Abstract
SAP (also named SH2D1A) is an intracellular adaptor molecule expressed in T cells, natural killer (NK) cells, and some B cells. The SAP gene is mutated in X-linked lymphoproliferative (XLP) disease, a human immunodeficiency characterized by a faulty immune response to Epstein–Barr virus infection. Previous reports documented severe defects in antibody production and germinal center (GC) formation in SAP-deficient humans and mice genetically engineered to lack SAP expression. However, in vitro studies and adoptive transfer experiments provided conflicting data as to whether this phenotype is caused by a functional defect resulting from SAP deficiency in T cells, B cells, or both. Here, we ascertained which cell types are responsible for this humoral immunity defect by using a conditional gene targeting approach. We also thoroughly examined the expression pattern of SAP in normal immune cells by using intracellular flow cytometry. The results showed that expression of SAP in T cells, but not in B cells or NK cells, is required and sufficient for SAP-dependent antibody production and GC formation. These data provide a critical insight into the mechanism by which SAP regulates humoral immunity. They also help elucidate the basis of a severe human immunodeficiency.
Keywords: antibodies, conditional knockout, lymphocytes, X-linked lymphoproliferative, SH2D1A
The intracellular adaptor molecule SAP [SLAM (signaling lymphocytic activation molecule)-associated protein)] is composed almost exclusively of a Src homology 2 (SH2) domain (1, 2). It is expressed in T cells, natural killer (NK) cells, NK-T cells, and some B cells, including transformed B cell lines and, perhaps, normal B cells. SAP operates in immune regulation through its capacity to bind, by way of its SH2 domain, the cytoplasmic domain of members of the SLAM family of immune cell-specific receptors. It couples SLAM-related receptors to tyrosine phosphorylation signals by its capacity to recruit Src-related kinase FynT. Additional FynT-independent mechanisms of SAP signaling may exist (3, 4).
The SAP gene is mutated in X-linked lymphoproliferative (XLP) disease, a human immune deficiency characterized by a faulty immune response to Epstein–Barr virus (EBV) (1, 2, 5). In general, patients with XLP present a fulminant lymphoproliferative illness in reaction to EBV. These patients also develop severe hypogammaglobulinemias and malignant lymphomas. XLP patients exhibit defects in several immune cell lineages (1, 2, 5). They show compromised CD4+ T cell differentiation, diminished NK cell cytotoxicity, reduced humoral responses with low levels of Igs, impaired isotype switching and absent germinal center (GC) formation, a severe deficiency in memory B cell numbers, and a near absence of NK-T cells. Although it is unclear which abnormalities are responsible for the various clinical syndromes of XLP, defects involving multiple lineages are believed to be implicated. Most of the immune alterations seen in XLP patients were also observed in SAP-deficient mice (6–8).
A major question arising with SAP-deficient humans and mice is whether their severely impaired antibody responses and memory B cell generation are caused by defects in T cell help toward B cells, intrinsic defects in B cell functions, or both. In humans, it was observed that purified CD4+ T cells from XLP patients had a reduced ability to provide help to B cells in vitro, with diminished IL-10 secretion and decreased expression of inducible costimulator (ICOS) (9). By opposition, purified memory B cells from these patients, while dramatically reduced in numbers (10), had a normal capacity to undergo somatic hypermutation and secrete Ig in vitro. These limited data with human cells suggested that the humoral immunity dysfunctions are largely caused by a T cell help defect.
More extensive studies, including in vivo analyses, were conducted with SAP-deficient mice. Purified CD4+ T cells from these animals had altered activation responses in vitro, including decreased production of T helper 2 (TH2)-type cytokines (such as IL-4 and IL-13), reduced expression of ICOS, and increased levels of CD40 ligand (CD40L) (3, 6, 7, 11, 12). SAP-deficient mice also had defective antibody responses to T cell-dependent antigens, but not T cell-independent antigens, implying a T cell-derived anomaly (3, 13, 14). However, purified naïve B cells from SAP-deficient mice exhibited reduced Ig secretion and Ig switch recombination in vitro, suggesting an additional intrinsic B cell problem (15). Conflicting data were also obtained with adoptive transfer experiments. Three groups using the same SAP-deficient mouse strain showed that transfer of CD4+ T cells, but not B cells, from SAP-deficient mice recapitulated the antibody production and GC formation impairment seen in sap−/− mice (3, 4, 13, 16). Nevertheless, another team using a distinct SAP-deficient mouse strain reported that transfer of either naïve B cells or CD4+ T cells from sap−/− mice was apt at recreating the compromise in humoral immunity, implying that SAP expression in both cell types was actually important (17).
Here, we tested the relative contribution of SAP expression in T cells and B cells to antibody production, using a conditional gene targeting approach in the mouse. We also carefully examined the expression pattern of SAP at the single cell level, using intracellular flow cytometry. These analyses provide formal evidence that the humoral defect seen in SAP-deficient mice and, presumably, humans is entirely caused by an intrinsic T cell dysfunction.
Results and Discussion
Creation of a Conditionally Targeted Allele of sap.
To test genetically the contribution of intrinsic T cell and B cell dysfunctions to the humoral immunity defects seen in SAP-deficient mice and humans, we engineered a conditionally targeted allele of the sap gene in the mouse. The targeting vector and strategy used are depicted in supporting information (SI) Fig. 5A. This vector was designed to delete exon 2 of sap. Based on a similar construct used to create conventional SAP-deficient mice (8), we expected to generate a null allele once the floxed allele had been deleted by the Cre recombinase. After transfection in ES cells, two clones showing evidence of homologous recombination (SI Fig. 5B; data not shown) were injected into blastocysts (SI Fig. 5C, lane 2). Mice carrying the floxed sap allele (sapfl) were first bred with transgenic mice expressing the Flpe recombinase, to excise the neo resistance cassette. They were subsequently back-crossed for five to seven generations with C57BL/6 mice.
To delete sap in either T or B cells, sapfl/fl mice (as the sap gene is X-linked, these mice were either sapfl/fl females or sapfl/fl males) were crossed with transgenic mice expressing Cre under the control of the cd4 promoter (active in T cells) (18), the proximal lck promoter (active in T cells) (18), or the cd19 promoter (active in B cells) (19). sapfl/fl mice were also bred with mice expressing Cre under the control of the ubiquitous IIE promoter (20), to delete sap in the germ line and obtain a conventional SAP-deficient mouse (sap−/−) (SI Fig. 5C, lane 3).
Expression of SAP in Normal Mouse Immune Cells.
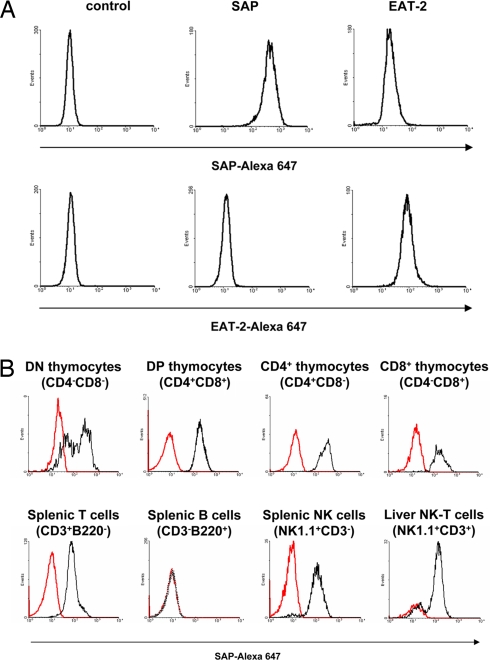
To examine the distribution of SAP in immune cells from WT and conditionally targeted sapfl/fl mice, we developed an intracellular flow cytometry assay to study SAP expression at the single cell level. The antibody used recognized transfectants of a T cell line expressing SAP, but not those expressing the closely related adaptor EAT-2 (Fig. 1A). We first examined the expression pattern of SAP in immune cells from WT C57BL/6 mice (Fig. 1B). To confirm that the antibody was specifically recognizing SAP, parallel assays were conducted with cells from sap−/− mice. SAP was expressed in thymus (including CD4−CD8−, CD4+CD8+, CD4+CD8−, and CD4−CD8+ thymocytes), spleen and lymph node T cells (CD3+B220− cells), spleen NK cells (NK1.1+CD3− splenocytes), and liver and spleen NK-T cells (NK1.1+CD3+ cells) (Fig. 1B; data not shown). The expression of SAP in these various populations was generally uniform with the exception of CD4−CD8− thymocytes and NK1.1+CD3+ liver cells, which contained a subpopulation that expressed little or no SAP. The identity of these SAP-negative cells remains to be clarified. In contrast, SAP was not detected in splenic or lymph nodes B cells (B220+CD3− cells) (Fig. 1B; data not shown). Because we roughly estimated that the anti-SAP antibody would detect levels of SAP as low as 5–10% of those seen in T cells, this observation implied that most peripheral B cells express little or no SAP. We could not exclude the possibility that SAP is expressed in higher amounts in a very small B cell subset.
Fig. 1.
Detection of SAP protein in normal immune cells by intracellular flow cytometry. (A) Specificity of anti-SAP antibody. (Upper) Anti-SAP mAb 12C4 was coupled to Alexa 647 and tested by intracellular flow cytometry using BI-141 T cell transfectants expressing SAP, EAT-2, or neither of the two (control). (Lower) Anti-EAT-2 mAb 15G3 coupled to Alexa 647 was used to assess expression of EAT-2. (B) Expression of SAP in normal immune cells. Immune cells were isolated from WT C57BL/6 mice and used for multicolor flow cytometry analyses. The presence of SAP in various cell populations was ascertained by gating on the appropriate surface markers and measuring SAP intracellular staining (black lines). As negative control, intracellular staining was performed on immune cells derived from SAP-deficient mice (red lines).
Expression of SAP in Conditionally Targeted sapfl/fl Mice.
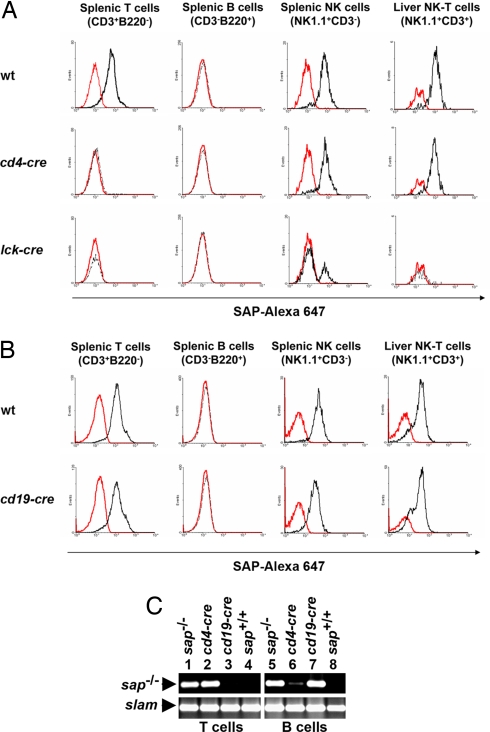
Next, SAP expression was studied in conditionally targeted sapfl/fl mice (Fig. 2). In sapfl/fl;cd4-cre mice (Fig. 2A Middle), SAP expression was eliminated in T cells (Fig. 2A Left), but not in NK cells (Fig. 2A Third from Left). It was not abrogated in NK-T cells (Fig. 2A Right), although NK-T cells were reduced in number by ≈30–50% (data not shown). This last observation suggested that the cd4-cre transgene was causing partial deletion of the sap gene in NK-T cells, in agreement with the fact that NK-T cells are derived from a CD4+CD8+ T cell precursor (21). As reported (22–24), a severe reduction (≈90%) of NK-T cell numbers was seen in conventional sap−/− mice (data not shown). In sapfl/fl;lck-cre mice (Fig. 2A Bottom), the accumulation of SAP was abrogated in T cells (Fig. 2A Left). It was also eliminated in ≈60% of NK cells (Fig. 2A Third from Left), consistent with the known expression of Lck in NK cells (25). In addition, it was eliminated in NK-T cells (Fig. 2A Right). The abundance of NK-T cells was also severely reduced (≈90%; data not shown), to the same degree as that noted in sap−/− mice. In contrast, in sapfl/fl;cd19-cre mice (Fig. 2B Lower), there was no alteration of the expression of SAP in T cells (Fig. 2B Left), NK cells (Fig. 2B Third from Left) and NK-T cells (Fig. 2B Right). Moreover, the presence of NK-T cells was not affected (data not shown). These findings were in line with the idea that cd19-cre is not active in T cells, NK cells, or NK-T cells (19).
Fig. 2.
SAP expression in conditionally targeted sapfl/fl mice. (A) Detection of SAP protein in sapfl/fl;cd4-cre and sapfl/fl;lck-cre mice. SAP protein expression was detected in cells from WT, sapfl/fl;cd4-cre (cd4-cre), and sapfl/fl;lck-cre (lck-cre) mice (black lines), as detailed in the legend of Fig. 1B. For negative control, intracellular staining was performed by using cells from a conventional SAP-deficient mouse (red lines). Absolute cell numbers in this experiment were: for total splenocytes, WT, 3.6 × 107; sap−, 2.0 × 107; cd4-cre, 2.0 × 107; lck-cre, 2.0 × 107; for splenic T cells, WT, 8.3 × 106; sap−, 3.6 × 106; cd4-cre, 4.1 × 106; lck-cre, 2.3 × 106; for splenic B cells, WT, 2.3 × 107; sap−, 1.4 × 107; cd4-cre, 1.3 × 107; lck-cre, 1.5 × 107; for splenic NK cells, WT, 1.9 × 106; sap−, 1.0 × 106; cd4-cre, 1.0 × 106; lck-cre, 1.0 × 106; for liver NK-T cells, WT, 1.5 × 106; sap−, 0.2 × 106; cd4-cre, 0.9 × 106; lck-cre, 0.2 × 106. (B) Detection of SAP in sapfl/fl;cd19-cre mice. SAP protein expression was detected in cells from WT and sapfl/fl;cd19-cre (cd19-cre) mice (black lines), as detailed in the legend of Fig. 1B. For negative control, intracellular staining was performed by using cells from a conventional SAP-deficient mouse (red lines). Absolute cell numbers in this experiment were: for total splenocytes, WT, 3.0 × 107; sap−, 3.2 × 107; cd19-cre, 2.9 × 107; for splenic T cells, WT, 7.6 × 106; sap−, 7.2 × 106; cd19-cre, 6.6 × 106; for splenic B cells, WT, 1.8 × 107; sap−, 2.0 × 107; cd19-cre, 1.7 × 107; for splenic NK cells, WT, 1.1 × 106; sap−, 0.9 × 106; cd19-cre, 1.2 × 106; for liver NK-T cells, WT, 2.2 × 106; sap−, 1.2 × 106; cd19-cre, 3.0 × 106. (C) Deletion of the sap gene in conditionally targeted sapfl/fl mice. Splenic T cells or B cells were purified by negative selection from WT (sap+/+), sapfl/fl;cd4-cre (cd4-cre), sapfl/fl;cd19-cre (cd19-cre), and conventional SAP-deficient (sap−/−) mice. DNA from mouse tail was screened by PCR using oligos 2 and 3 (Upper) depicted in SI Fig. 5A. Using these oligos, a ≈300-nt fragment was observed with the deleted allele. slam was amplified as control (Lower).
Because SAP was not detected in normal B cells (Fig. 1B), it was impossible to determine with this approach whether the cre transgenes, in particular cd19-cre, were causing sap deletion in B cells. To address this issue adequately, we used a PCR assay to detect deletion of the sap gene at the genomic DNA level (Fig. 2C). T cells or B cells were purified from sapfl/fl;cd4-cre and sapfl/fl;cd19-cre mice, and genomic DNA was tested by PCR. Cell purity was determined to be >98% (data not shown). In sapfl/fl;cd4-cre mice, sap was deleted in T cells (Fig. 2C Upper, lane 2). There was weak (<5%) deletion in the B cell preparation (Fig. 2C Upper, lane 6), possibly caused by contaminating T cells. This finding confirmed that the cd4-cre transgene was active in T cells, but not in most, if not all, B cells. By opposition, sapfl/fl;cd19-cre mice exhibited efficient sap deletion (>90%) in B cells (Fig. 2C Upper, lane 7). Together, these results supported the idea that cd19-cre was highly effective in B cells, albeit not in T cells.
Antibody Production in Conditionally Targeted sapfl/fl Mice.
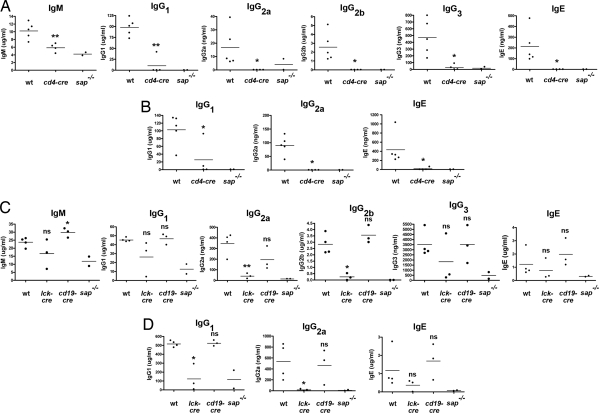
SAP-deficient mice exhibit a marked impairment of antibody production after immunization with antigens such as trinitrophenyl (TNP)-chicken gamma globulin (CGG) or nitrophenyl (NP)-keyhole limpet hemocyanin (3, 11, 14, 17). Thus, mice were immunized with TNP-CGG in the presence of alum, and high-affinity TNP-specific antibodies were measured in the serum by ELISA (Fig. 3 and SI Fig. 6). sapfl/fl;cd4-cre mice, in which sap was deleted in T cells but not in B cells or NK cells, exhibited a marked antibody defect in response to TNP-CGG, both after primary immunization (Fig. 3A) and after secondary immunization (Fig. 3B). The defect was seen for essentially all Ig classes and isotypes, namely IgE, IgG1, IgG2a, IgG2b, and IgG3, and at all time points after immunization (7, 14, and 21 days; Fig. 3A and data not shown). It was essentially of the same magnitude as that seen in conventional SAP-deficient animals. Likewise, sapfl/fl;lck-cre mice, in which sap was deleted in T cells and some NK cells, had a severe antibody production defect (Fig. 3 C and D). However, whereas most sapfl/fl;lck-cre mice had a defect analogous to that seen in conventional SAP-deficient mice, some were able to mount a partial antibody response. We presume that this observation was caused by partial deletion of sap gene in these mice, a phenomenon occasionally noted with lck-cre but not in cd4-cre or cd19-cre mice (data not shown). Contrary to sapfl/fl;cd4-cre and sapfl/fl;lck-cre, the sapfl/fl;cd19-cre mice, in which sap was deleted in B cells, although not in T cells or NK cells, had no defect in antibody production (Fig. 3 C and D and SI Fig. 6). This result was true for primary (Fig. 3C and SI Fig. 6) or secondary (Fig. 3D) immunization. Together, these data implied that antibody production in response to protein antigens necessitate the expression of SAP in T cells, but not in B cells or NK cells.
Fig. 3.
Antibody responses in conditionally targeted sapfl/fl mice. (A) Primary immunization of sapfl/fl;cd4-cre mice. sapfl/fl;cd4-cre (cd4-cre) mice were immunized with TNP-CGG in the presence of alum, as outlined in Methods. After 14 days, anti-TNP-specific antibodies were measured in the serum by ELISA. WT and conventional SAP-deficient (sap−/−) mice were used as controls. Values for individual mice are shown as dots, and the mean of all values is represented as a horizontal line. Similar results were obtained when antibody titers were measured 7 or 21 days after immunization (data not shown). Similar results were seen with at least six mice in each group (data not shown). P values (compared with WT mice) are: *, < 0.05; **, < 0.01; ns, not significant (> 0.05). No anti-TNP antibodies were detected in nonimmunized (naïve) mice (data not shown). (B) Secondary immunization of sapfl/fl;cd4-cre mice. The immunized mice shown in A were boosted by a second injection of TNP-CGG, in the absence of any adjuvant. Antibody titers were measured after 7 days. Similar results were seen with at least six mice in each group (data not shown). P values (compared with WT mice) are: *, < 0.05; **, < 0.01; ns, not significant (> 0.05). (C) Primary immunization of sapfl/fl;lck-cre and sapfl/fl;cd19-cre mice. sapfl/fl;lck-cre (lck-cre) and sapfl/fl;cd19-cre (cd19-cre) mice were immunized and tested as described for A. Similar results were seen with at least six mice in each group (SI Fig. 6; data not shown). P values (compared with WT mice) are: *, < 0.05; **, < 0.01; ns, not significant (> 0.05). No anti-TNP antibodies were detected in nonimmunized (naïve) mice (data not shown). (D) Secondary immunization of sapfl/fl;lck-cre and sapfl/fl;cd19-cre mice. The immunized mice depicted in C were boosted by a second injection of TNP-CGG, in the absence of any adjuvant. Antibody titers were measured after 7 days. P values (compared with WT mice) are: *, < 0.05; **, < 0.01; ns, not significant (> 0.05).
GC Formation in Conditionally Targeted sapfl/fl Mice.
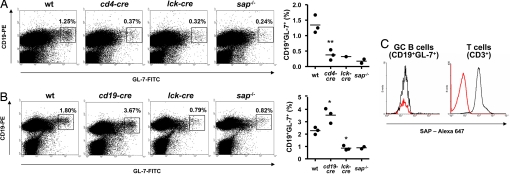
sap−/− mice also exhibit a pronounced defect in the development of GC B cells, a B cell subset implicated in T cell-dependent antibody production (3, 14, 17). To assess whether GC B cell development was altered in sapfl/fl mice, animals were immunized by i.p. injection of sheep red blood cells (SRBCs). Nine days later, GC B cells were detected in spleen by staining with anti-CD19 and anti-GL-7 antibodies (Fig. 4 and SI Fig. 7). GC B cells are CD19+GL-7+ (3, 26).
Fig. 4.
GC formation in conditionally targeted sapfl/fl mice. (A) GC formation in sapfl/fl;cd4-cre mice. sapfl/fl;cd4-cre (cd4-cre) mice were immunized with SRBCs. After 9 days, splenic GC B cells were detected by staining with anti-CD19 and anti-GL-7 antibodies. WT and conventional SAP-deficient (sap−/−) mice were used as controls. One sapfl/fl;lck-cre (lck-cre) was also included in the experiment and is shown in the analysis. (Left) Dot plots of representative mice. GC B cells are boxed and the percentage of CD19+GL-7+ cells is indicated at the top right of each dot plot. (Right) A schematic representation of the values obtained for the various mice is included. Values for individual mice are shown as dots, and the mean of all values is represented by a horizontal line. Similar results were seen with at least five mice in each group (SI Fig. 7; data not shown). P values (compared with WT mice) are: *, < 0.05; **, < 0.01; ns, not significant (> 0.05). In nonimmunized (naïve) mice, the proportions of CD19+GL-7+ cells were between 0.3% and 0.4% and did not differ between the various genotypes (data not shown). (B) GC formation in sapfl/fl;cd19-cre and sapfl/fl;lck-cre mice. This experiment was performed as detailed for A, except that sapfl/fl;cd19-cre (cd19-cre) and sapfl/fl;lck-cre (lck-cre) mice were studied. Similar findings were made with 11 sapfl/fl;cd19-cre mice (data not shown). P values are: *, < 0.05; **, < 0.01; not significant, > 0.05. In nonimmunized (naïve) mice, the proportions of CD19+GL-7+ cells were between 1.0% and 1.1% and did not differ between the various genotypes (data not shown). (C) Detection of SAP expression in GC B cells. WT C57BL/6 mice were injected with SRBCs. After 9 days, spleen cells were isolated and used for multicolor flow cytometry analyses. (Left) The presence of SAP in GC B cells was ascertained by gating on CD19+GL-7+ cells and measuring SAP intracellular staining (black lines). (Right) CD3+ T cells were evaluated as positive control. Cells from SAP-deficient mice were used as negative control (red lines).
Immunization of WT mice with SRBCs resulted in a prominent accumulation of CD19+GL-7+ cells (i.e., GC B cells) in spleen (Fig. 4 A and B and SI Fig. 7). However, there was a marked paucity of these cells in sap−/− mice, in agreement with earlier reports (3, 14, 17). Likewise, a severe defect in GC B cell development was seen in sapfl/fl;cd4-cre (Fig. 4A) and sapfl/fl;lck-cre mice (Fig. 4 A and B). In contrast, it was not observed in sapfl/fl;cd19-cre mice (Fig. 4B and SI Fig. 7). Hence, development of GC B cells in response to immunization required the presence of SAP in T cells, but not in B cells or NK cells.
Considering this notion, we examined by intracellular flow cytometry if SAP was detectable in GC B cells, as had been reported by Morra et al. (17) and Nichols et al. (27) using alternative detection methods (Fig. 4C). This experiment showed that the intensity of staining of CD19+GL-7+ cells from immunized WT mice with anti-SAP antibodies was similar to that seen in the corresponding small population observed in SAP-deficient mice (mean fluorescence values of 19.0 and 14.0, respectively). Although this finding did not exclude the possibility that small amounts of SAP exist in normal GC B cells, it nonetheless implied that these levels would be very low.
Conclusions
The data reported herein yielded two major conclusions. First, they show that SAP expression in T cells is required for a normal humoral immune response. This notion was most convincingly demonstrated by the finding that sapfl/fl;cd4-cre mice, which were devoid of SAP in T cells, but not in B cells or NK cells, faithfully recreated the humoral defects seen in sap−/− mice. It was also consistent with the phenotype of sapfl/fl;lck-cre mice, although one limitation with these mice was that sap was deleted also in some NK cells. At this time, it is not known whether conventional CD4+ T cells, NK-T cells (a population of CD4+ T cells that bears NK cell markers and responds to lipid antigens presented by CD1d) (21), or both are responsible for the SAP-dependent function in humoral immunity. This issue is especially important, as NK-T cells are absent in SAP-deficient mice and can play a role in regulating antibody production (21). Nevertheless, our results suggested that NK-T cells may not be relevant, given that sapfl/fl;cd4-cre mice had a severe humoral defect while exhibiting only partially reduced numbers of NK-T cells. This notion is also in keeping with studies of Fyn-deficient mice (28, 29). Although Fyn-deficient mice lack NK-T cells, they exhibited normal antibody responses and GC formation in response to protein antigens (3, 4). Hence, conventional CD4+ T cells, rather than NK-T cells, are likely implicated in this process.
Second, our results implied that expression of SAP in B cells is not necessary for antibody production and GC formation. This concept stemmed largely from the observation that sapfl/fl;cd19-cre mice, in which the sap gene was deleted only in B cells, had normal humoral immune responses. Furthermore, it was supported by our inability to document that SAP is expressed in B cells, including GC B cells, by using intracellular flow cytometry. The latter result was at odds with earlier reports showing expression of SAP in some GC B cells (17, 27). It also contrasted with a study demonstrating SAP expression in purified (>99%) mouse B cells using protein immunoblot analysis (15). Although the basis for this discrepancy is not known, it is possible that some B cells express small amounts of SAP that were below the sensitivity level of our intracellular flow cytometry assay. Alternatively, it is conceivable that a very small B cell population that was not detected by intracellular flow cytometry expresses SAP. In any case, even if some B cells were to express SAP, our experiments with the conditionally targeted sap allele provided evidence that this expression would not be critical for SAP-dependent humoral immunity in vivo.
The reason, in an earlier study (17), adoptive transfer of SAP-deficient B cells in the presence of normal CD4+ T cells was sufficient to recapitulate the impaired humoral immunity observed in sap−/− mice is uncertain. Morra et al. (17) appear to have taken great care at purifying B cells from nonimmunized SAP-deficient mice, to ensure that naïve non-T cell-primed B cells were used in the adoptive transfer. However, it is possible that even naïve B cells are subjected to SAP-dependent T cell activities in vivo that promote their subsequent capacity, when activated, to produce antibodies. This notion may also explain the reduced Ig secretion and Ig switch recombination in vitro observed by others (15) using purified naïve B cells from SAP-deficient mice. An alternative explanation relates to the use of recombinase-activating gene (RAG)-deficient mice and irradiated mice as adoptive transfer recipients in the previous report (17). It is possible that these experimental settings revealed a cryptic function of SAP expression in B cells. Indeed, immune cells can undergo important changes in their characteristics when placed in a lymphopenic environment or exposed to irradiation. Future studies addressing these possibilities may provide useful information.
The SAP-dependent mechanisms by which CD4+ T cells influence humoral immunity remain nebulous despite intense investigation by several laboratories. CD4+ T cells from SAP-deficient mice exhibited a marked defect in TH2 cytokine production in vitro, suggesting a possible basis for this function (6, 7, 9, 11, 12). However, correction of this anomaly did not appear to alleviate the humoral deficit in vivo, implying that it may not play a central role in the humoral dysfunction (3). In one report, SAP-deficient CD4+ T cells also had altered expression of ICOS and CD40L, two receptors necessary for the aptitude of T cells to promote antibody production by B cells (3). Defects in ICOS expression were also documented in CD4+ T cells from XLP patients (9). Hence, SAP may function by regulating the expression of cell surface molecules critical for productive T cell–B cell interactions. Lastly, it is conceivable that SAP stimulates other T cell functions such as homing or migration in GCs, adhesion to B cells, or secretion of other cytokines regulating B cell proliferation or maturation.
In summary, we provide compelling evidence in mice that expression of SAP in T cells, but not in B cells, is required and sufficient for the aptitude of SAP to promote humoral immunity. In addition to improving our understanding of the role of SAP in normal immunity, these results help explain the severe defects in antibody production and memory B cell generation observed in patients suffering from XLP.
Methods
Mice.
The conditionally targeted allele of sap was generated by inserting loxP sites on either side of exon 2 of sap. Based on previous data (8), deletion of this loxP site-flanked fragment by the Cre recombinase was expected to produce a null allele. Once obtained, female sapfl/+ mice were bred to C57BL/6 mice for five to seven generations, and subsequently crossed with transgenic mice expressing the Cre recombinase under the control of the cd4 promoter (Taconic) (18), lck promoter (Taconic) (18), or cd19 promoter (Artemis Pharmaceuticals) (19). In our functional studies, two negative control (WT) mice were used: sap+/+;cre+ and sapfl/fl;cre−. Both exhibited immune responses that were comparable with those of C57BL/6 mice. All mouse experimentation was approved by the Institut de Recherches Cliniques de Montréal Animal Ethics Committee and done according to the guidelines of the Canadian Council for Animal Care. Additional details are provided in SI Text.
Cells, Antibodies, and Flow Cytometry.
For intracellular staining of SAP, anti-SAP mAb 12C4 (30) was coupled to Alexa 647 (Molecular Probes/Invitrogen), as detailed by the manufacturer. Anti-EAT-2 mAb 15G3 (30) coupled to Alexa 647 was used as control in some experiments. These antibodies were used to stain cells previously fixed and permeabilized with BD Cytofix/Cytoperm buffer (BD Biosciences). After staining, cells were washed with BD Perm/Wash buffer (BD Biosciences). The expression of SAP in a given cell population was determined by gating cells expressing the appropriate surface markers. Additional details are provided in SI Text.
Antibody Responses.
For primary immunization, naïve mice (6–12 weeks old) were injected i.p. with TNP(12)-CGG (100 μg in 100 μl of PBS; Biosearch Technologies), in the presence of an equivalent volume of alum (Pierce). For secondary immunization, previously immunized mice were injected i.p. with TNP(12)-CGG (100 μg) in the absence of adjuvant. In both cases, serum was collected weekly after immunization. High-affinity TNP-specific antibodies were measured by ELISA using serial dilutions of mouse serum and TNP(3)-BSA (Biosearch Technologies) as capture antigen. IgM, IgE, and the various classes of IgG were detected by using the SBA Clonotyping System/HRP kit from Southern Biotechnology Associates or anti-mouse IgE mAb R35-118 (BD Biosciences). IgM, IgE, and isotype-specific IgG standards were obtained from Southern Biotechnology Associates.
GC Formation.
Mice (6–12 weeks old) were injected i.p. with SRBCs (200 μl of 10% suspension in PBS; Cappel MP Biomedicals). After 9 days, they were killed, and GC B cells were detected in spleen by flow cytometry using anti-CD19 and anti-GL-7 antibodies. GC B cells are CD19+GL-7+ (3, 26).
Statistics.
Statistical significance was determined by using unpaired, two-tailed Student's t tests.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Sylvain Latour for critical reading of the manuscript and Luo Yin (International Agency for Research on Cancer, Lyon, France) for providing the conventional SAP-deficient mouse. This work was supported by grants from the Canadian Institutes of Health Research, the National Cancer Institute of Canada, and the Howard Hughes Medical Institute (to A.V.). A.V. holds the Canada Research Chair in Signaling in the Immune System and is a Howard Hughes Medical Institute International Scholar. Z.D. is supported in part by a Pizzagalli Fellowship from the Institut de Recherches Cliniques de Montréal. M.-C.Z. held a fellowship from the Canadian Institutes of Health Research-funded Institut de Recherches Cliniques de Montréal Training Program in Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710698105/DC1.
References
- 1.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 2.Ma CS, Nichols KE, Tangye SG. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol. 2007;25:337–379. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- 3.Cannons JL, et al. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med. 2006;203:1551–1565. doi: 10.1084/jem.20052097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCausland MM, et al. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J Immunol. 2007;178:817–828. doi: 10.4049/jimmunol.178.2.817. [DOI] [PubMed] [Google Scholar]
- 5.Morra M, et al. X-linked lymphoproliferative disease: A progressive immunodeficiency. Annu Rev Immunol. 2001;19:657–682. doi: 10.1146/annurev.immunol.19.1.657. [DOI] [PubMed] [Google Scholar]
- 6.Wu C, et al. SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nat Immunol. 2001;2:410–414. doi: 10.1038/87713. [DOI] [PubMed] [Google Scholar]
- 7.Czar MJ, et al. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci USA. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin L, et al. Mice deficient in the X-linked lymphoproliferative disease gene sap exhibit increased susceptibility to murine gammaherpesvirus-68 and hypo-gammaglobulinemia. J Med Virol. 2003;71:446–455. doi: 10.1002/jmv.10504. [DOI] [PubMed] [Google Scholar]
- 9.Ma CS, et al. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J Clin Invest. 2005;115:1049–1059. doi: 10.1172/JCI23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma CS, et al. Selective generation of functional somatically mutated IgM+CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J Clin Invest. 2006;116:322–333. doi: 10.1172/JCI25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson D, et al. Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in T(H)2 cytokine regulation. Immunity. 2004;21:707–717. doi: 10.1016/j.immuni.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Cannons JL, et al. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. SAP is required for generating long-term humoral immunity. Nature. 2003;421:282–287. doi: 10.1038/nature01318. [DOI] [PubMed] [Google Scholar]
- 14.Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL. SH2D1A regulates T-dependent humoral autoimmunity. J Exp Med. 2004;200:261–266. doi: 10.1084/jem.20040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al Alem U, et al. Impaired Ig class switch in mice deficient for the X-linked lymphoproliferative disease gene Sap. Blood. 2005;106:2069–2075. doi: 10.1182/blood-2004-07-2731. [DOI] [PubMed] [Google Scholar]
- 16.Kamperschroer C, Dibble JP, Meents DL, Schwartzberg PL, Swain SL. SAP is required for Th cell function and for immunity to influenza. J Immunol. 2006;177:5317–5327. doi: 10.4049/jimmunol.177.8.5317. [DOI] [PubMed] [Google Scholar]
- 17.Morra M, et al. Defective B cell responses in the absence of SH2D1A. Proc Natl Acad Sci USA. 2005;102:4819–4823. doi: 10.1073/pnas.0408681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 19.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommers CL, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002;296:2040–2043. doi: 10.1126/science.1069066. [DOI] [PubMed] [Google Scholar]
- 21.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 22.Nichols KE, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasquier B, et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 25.Olszowy MW, Leuchtmann PL, Veillette A, Shaw AS. Comparison of p56lck and p59fyn protein expression in thymocyte subsets, peripheral T cells, NK cells, and lymphoid cell lines. J Immunol. 1995;155:4236–4240. [PubMed] [Google Scholar]
- 26.Naito Y, et al. Germinal center marker GL7 probes activation-dependent repression of N-glycolylneuraminic acid, a sialic acid species involved in the negative modulation of B-cell activation. Mol Cell Biol. 2007;27:3008–3022. doi: 10.1128/MCB.02047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols KE, et al. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc Natl Acad Sci USA. 1998;95:13765–13770. doi: 10.1073/pnas.95.23.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn- deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- 30.Roncagalli R, et al. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nat Immunol. 2005;6:1002–1010. doi: 10.1038/ni1242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.