeiss LSM510META laser confocal microscope. Serial sections of the embryo at 1.0 μm were processed to make images.
Supplementary Material
ACKNOWLEDGMENTS.
We thank A. Craig, J. Kuwada, S. Sweeney, and R. Tsien for plasmids; T. Uematsu, N. Mouri, S. Takeda, R. Mimura, and M. Mizushina for maintenance of fish; M. Okamura for technical assistance; and the Zebrafish International Resource Center (supported by National Institutes of Health/National Center for Research Resources Grant NCRRRR12546) for zn-12. This work was supported by postdoctoral fellowships from the Japan Society for the Promotion of Science (to K.A. and M.L.S.), a Sasakawa Scientific Research Grant from the Japan Science Society (to K.A.), National Institutes of Health/National Institute of General Medical Sciences Grant R01GM069382, the National BioResource Project, and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704963105/DC1.
References
- 1.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 2.Schiavo G, et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 4.Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- 5.Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–1107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- 6.Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- 7.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 8.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 9.Scott EK, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami K, Koga A, Hori H, Shima A. Excision of the Tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami K, Shima A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene. 1999;240:239–244. doi: 10.1016/s0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Nagayoshi S, et al. Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes, tcf7 and synembryn-like. Development. 2008;135:159–169. doi: 10.1242/dev.009050. [DOI] [PubMed] [Google Scholar]
- 15.Davison JM, et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron U, Gossen M, Bujard H. Tetracycline-controlled transcription in eukaryotes: Novel transactivators with graded transactivation potential. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto M, et al. Reversible suppression of glutamatergic neurotransmission of cerebellar granule cells in vivo by genetically manipulated expression of tetanus neurotoxin light chain. J Neurosci. 2003;23:6759–6767. doi: 10.1523/JNEUROSCI.23-17-06759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellefroid EJ, et al. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87:1191–1202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- 20.Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- 21.Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- 22.Sagasti A, Guido MR, Raible DW, Schier AF. Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr Biol. 2005;15:804–814. doi: 10.1016/j.cub.2005.03.048. [DOI] [PubMed] [Google Scholar]
- 23.Saint-Amant L, Drapeau P. Time course of the development of motor behaviors in the zebrafish embryo. J Neurobiol. 1998;37:622–632. doi: 10.1002/(sici)1097-4695(199812)37:4<622::aid-neu10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 24.Downes GB, Granato M. Supraspinal input is dispensable to generate glycine-mediated locomotive behaviors in the zebrafish embryo. J Neurobiol. 2006;66:437–451. doi: 10.1002/neu.20226. [DOI] [PubMed] [Google Scholar]
- 25.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 26.Higashijima S, Masino MA, Mandel G, Fetcho JR. Engrailed-1 expression marks a primitive class of inhibitory spinal interneuron. J Neurosci. 2004;24:5827–5839. doi: 10.1523/JNEUROSCI.5342-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura Y, Okamura Y, Higashijima S. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J Neurosci. 2006;26:5684–5697. doi: 10.1523/JNEUROSCI.4993-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke JD, Hayes BP, Hunt SP, Roberts A. Sensory physiology, anatomy and immunohistochemistry of Rohon-Beard neurones in embryos of Xenopus laevis. J Physiol. 1984;348:511–525. doi: 10.1113/jphysiol.1984.sp015122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halloran MC, et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 31.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harms KJ, Tovar KR, Craig AM. Synapse-specific regulation of AMPA receptor subunit composition by activity. J Neurosci. 2005;25:6379–6388. doi: 10.1523/JNEUROSCI.0302-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- 34.Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 35.Granato M, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.