Abstract
Upon encountering antigens, B-lymphocytes can adapt to produce a highly specific and potent antibody response. Somatic hypermutation, which introduces point mutations in the variable regions of antibody genes, can increase the affinity for antigen, and antibody effector functions can be altered by class switch recombination (CSR), which changes the expressed constant region exons. Activation-induced cytidine deaminase (AID) is the mutagenic antibody diversification enzyme that is essential for both somatic hypermutation and CSR. The mutagenic AID enzyme has to be tightly controlled. Here, we show that engagement of the membrane-bound antibodies of the B-cell receptor (BCR), which signals that good antibody affinity has been reached, inhibits AID gene expression and that calcium (Ca2+) signaling is essential for this inhibition. Moreover, we show that overexpression of the Ca2+ sensor protein calmodulin inhibits AID gene expression, and that the transcription factor E2A is required for regulation of the AID gene by the BCR. E2A mutated in the binding site for calmodulin, and thus showing calmodulin-resistant DNA binding, makes AID expression resistant to the inhibition through BCR activation. Thus, BCR activation inhibits AID gene expression through Ca2+/calmodulin inhibition of E2A.
Keywords: activation-induced cytidine deaminase, BCR, calcium, E2A
Processes that diversify antibodies play a major role in protecting higher organisms from pathogens. Upon encountering antigens, antibody-expressing B-lineage lymphocytes adapt to produce a highly specific and potent antibody response. Somatic hypermutation, which introduces point mutations in the variable regions of antibody genes, and gene conversion can increase the affinity for antigen, and antibody effector functions can be altered by class switch recombination (CSR), which changes the expressed constant region exons (1, 2). Activation-induced cytidine deaminase (AID) is the antibody diversification enzyme that is essential for somatic hypermutation, gene conversion, and CSR (3–5). The mutagenic AID enzyme has to be tightly controlled, and both aberrant AID activity and chromosomal translocations involving switch regions of antibody genes are hallmark features of B-cell malignancies (2, 6–9).
AID is specifically expressed in a subset of germinal center B-lymphocytes (10, 11). Specific stimulatory signals activate their proliferation and the diversification of the variable regions of their Ig genes, followed by expression of mutated surface Ig and selection of the mutated B-cell receptors (BCRs) for reactivity with antigen (12–14). E-protein, NF-κB, Pax5, STAT6, and IRF-8 transcription factors participate in expression of the AID gene (15–18). The gene is induced by IL-4 and ligation of CD40, and, in mice, other inducers have also been identified, including LPS (11, 17, 19). Only ligation of CD45 has been reported to lead to signaling that inhibits AID gene expression, and this was shown to involve inhibition of signaling to STAT6 (19).
In the present study, we show that activation of the B-cell receptor, which signals that good antibody affinity has been reached, can inhibit AID gene expression, and we analyze the mechanism of this inhibition of AID gene expression. We report that calcium (Ca2+) signaling is essential for this inhibition. Moreover, we show that overexpression of the Ca2+ sensor protein calmodulin inhibits AID gene expression and that the transcription factor E2A is required for regulation of the AID gene by the BCR. We further show that E2A mutated in the binding site for calmodulin, and thus showing calmodulin-resistant DNA binding, makes AID expression resistant to the inhibition through BCR activation. Taken together, our findings show that BCR activation inhibits AID gene expression through Ca2+/calmodulin inhibition of E2A.
Results
BCR Activation Can Inhibit Expression of AID and the Inhibition Is Ca2+ Signaling-Dependent.
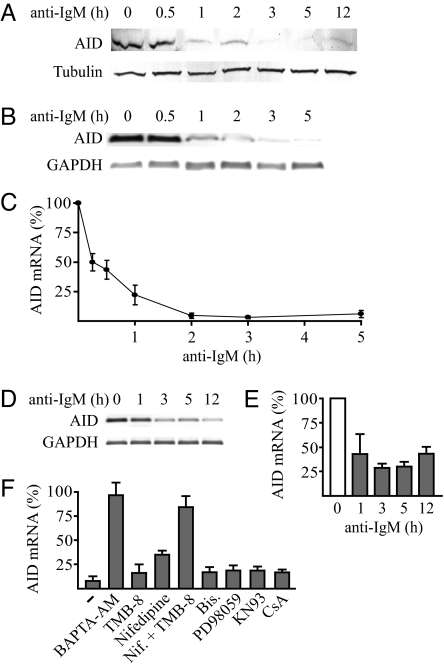
To determine whether BCR activation can inhibit expression of AID, we used purified mouse splenic B cells that were stimulated with LPS and IL-4. The stimulation increased the expression level of AID mRNA ≈100-fold within 48 h, and the AID protein became readily detectable by western analysis. Activation of the BCR with anti-IgM rapidly reduced the level of AID protein (Fig. 1A). The decrease after 1 h was in average 73% with a standard deviation of 6% (n = 3). A few hours later, the AID protein level was close to background (Fig. 1A). This shows that strong engagement of the BCR, which signals that good antibody affinity has been achieved, rapidly and dramatically inhibits expression of the mutagenic AID protein.
Fig. 1.
BCR activation with anti-IgM down-regulates AID expression, and the reduction is Ca2+ signaling-dependent. (A) Expression of AID protein in B lymphocytes from mouse spleen after activation of the gene with IL-4 (5 ng/ml) and LPS (10 μg/ml) for 48 h followed by addition of anti-IgM for the indicated times. The levels of AID and of α-tubulin used as loading control were determined by Western blot analysis. (B) AID mRNA levels of mouse B lymphocytes activated as in A and anti-IgM-treated for the indicated times determined by PCR and agarose gel electrophoresis of synthesized cDNA strands. The mRNA levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown as controls. (C) AID mRNA levels of mouse B lymphocytes activated as in A and anti-IgM-treated for the indicated times determined by quantitative RT-PCR and normalized by using the mRNA levels of GAPDH. The level before addition of anti-IgM was set at 100%. Results are mean ± SD (n = 4). (D) Expression level of AID mRNA during 12 h of treatment of the AID-expressing human lymphoma cell line DG75 with anti-IgM determined by PCR and agarose gel electrophoresis of synthesized cDNA strands. GAPDH is shown as control. (E) AID mRNA levels of the anti-IgM-treated DG75 cells determined by quantitative RT-PCR and normalized by using the mRNA levels of GAPDH. Gray bars represent cells treated with anti-IgM and the open bar represents cells not treated with anti-IgM. The level before addition of anti-IgM was set at 100%. Results are mean ± SD (n = 3). (F) Effects of different inhibitors of BCR signaling pathways on expression of AID mRNA in B lymphocytes from mouse spleen activated by IL-4 and LPS. Anti-IgM was added to one-half of cells treated with BAPTA-AM (12.5 μM), TMB-8 (50 μM), Nifedipine (50 μM), TMB-8 plus Nifedipine (Nif.), Bisindolylmaleimide I (Bis.; 0.1 μM), PD98059 (100 μM), KN93 (20 μM), or cyclosporin A (CsA)-treated (0.2 μM) and untreated (-) B lymphocytes for 3 h, followed by quantitative RT-PCR as in C. Values represent normalized AID mRNA levels with anti-IgM, as percentage of the levels without anti-IgM. Results are mean ± SD (n ≥ 3).
To determine whether the rapid reduction in AID protein after activation of the BCR was posttranscriptional or due to regulation at the mRNA level, we analyzed the levels of AID mRNA. The levels, compared with those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, are shown by PCR and agarose gel electrophoresis of synthesized cDNA strands in Fig. 1B, and the levels determined by quantitative real-time PCR are shown in Fig. 1C. The activation of the BCR with anti-IgM reduced the level of AID mRNA 2-fold in 15 min and 23-fold on average in 2 h (Fig. 1C). This result shows that engagement of the BCR inhibits expression of the mutagenic AID through a rapid reduction of the mRNA. The reduction could either be through anti-IgM-induced degradation of AID mRNA or, alternatively, through inhibition of transcription from the AID gene provided that the AID mRNA is constitutively very short-lived. The effect of anti-IgM was therefore compared with that of inhibition of transcription with actinomycin D. The AID mRNA level fell precipitously also after this treatment [supporting information (SI) Fig. 6A], showing that the AID mRNA is very short-lived. Thus, the dramatic reduction in AID mRNA appears to be through a halt in transcription of AID mRNA, which constitutively has a high degradation rate.
To investigate the mechanism of the reduction by BCR activation of the mRNA transcribed from the AID gene, both splenic B cells and the readily manipulated human lymphoma cell line DG75 were used. Like many other B-cell lymphomas (6–9), DG75 cells strongly express the AID gene. Like in stimulated splenic B cells, activation of the BCR with anti-IgM rapidly reduced the level of AID mRNA in the cell line (Fig. 1 D and E). The decrease was ≈60% within 1 h and ≈75% within 3 h (Fig. 1E).
Activation of the BCR leads to formation of a protein complex below the receptor that results in an intracellular increase in Ca2+ and a combination of Ca2+ signaling and a cascade of phosphorylations (20, 21). To investigate whether Ca2+ signaling and/or serine protein kinases were essential for the reduction in AID mRNA, the activation of the BCR with anti-IgM was performed in the presence of various inhibitors of the signaling pathways (Fig. 1F and SI Fig. 6 B–D). BCR activation still reduced AID expression in the presence of protein kinase C (bisindolylmaleimide) or MAP kinase (PD98059) inhibitor in both stimulated splenic B cells (Fig. 1F) and DG75 cells (SI Fig. 6B). Inhibition of L-type Ca2+ channels with nifedipine or IP3 receptor Ca2+ channels with TMB-8 had little effect on the reduction in AID mRNA by anti-IgM, whereas inhibition of all Ca2+ signaling, either by the Ca2+ chelator BAPTA-AM or by combined treatment with TMB-8 and nifedipine, abolished reduction of the AID mRNA level by anti-IgM (Fig. 1F and SI Fig. 6B). Interestingly, although Ca2+ signaling was essential for reduction of the AID mRNA level by anti-IgM, this reduction could still occur in the presence of cyclosporin A (CsA), which inhibits the Ca2+/calmodulin-dependent phosphatase calcineurin, or the calmodulin-dependent protein kinase inhibitor KN93 (Fig. 1F and SI Fig. 6B). This indicates that the inhibition was through a Ca2+ signaling pathway not involving any of these enzymes.
Inhibition of AID Gene Expression by Calmodulin.
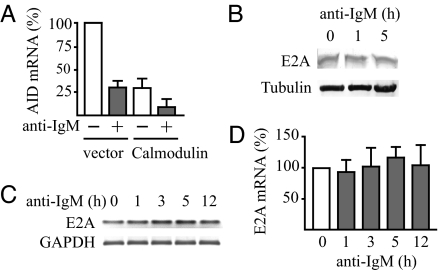
To examine whether AID gene expression could be inhibited through the main Ca2+-sensor protein calmodulin, the effect of overexpression of calmodulin was investigated. This overexpression of calmodulin in DG75 cells was verified by Western blot analysis (SI Fig. 7A). The overexpression reduced AID mRNA levels 3-fold compared with the vector control, and calmodulin reduced also the level of AID mRNA further compared with BCR activation alone (Fig. 2A).
Fig. 2.
Inhibition of AID gene expression by calmodulin and constant E2A mRNA and protein expression after engagement of the BCR with anti-IgM. (A) Down-regulation of the AID mRNA level by calmodulin overexpression with and without inhibition of the gene by BCR engagement. DG75 cells were transfected with 2 μg of pcDNAI/amp (vector) or pcDNAI/amp mCaM (Calmodulin) plasmid. Eight hours later, live cells were separated by using Lymphoprep (Axis-Shield), and, 12 h later, anti-IgM was added to one-half of the cells followed by continued culture for 3 h. Expression levels of AID mRNA were determined by quantitative RT-PCR and normalized by using GAPDH as in Fig. 1. The level before addition of anti-IgM in cells transfected with the pcDNAI/amp vector was set at 100%. Results are mean ± SD (n ≥ 3). (B) Levels of E2A protein in anti-IgM-treated DG75 cells determined by Western blot analysis. Levels of α-tubulin were used as loading controls. (C) E2A mRNA levels of anti-IgM-treated DG75 cells determined by PCR and agarose gel electrophoresis of synthesized cDNA strands as in Fig. 1. (D) E2A mRNA levels of the anti-IgM-treated DG75 cells determined by quantitative RT-PCR as in Fig. 1. Results are mean ± SD (n = 3).
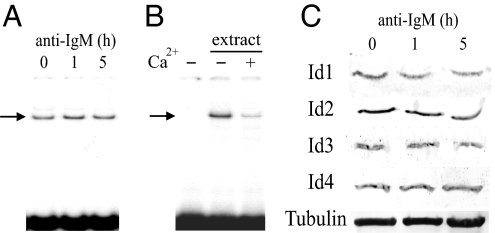
Among the transcription factors involved in AID gene expression, no Ca2+-dependent inhibitory pathway involving calmodulin without involving calcineurin or calmodulin-dependent protein kinase has been ascribed to any of NF-κB, Pax5, STAT6, or IRF-8. We have shown, however, that Ca2+-loaded calmodulin can inhibit DNA binding of E2A and other E-proteins by directly binding to the DNA-binding basic sequences in their basic helix–loop–helix domains (22–24). Thus, we examined the possible participation of E2A in the inhibition of AID gene expression. We first determined whether activation of the BCR affected the amount of E2A. Activation of the BCR with anti-IgM did not change the level of E2A protein or mRNA significantly (Fig. 2 B–D). The amount of E-protein capable of binding to an AID enhancer probe was followed by electrophoretic mobility shift assay (EMSA) of nuclear extracts in the absence of Ca2+. The EMSA gave a protein-DNA complex (Fig. 3 A and B) with the nucleotide sequence specificity of E-protein DNA complexes (data not shown). It was identified as an E2A-DNA complex by Western blot analysis with anti-E2A (SI Fig. 8A) and shown with neutralizing antibodies to contain both the E12 and the E47 splice form (SI Fig. 8B). No decrease in the amount of the E2A-DNA complex was seen during 5 h of anti-IgM treatment (Fig. 3A). The binding of E2A protein to the site in the AID enhancer was sensitive to Ca2+, because the amount of the complex was much reduced when the DNA binding analysis was performed in the presence of Ca2+ (Fig. 3B).
Fig. 3.
Constant DNA binding activity of E-proteins in the absence of Ca2+ and constant Id protein expression levels after engagement of the BCR with anti-IgM. (A) Amounts of E-protein DNA complex (indicated with arrow), using a probe containing an E-protein binding site from the AID enhancer, before anti-IgM treatment and after 1 and 5 h of treatment of DG75 cells. Nuclear extracts were analyzed by electrophoretic mobility shift assay (EMSA) in the presence of the chelator EDTA (0.5 mM). (B) Inhibition of formation of the E-protein DNA complex in the presence of Ca2+ (0.5 mM CaCl2) in EMSA as in A. The first lane was without addition of nuclear extract. (C) Id1, Id2, Id3, and Id4 protein expression levels during 5 h of BCR activation of DG75 with anti-IgM, determined by Western blot analysis. Levels of α-tubulin were used as loading controls.
Id proteins, which inhibit DNA binding of E-proteins by binding to their helix–loop–helix dimerization domain, participate at several regulatory steps in lymphocyte development (25, 26), and enforced overexpression of either Id2 or Id3 can block AID gene expression (16, 18). However, during 5 h of BCR activation, we saw no decrease in the amount of E-protein able to bind to the AID enhancer probe in the absence of Ca2+ (Fig. 3A), suggesting that no increase in inhibition of DNA binding of E-proteins by Id proteins contributed to the rapid inhibition of AID gene expression. Even so, we also examined whether anti-IgM affected the amounts of the four Id mRNA and Id proteins. None of the Id mRNA levels showed any large increase that could explain the large decrease in AID mRNA (SI Fig. 9), and no significant change was seen in the protein expression level of any of Id1, Id2, Id3, or Id4 during the 5 h of BCR activation when expression of the AID gene decreased (Fig. 3C).
Inhibition of AID Gene Expression upon BCR Activation Through Ca2+/Calmodulin Inhibition of E2A.
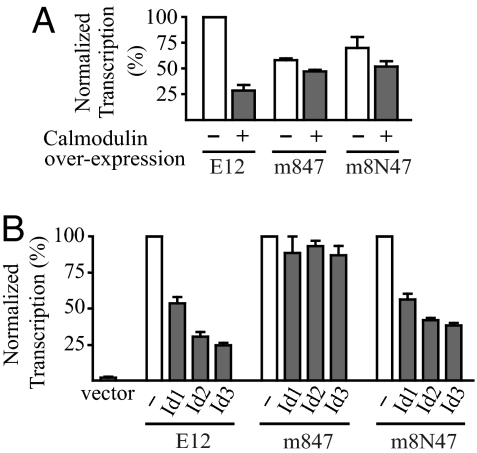
We have reported a series of mutants in the basic DNA and calmodulin binding sequence of the E2A isoform E12 that, through combinations of mutations, are resistant to calmodulin to different extents (24). To investigate the possible role of Ca2+/calmodulin inhibition of E2A in the inhibition of AID mRNA expression, we used the most calmodulin-resistant one of these mutants: the m847 mutant, which has three amino acid substitutions. Overexpression of calmodulin resulted in a fourfold decrease in activation of an E-protein responsive luciferase reporter by the wild-type E12, whereas it had very little inhibitory effect on activation by the m847 mutant (Fig. 4A). However, this mutant also renders the transcriptional activation of the reporter almost completely resistant to overexpression of Id1, Id2, or Id3 (Fig. 4B). The Id-resistance of m847 was due to the mutation of aspartate 561 to alanine, because a mutant, m8N47, with mutation of this position to an asparagine (N) instead, while keeping the other two mutations, was almost as sensitive as the wild type to overexpression of Id proteins (Fig. 4B), although it was almost as resistant as the m847 mutant to overexpression of calmodulin (Fig. 4A).
Fig. 4.
Differential sensitivity of E12 mutants to Id proteins and calmodulin. (A) Transcriptional activity of wild-type mouse E12 and the E12 mutants m847 and m8N47, with and without overexpression of calmodulin in a reporter assay. Expression vectors for E12 proteins (2.5 μg) were transiently expressed in DG75 cells together with 2.5 μg of a luciferase reporter plasmid for E-proteins, 6× (μE5+μE2)-luciferase, and 2.5 μg of the calmodulin expression plasmid or empty vector. hCMV-βgal plasmid (2.5 μg) was included to normalize each assay. The luciferase reporter activities of cells harvested after 24 h were measured and normalized to the β-galactosidase activities. The transcriptional activity of wild-type E12 without calmodulin overexpression was set at 100%. Results are mean ± SD (n = 3). (B) Transcriptional activity of wild-type and mutant E12 with and without overexpression of Id proteins in a reporter assay. DG75 cells were transiently transfected as in A, except by using expression vector for the indicated Id protein (0.25 μg) instead of calmodulin. The activity of each E12 protein together with empty Id expression vector was set at 100%. Results are mean ± SD (n = 3).
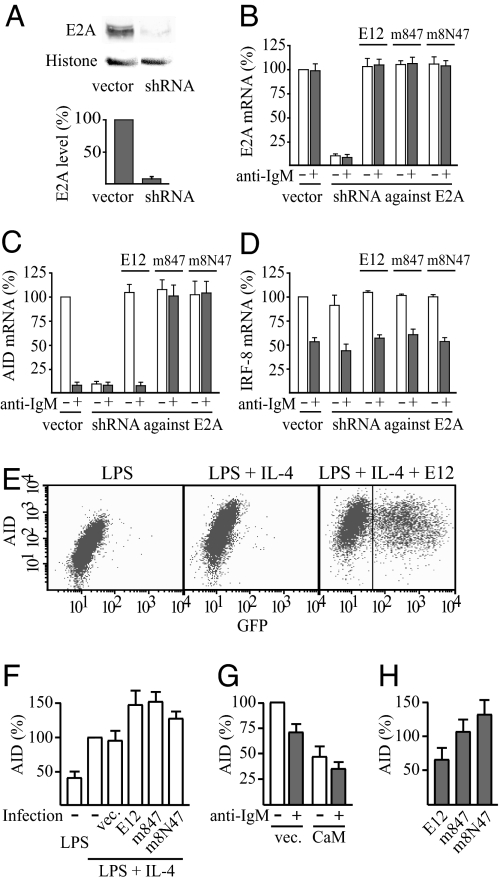
To examine whether the Ca2+ signaling from the BCR inhibited AID mRNA expression through calmodulin-mediated inhibition of E2A, we constructed derivatives of the hygromycin-selectable EBV-based shuttle vector pMEP4 that stably directs synthesis of short hairpin RNA (shRNA) interfering with both the E12 and E47 isoforms of human E2A mRNA. The most efficiently interfering shRNA plasmid reduced the expression of both E2A protein and E2A mRNA by >90% (Fig. 5 A and B). This shRNA expression plasmid reduced the expression of the AID gene in DG75 B-cells by 90.6 ± 2.6% (Fig. 5C), confirming previous reports that AID gene expression is E2A-dependent. The reduction of the E2A and AID mRNA was specific, because the shRNA expression plasmid did not reduce expression of a number of other BCR-regulated genes that have not been reported to be E2A-regulated (Fig. 5D, SI Fig. 10 A–C, and data not shown).
Fig. 5.
Loss of anti-IgM sensitivity of AID expression by expression of calmodulin-resistant E12. (A) Reduced expression of E2A in DG75 cells by a shuttle vector that expresses short hairpin RNA (shRNA) targeting both E12- and E47-splice forms of human E2A mRNA. A typical Western blot and mean ± SD of quantification (n = 3) are shown. (B–D) Effects of the shuttle vector that expresses short hairpin RNA (shRNA) interfering with human E2A mRNA, and complementation by expression of wild-type or mutant mouse E12, on the expression of E2A (B), AID (C), and IRF-8 (D) mRNA. DG75 (1 × 107 cells) was transfected with 5 μg of pMEP4 shuttle vector expressing shRNA against human E2A mRNA or the empty vector or with 5 μg of the shRNA expression vector plus 0.5 μg of the corresponding pMEP4 vector expressing wild-type or calmodulin-resistant mutant of mouse E12, all added together with 25 μg of EBNA expression plasmid. Where indicated, cells were treated with anti-IgM for 3 h before the harvest. The expression level of E2A, AID, or IRF-8 mRNA, respectively, in cells transfected with empty pMEP4 vector and not treated with anti-IgM was set at 100%. Results are mean ± SD (n = 3). (E) Expression of AID protein in B lymphocytes from mouse spleen after activation with LPS (10 μg/ml) and where indicated also with IL-4 (5 ng/ml) for 75 h. (Right) Cells infected with retrovirus encoding E12 followed by an internal ribosome entry site (IRES) and green fluorescent protein (GFP) after 24 h of the incubation with the activators. The levels of AID and GFP were determined by intracellular immunostaining and flow cytometry. (F–H) Average AID protein expression levels of cells from experiments such as those shown in E and corresponding infections with retrovirus encoding wild-type E12, m847, or the m8N47 mutant, calmodulin, or the empty retrovirus vector. Cells with a GFP fluorescence exceeding 43 (indicated in E Right) were considered as retrovirus-infected and used to calculate average AID expression levels of infected cells. Results are mean AID immunostaining fluorescence intensity ± SD. (n = 3). (F) The level in noninfected cells activated with both LPS and IL-4 was set at 100%. (G) The level in control cells infected with empty retrovirus vector (vec.) and not treated with anti-IgM was set at 100%. (H) AID expression levels of infected cells treated with anti-IgM for 3 h are expressed as percentage of the levels of the corresponding infected cells not treated with anti-IgM.
Importantly, anti-IgM had no further inhibitory effect on the reduced AID gene expression in the presence of the shRNA against E2A (Fig. 5C). In contrast to the ≈10-fold reduction in AID mRNA by anti-IgM in the presence of vector control plasmid, the expression of the AID gene was ≈10% of the uninhibited control both with and without anti-IgM treatment in the presence of this shRNA expression plasmid (Fig. 5C). This loss of effect of anti-IgM was specific, because expression of the shRNA against E2A did not affect the result of anti-IgM treatment for any gene analyzed that was induced or inhibited by BCR activation and not reported to be E2A-regulated (Fig. 5D, SI Fig. 10 A–C, and data not shown). The expression of the genes coding for these proteins, including those for the cytokine IL-2 (SI Fig. 10C) and the transcription factors IRF-8, IRF-4, and Oct-2 (Fig. 5D and SI Fig. 10 A–B), were all increased or reduced as normal by anti-IgM in presence of the shRNA expression plasmid.
Complementation by cotransfection with expression plasmid for the mouse E12 isoform of E2A could reverse the decrease in E2A mRNA (Fig. 5B), because the shRNA does not interfere with the mouse E2A. This complementation effect did not change the expression of any of the control genes significantly, confirming that they are not E-protein-regulated (Fig. 5D, SI Fig. 10 A–C, and data not shown). For the AID gene, this complementation fully restored both the mRNA expression level and the sensitivity of this expression to anti-IgM treatment (Fig. 5C). Importantly, in contrast to the ≈10-fold reduction in AID mRNA by anti-IgM in the presence of vector control plasmid or after complementation with wild-type E12, anti-IgM had no inhibitory effect after complementation with the m847 mutant of E12 (Fig. 5C). This loss of effect of anti-IgM on the expression of the AID gene could be due to either the calmodulin-resistance (Fig. 4A) or the Id-resistance (Fig. 4B) of this mutant. The m8N47 mutant allows one to discriminate between these two alternatives, because it is approximately as calmodulin-resistant as the m847 mutant but almost as Id-sensitive as the wild-type E12. Complementation by expression of this mutant resulted in complete loss of the reduction in AID mRNA in the presence of anti-IgM (Fig. 5C), showing that the sensitivity to BCR activation was due to the calmodulin sensitivity of the protein and not to the Id sensitivity. This loss of effect of anti-IgM was specific for the E-protein-regulated AID gene, because the complementation of the shRNA against E2A by expression of calmodulin-resistant m847 or m8N47 mutant of E12 did not affect the result of anti-IgM treatment for any of the control genes stimulated or inhibited by BCR activation (Fig. 5D, SI Fig. 10 A–C, and data not shown).
To further examine whether the Ca2+ signaling from the BCR inhibited AID mRNA expression through calmodulin-mediated inhibition of E2A, we performed FACS analyses of primary splenic B-cell cultures after intracellular immunostaining. The activation of AID protein expression was efficient, and it became 2- to 3-fold higher by activation with both LPS and IL-4 compared with activation with LPS alone (Fig. 5 E and F). We constructed retroviruses encoding calmodulin or wild-type or mutant E12 followed by an internal ribosome entry site (IRES) and green fluorescent protein (GFP). The efficiency of retrovirus infections, detected as GFP immunostaining, was ≈40–50% (Fig. 5E and data not shown). Infection of the splenic B-cell cultures with E12 expressing retroviruses resulted in ≈1.5-fold increased activation of AID protein expression (Fig. 5 E and F). Infection with calmodulin expressing retrovirus showed that calmodulin overexpression, verified by Western blot analysis (SI Fig. 7B), inhibits AID expression in primary splenic B-cells (Fig. 5G), in agreement with the results in DG75 cells (Fig. 2A). Treatment of primary splenic B-cells infected with retrovirus expressing wild-type E12 or the vector control with anti-IgM for 3 h before harvest to activate the BCR resulted in clear inhibition of AID expression (Fig. 5 G and H). The inhibition was somewhat less pronounced than in the experiments of Fig. 1A because of the longer protocol needed with harvest at 75 h. By that time, many B-cells are not as sensitive to the anti-IgM treatment, presumably because they class switch away from IgM. Nevertheless, a clear difference was obtained between primary splenic B-cells infected with wild-type E12 that showed AID expression sensitive to anti-IgM treatment and cells infected with calmodulin resistant m847 or m8N47 mutant of E12 that showed AID expression clearly resistant to the anti-IgM (Fig. 5H). Thus, we conclude that calmodulin-resistant E12 renders the expression of the AID gene resistant to activation of the BCR.
Discussion
The results presented here show that activation of the B-cell receptor (BCR) can inhibit AID gene expression. This is a very functional response, because it is essential that mutagenesis of the antibody genes ceases when good antibody affinity has been reached, and the BCR with its membrane bound antibody is the receptor that can give such a signal. We show also that the BCR activation inhibits AID gene expression by Ca2+ signaling through inhibition of E2A by the major Ca2+ sensor calmodulin. Mutations in the DNA binding sequence of E2A, which makes it resistant to calmodulin without spoiling its DNA binding, make AID gene expression resistant to BCR activation. This resistance of the AID gene expression when E2A is mutated (Fig. 5C) does not exclude that the other E-proteins, E2-2 and HEB, may also contribute to the AID expression in vivo and become calmodulin inhibited like E2A when the BCR is activated. However, the 10-fold reduction in AID mRNA by shRNA against E2A in the DG75 cells (Fig. 5C) suggests that E2A is the dominant E-protein in AID expression. The stable levels of the Id proteins that we observe during the inhibition of AID mRNA expression (Fig. 3C) do not contradict previous reports that overexpression of Id2 or Id3 inhibits AID expression (16, 18). Instead, our findings imply that the E-protein activity has to be carefully fine-tuned to maintain the right levels of AID activation and sensitivity toward Ca2+/calmodulin after BCR activations of different strengths, and it is likely that Id proteins have a major role in this optimization of the regulatory system. In this context, it is also interesting that the decrease in the humoral immune response in aging mice and humans is paralleled by decreases in expression of both E2A and AID with age (27).
It is notable that anti-IgM reduced expression of mRNA for the IRF-8 transcription factor (Fig. 5D), because IRF-8 has recently been reported to participate in the activation of AID gene expression (15). It is also interesting that anti-IgM reduced the expression of IRF-4 mRNA, because IRF-4 has been shown to synergize with the E2A protein E47 at another promoter, the Iμ promoter activating germ line Iμ transcription, which is required for isotype class switching (28, 29). In addition, IRF4-deficient B cells have been shown to display impaired expression of AID and lack class-switch recombination (30). Neither the IRF-8 inhibition nor the IRF-4 inhibition was due to the Ca2+/calmodulin inhibition of E2A (Fig. 5D and SI Fig. 10A), showing that the inhibition of their expression by BCR activation is through another signaling pathway. The decreases in IRF-4 and IRF-8 mRNA do not appear to have contributed in the initial phase of the inhibition of AID expression, because most of the decrease in AID mRNA levels by activation of the BCR has occurred before any reduction in IRF-4 or IRF-8 mRNA (Fig. 1B and SI Fig. 10 D–E). However, this result does not exclude the possibility of a role for IRF-4 and/or IRF-8 at a later stage in the process of shutting off AID expression. The Ca2+ signaling from the BCR will last for a limited time, and the inhibition, the start of which has been studied here, ultimately leads to the AID gene being stably and completely shut off. It is therefore possible that inhibition of IRF-4 and/or IRF-8 is involved in a later step of this shut-off process. Our discovery that Ca2+ inhibits AID gene expression upon BCR activation through Ca2+/calmodulin-mediated inhibition of E2A provides a starting point for detailed analysis of the further steps of this process.
Materials and Methods
Plasmids and Mutagenesis.
The luciferase reporter for E12, 6× (μE5+μE2)-luciferase, and the hCMV-β-gal plasmid for normalization of transfections by dividing the enzymatic activity from the luciferase reporters with the β-galactosidase activity are described in ref. 22. The expression vector for mammalian calmodulin (CaM), pcDNAI/amp mCaM, is also described in ref. 31. The cDNAs coding for rat Id1, Id2, and Id3, all inserted in pcDNA3 (Invitrogen), were kindly provided by M. D. Walker (Weizmann Institute of Science, Rehovot, Israel). The calmodulin-resistant E12 basic sequence mutant m847 is described in ref. 24, and the m8N47 mutant was generated correspondingly by standard PCR mutagenesis and substituted into full-length E12 in pcDNAI/amp E12 as a DrdI-StuI fragment. The DNA encoding wild-type and modified E12 and a DNA sequence (5′-CAGCAGCCTCTCTTCATCCTTCAAGAGAGGATGAAGAGAGGCTGCTG-3′) encoding a short hairpin RNA (shRNA) that interferes with E12/E47-specific human RNA were inserted into a derivative of the replicating EBV-based shuttle vector pMEP4 (32) as a BglII-HindIII fragment. The DNA encoding wild-type and modified E12 and the calmodulin was also cloned as EcoRI-XhoI fragments into MSCV-IRES-GFP vector for production of retroviruses in 293T cells.
Cell Culture and Transfections.
The human B-cell lymphoma line DG75 (33) was maintained in RPMI medium 1640 supplemented with 5% FCS and antibiotics. DG75 cells were transfected by electroporation as described in ref. 34. Four hours after transfection of DG75 with the empty pMEP4 derivative or pMEP4 derivative encoding the shRNA, E12, or m847 or m8N47 mutant of E12, Hygromycin B (Roche) was added to the medium at a concentration of 0.75 mg/ml for 3 days, followed by 0.6 mg/ml for another 2 days.
For BCR activation experiments, 1 × 107 cells were stimulated for 3 h (unless otherwise specified) with 5 μg of goat F(ab′)2 anti-human IgM (Southern Biotechnologies) in 1 ml of complete RPMI medium 1640 supplemented with 5% FCS.
Western Blot and DNA Binding Assays.
Nuclear and cytoplasmic extracts were prepared as described in ref. 24, and electrophoretic mobility shift assays were performed as described in ref. 23, using an end-labeled DNA probe (5′-GGGAGCACAGCTGTCTCAGC-3′) containing the E-protein binding sequence from the AID enhancer (16, 18). Western blot was performed by using the WesternBreeze immunodetection system (Invitrogen) according to the manufacturer's instructions, using antibodies to E2A (V-18), Id1 (Z-8), Id2 (C-20), Id3 (H-70), or Id4 (H-70) (all from Santa Cruz Biotechnology) or α-tubulin (clone B-5-1-2; Sigma–Aldrich).
Purification and Analysis of B-Lymphocytes from Mouse Spleens.
Primary B-lymphocytes were purified from spleens of C57BL/6 mice, using the B Cell Isolation Kit, MidiMACS Separator and LS Columns (all from Miltenyi Biotech) according to the manufacturer's recommendations. Purified B-cells (1 × 106 cells per ml) were maintained in RPMI medium supplemented with 10% heat-inactivated FCS, antibiotics and 50 μM mercaptoethanol for 48 h, with or without LPS (10 μg/ml) and IL-4 (5 ng/ml). Stimulated cells (1 × 106 cells per ml) were then incubated in a 24-well cell culture plate with anti-mouse IgM antibody (2.5 μg/ml) for the indicated times. For infections, retrovirus concentrated by centrifugation was added with polybrene (5 μg/ml) to 0.5 × 106 B-cells after activation with LPS plus IL-4 for 24 h. After 8 h incubation, the infection was repeated, this time for 16 h, followed by incubation for a further 27 h after infection in fresh complete medium with the stimulants to allow for expression of GFP and E12 or Calmodulin. Where indicated, goat F(ab′)2 anti-mouse IgM (2.5 μg/ml; Southern Biotechnologies) was added for 3 h before harvest. Intracellular immunostaining was with 2% paraformaldehyde and ethanol, as described in ref. 35, using rabbit anti-AID (ProSci; 1 μg/250 μl staining volume), APC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch; 1 μg/250 μl) and FITC-conjugated goat anti-GFP (Novus Biologicals; 0.5 μg/250 μl). Stainings were for 30 min at RT in the dark. Flow Cytometry was with a FACSCalibur instrument and analysis with CellQuest software (Becton Dickinson).
PCR.
Total RNA was extracted by using TRIzol Reagent (Invitrogen) according to the instruction manual. First-strand cDNA was synthesized from 1 μg of total RNA, using a cDNA synthesis kit with random hexamers (Fermentas) according to the manufacturer's instructions. Real-time PCR analysis was performed in triplicate in 25-μl reaction volumes, using iTaq or iTaq SYBR Green supermix (BioRad) and the iCycler iQ Real-Time PCR Detection System (BioRad). Mouse AID mRNA was quantified with the iScript One-Step real-time PCR kit with SYBR Green (BioRad). Primer sets were designed by using Beacon Designer software (BioRad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Real-time PCR values were determined by Comparative Quantification (36). The specificity of the Real-time PCR was analyzed by melt-curve analysis as described by the manufacturer, and the sizes of the PCR products with the different primer pairs were verified by 2.5% agarose gel electrophoresis. The primer pairs used in the PCR are specified in SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by grants from the Swedish Research Council and the Swedish Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708220105/DC1.
References
- 1.Maizels N. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 2.Frieder D, Larijani M, Tang E, Parsa JY, Basit W, Martin A. Immuol Res. 2006;35:75–88. doi: 10.1385/IR:35:1:75. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa H, Hauschild J, Buerstedde JM. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 6.Kuppers R. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 7.Luo Z, Ronai D, Scharff MD. J Allergy Clin Immunol. 2004;114:726–735. doi: 10.1016/j.jaci.2004.07.049. quiz 736. [DOI] [PubMed] [Google Scholar]
- 8.Xu Z, Fulop Z, Zhong Y, Evinger AJ, III, Zan H, Casali P. Ann NY Acad Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jardin F, Sahota SS. Hematology. 2005;10:115–129. doi: 10.1080/10245330400026105. [DOI] [PubMed] [Google Scholar]
- 10.Cattoretti G, Buttner M, Shaknovich R, Kremmer E, Alobeid B, Niedobitek G. Blood. 2006;107:3967–3975. doi: 10.1182/blood-2005-10-4170. [DOI] [PubMed] [Google Scholar]
- 11.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 12.Wolniak KL, Shinall SM, Waldschmidt TJ. Crit Rev Immunol. 2004;24:39–65. doi: 10.1615/critrevimmunol.v24.i1.20. [DOI] [PubMed] [Google Scholar]
- 13.MacLennan IC. Immunity. 2005;22:656–657. doi: 10.1016/j.immuni.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Cyster JG. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 15.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, et al. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, Mori KJ, Yokota Y, Shimizu A. J Exp Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Int Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 18.Sayegh CE, Quong MW, Agata Y, Murre C. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 19.Zhou C, Saxon A, Zhang K. J Immunol. 2003;170:1887–1893. doi: 10.4049/jimmunol.170.4.1887. [DOI] [PubMed] [Google Scholar]
- 20.Jun JE, Goodnow CC. Nat Immunol. 2003;4:1057–1064. doi: 10.1038/ni1001. [DOI] [PubMed] [Google Scholar]
- 21.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Corneliussen B, Holm M, Waltersson Y, Onions J, Hallberg B, Thornell A, Grundström T. Nature. 1994;368:760–764. doi: 10.1038/368760a0. [DOI] [PubMed] [Google Scholar]
- 23.Onions J, Hermann S, Grundström T. J Biol Chem. 1997;272:23930–23937. doi: 10.1074/jbc.272.38.23930. [DOI] [PubMed] [Google Scholar]
- 24.Saarikettu J, Sveshnikova N, Grundström T. J Biol Chem. 2004;279:41004–41011. doi: 10.1074/jbc.M408120200. [DOI] [PubMed] [Google Scholar]
- 25.Lazorchak A, Jones ME, Zhuang Y. Trends Immunol. 2005;26:334–338. doi: 10.1016/j.it.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Murre C. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 27.Frasca D, Riley RL, Blomberg BB. Semin Immunol. 2005;17:378–384. doi: 10.1016/j.smim.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Nagulapalli S, Atchison ML. Mol Cell Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagulapalli S, Goheer A, Pitt L, McIntosh LP, Atchison ML. Mol Cell Biol. 2002;22:7337–7350. doi: 10.1128/MCB.22.20.7337-7350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 31.Hughes K, Antonsson A, Grundström T. FEBS Lett. 1998;441:132–136. doi: 10.1016/s0014-5793(98)01537-3. [DOI] [PubMed] [Google Scholar]
- 32.Holmfeldt P, Stenmark S, Gullberg M. EMBO J. 2004;23:627–637. doi: 10.1038/sj.emboj.7600076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey JM, Cohen MM, Bentwich Z, Ramot B, Klein E, Klein G. Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 34.Hughes K, Saarikettu J, Grundström T. Methods Mol Biol. 2002;173:355–363. doi: 10.1385/1-59259-184-1:355. [DOI] [PubMed] [Google Scholar]
- 35.Krutzik PO, Nolan GP. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 36.Applied Biosystems. [Accessed 10 January 10, 2003];ABI Prism 7700 Sequence Detection System User Bulletin no. 2. 2001 Available at http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.