Abstract
The pandemic influenza of 1918 (Spanish flu) killed 21–50 million people globally, including in Iceland, where the characteristics and spread of the epidemic were well documented. It has been postulated that genetic host factors may have contributed to this high mortality. We identified 455 individuals who died of the Spanish flu in Iceland during a 6-week period during the winter of 1918, representing >92% of all fatal domestic cases mentioned by historical accounts. The highest case fatality proportion was 2.8%, and peak excess mortality was 162/100,000/week. Fatality proportions were highest among infants, young adults, and the elderly. A genealogical database was used to study relatedness and relative risk (RR) of the fatal influenza victims and relatives of their unaffected mates. The significance of these RR computations was assessed by drawing samples randomly from the genealogical database matched for age, sex, and geographical distribution. Familial aggregation of fatalities was seen, with RRs for death ranging from 3.75 for first-degree relatives (P < 0.0001) to 1.82 (P = 0.005), 1.12 (P = 0.252), and 1.47 (P = 0.0001) for second- to fourth-degree relatives of fatal influenza victims, respectively. The RRs within the families of unaffected mates of fatal influenza victims were 2.95 (P < 0.0001), 1.27 (P = 0.267), 1.35 (P = 0.04), and 1.42 (P = 0.001), for first- to fourth-degree relatives, respectively. In conclusion, the risk of death from the Spanish flu was similar within families of patients who succumbed to the illness and within families of their mates who survived. Our data do not provide conclusive evidence for the role of genetic factors in susceptibility to the Spanish flu.
Keywords: heritability, host factors, medical history, mortality, transmission
Influenza pandemics have occurred at least two to three times each century since the sixteenth century. Another pandemic of influenza is therefore considered to be inevitable (1). In the twentieth century, three influenza A pandemics occurred, the “Spanish influenza” caused by an H1N1 virus in 1918, the “Asian flu” caused by H2N2 in 1957, and the “Hong Kong flu” caused by H3N2 in 1968. Of these, the 1918 influenza virus was by far the most virulent, killing 21–50 million individuals globally (2). Recent molecular studies suggest that the 1918 pandemic strain was an avian-like virus that adapted to humans through gradual mutations (3, 4). At present, new strains of avian influenza bearing the H5N1 surface antigens have reached endemic levels among poultry in Southeast Asia and spread through birds to Europe and Africa. Although human infections with the H5N1 viruses are still rare, the severity and mortality associated with these infections is extremely high (5), although mild disease has also been described (6). Certain similarities between the H1N1 influenza virus of 1918 and the current avian H5N1 influenza virus have prompted governments all over the world to prepare against an influenza pandemic. Although clusters of cases in families are due to common exposures, it has been suggested that, for the H5N1 influenza, other factors, possibly genetic, may affect a host's susceptibility to disease (7).
The exceptionally high case fatality proportion among young adults clearly distinguishes the epidemic pattern of 1918 from subsequent pandemics, rather than an unusually high transmissibility of the influenza strain (8). The ability of the virus to induce severe hemorrhagic pneumonia in healthy young adults suggests that host factors other than immune status may have contributed to the severity of illness and outcome. A particularly intriguing possibility is that the 1918 strain may have exploited a weakness rooted in a relatively common variant(s) in the sequence of the human genome. Although epidemiological data on fatalities due to the Spanish flu have been published (9, 10), the details about the pandemic at the individual or familial level are largely unknown. Reykjavik, the capital of Iceland, was hit by the Spanish flu on October 19, 1918, and details of the epidemic are well described in historical newspaper and medical reports (11, 12). By using epidemiological data sources from 1918 and a nationwide genealogical database, we were able to identify 455 fatal cases of the influenza pandemic. The genealogical database has proven to be a valuable tool for studying familial aggregation of several disorders (13, 14). We used this tool to study familial aggregation of influenza-related deaths during the 1918 pandemic in Iceland in an attempt to discover genetic factors that might have contributed to outcomes from the Spanish flu.
Results
Historical Description of the 1918 Epidemic.
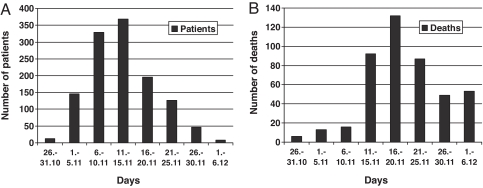
According to health reports, the Spanish flu killed 484 people in Iceland, thereof 476 Icelanders, with an attack rate of at least 63% in Reykjavik (9,016/15,176), and possibly as high as 80–90% (11). The case fatality proportion in Reykjavik was an estimated 2.8% (258/9,016), but lower in other parts of the country (11). A remarkably detailed account of the epidemic was written by a medical practitioner in Reykjavik who personally attended to 1,232 patients during a 40-day period (12). The time distribution of morbidity among his patients is shown in Fig. 1A.
Fig. 1.
Morbidity and fatality from the Spanish flu. (A) Morbidity in Reykjavik, Iceland, from October 28 to December 6, 1918. The number of new patient visits due to the illness is shown (n = 1,232), based on data from ref. 12. (B) Number of fatalities in Iceland from October 26 to December 6, 1918 (n = 448 with known date of death).
Reconstruction of Patient Cohort.
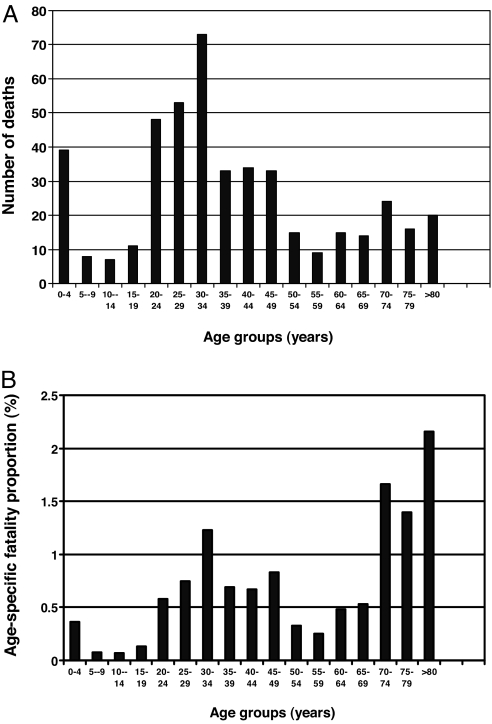
Through the use of relatively stringent criteria we were able to identify 455 individuals who died of influenza during the period October 26 to December 6, 1918. Initially, a list of 521 individuals was compiled on the potential victims of the Spanish flu. This number was lowered by 66 by using exclusion criteria. In comparison, 85 and 86 individuals died during October 26 to December 6, 1917 and 1919, respectively. It can therefore be approximated that of the 455 patients that were included in our familial aggregation analysis, close to 436 died of the Spanish flu, or 92% of all domestic fatal cases. The reconstructed cohort had 207 males and 248 females (1:1.20). The time distribution of fatalities is shown in Fig. 1B, peaking on November 17, 1918, with an excess mortality rate of 162 cases/week/100,000 inhabitants. The age distribution of fatal cases and age-specific mortality is shown in Fig. 2.
Fig. 2.
Age of fatal cases. (A) Age distribution of patients with fatal influenza in Iceland 1918. (B) Age-specific fatality proportion (%) of patients with fatal Spanish flu in Iceland, 1918 (n = 452). Three individuals had an unknown date of birth and are thus not included.
Transmissibility.
Estimates of transmissibility (reproduction ratio, R) based on increasing phases of the epidemic are shown for morbidity and mortality in Table 1. Sensitivity analysis using the entire morbidity epidemic curve yielded an estimate of R = 2.2 (95% C.I. 1.7–2.7). The extreme R was 3.5, based on the maximum log increase in weekly deaths and a serial interval of 4.1 days between symptoms in two successive cases.
Table 1.
Estimates of transmissibility of the Spanish flu epidemic in Iceland, 1918
*Estimates of transmissibility (R) based on the increasing phase of the Spanish flu epidemic (weeks 1–4) in Iceland, 1918.
Geographical Distribution.
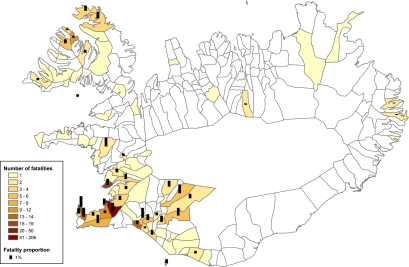
Of the 455 cases of fatal influenza, residency by county and parish could be determined for 437 cases; residency by county only could be determined for an additional 17 cases, leaving only one patient with no residency information. The incidence of fatalities differed notably within the southern and western parts of Iceland, both of which were known to be exposed (11, 12). Relative risk of death was calculated after correcting for residency by dividing Iceland into eight regions, based on percentage of inhabitants who died within the parishes. The number of fatal cases and fatality proportions, given as a percentage within different parishes, are shown in Fig. 3.
Fig. 3.
Geographical distribution by parish of fatalities (n = 437) due to Spanish influenza in Iceland in 1918. Parishes are color-coded to indicate the absolute number or range of fatalities. The black columns show the number of fatalities as a percentage of the total parish population (fatality proportion). For sake of clarity, columns were omitted in cases where there was only one fatality.
Familial Aggregation.
Familial aggregation analysis was performed for the 437 fatal influenza cases where residency could be established up to parish level and for the additional 17 cases where residency could be established up to county level. For both cohorts, familial aggregation analysis for the unaffected mates was also performed to help distinguish the impact of genetic factors from common exposure to infection in the household. Table 2 shows the risk of influenza fatalities for relatives of the 437 fatal influenza cases and the relatives of their unaffected mates. As shown, for almost all of these pairwise comparisons the risk of death fell within the 95% confidence intervals. When testing whether there was a difference in relative risk between relatives of affected victims and relatives of unaffected mates of the affected victims, we did find a nominally significant difference for parents (P = 0.026) and fathers (P = 0.016), in particular. However, this effect was not replicated when looking at more distantly related relatives. The nominally significant effect cannot therefore be considered to be a significant when considering the number of tests being performed. This lead us to conclude that, although we cannot refute that there may be a genetic component in susceptibility to the Spanish flu, the effect is too small to be separated from effects due to cohabilitation.
Table 2.
Familial risk of dying of the Spanish influenza according to relatedness within families in Iceland, 1918
| Relatives | RR affected, 95% C.I. | P value | RR unaffected mates, 95% C.I. | P value | P value affected RR vs. mate RR |
|---|---|---|---|---|---|
| Mates | 9.72 (5.23–16.95) | <0.001 | N/A | 0 | |
| Parents | 5.36 (3.15–10.99) | <0.001 | 0.79 (0.05–3.57) | 0.514 | 0.026 |
| Mother | 6.71 (3.68–12.89) | <0.001 | 1.21 (0.03–4.41) | 0.414 | 0.056 |
| Father | 3.54 (1.30–8.30) | 0.012 | 0.00 (0.00–1.00) | 0.211 | 0.016 |
| Children | 4.70 (2.74–10.03) | <0.001 | 5.23 (2.80–10.81) | <0.001 | 0.619 |
| All siblings | 2.37 (1.30–3.77) | 0.004 | 1.92 (1.07–3.31) | 0.017 | 0.253 |
| Full siblings | 2.83 (1.53–4.84) | 0.001 | 1.87 (0.93–3.63) | 0.036 | 0.177 |
| Maternal siblings | 2.66 (1.48–4.04) | 0.001 | 1.93 (1.04–3.75) | 0.022 | 0.225 |
| Paternal siblings | 2.50 (1.35–3.89) | 0.003 | 1.87 (0.98–3.69) | 0.027 | 0.202 |
| Half-siblings | 0.00 (0.00–1.00) | 0.568 | 2.19 (0.47–6.15) | 0.108 | 0.712 |
| Grandparents | 5.17 (2.32–10.72) | 0.002 | 0.00 (0.00–1.00) | 0.206 | 0.064 |
| Aunts and uncles | 1.68 (0.96–2.86) | 0.031 | 0.26 (0.00–2.29) | 0.972 | 0.047 |
| Nephews and nieces | 1.57 (0.83–2.61) | 0.077 | 1.35 (0.56–2.50) | 0.221 | 0.377 |
| Cousins | 1.06 (0.69–1.65) | 0.348 | 1.35 (0.90–1.99) | 0.081 | 0.744 |
| Relatives degree 1 | 3.75 (2.53–5.24) | <0.001 | 2.95 (2.01–4.49) | <0.001 | 0.198 |
| Relatives degree 2 | 1.82 (1.13–2.75) | 0.005 | 1.27 (0.65–1.95) | 0.267 | 0.121 |
| Relatives degree 3 | 1.12 (0.78–1.60) | 0.252 | 1.35 (0.97–1.81) | 0.04 | 0.735 |
| Relatives degree 4 | 1.47 (1.16–1.81) | 0.001 | 1.42 (1.11–1.74) | 0.001 | 0.411 |
| Relatives degree 5 | 1.16 (1.00–1.35) | 0.026 | 1.27 (1.03–1.44) | 0.012 | 0.837 |
| Relatives degrees 1 and 2 combined | 2.68 (1.99–3.45) | <0.001 | 2.02 (1.42–2.74) | <0.001 | 0.071 |
| Relatives degrees 3–5 combined | 1.24 (1.09–1.43) | <0.001 | 1.34 (1.14–1.50) | 0.057 | 0.747 |
Results for families of 437 affected patients are shown in the first column. Results for the families of unaffected mates (n = 218) of fatal influenza victims are shown in the third column. Comparison between the two groups is given in the fifth column. RR, relative risk; 95% C.I., 95% confidence intervals; N/A, not applicable.
Comparable results were obtained when we corrected for residency by dividing the country into three groups according to fatality proportions within counties (data not shown).
Discussion
We used historical sources and the deCODE Genetics genealogical database to identify cases of fatal influenza in Iceland during the Spanish flu epidemic of 1918. We feel that under such extreme circumstances, using death within a defined period as a surrogate for fatal influenza gives more accurate information about the impact of this illness than traditional medical records. Because the duration of the epidemic was very short, possibly because of the lack of public health measures in Reykjavik during the early stages, we were able to minimize the probability of including deaths from other causes by using a relatively short period (42 days), which corresponds well with historical descriptions of the epidemic. Most of the deaths occurred at home and diagnostic capabilities, other than history and physical examination, were virtually nonexistent. To reconstruct the cohort of fatal cases, all deaths of other known causes were excluded. Hence, the distribution of fatal cases in our study matches extremely well with historical descriptions (11).
One of the unique features of the Spanish flu epidemic was the so-called W-shaped mortality curve, with clear peaks among young children, young adults, and the elderly. Our study shows a similar W pattern. The cause of high mortality among young healthy people remains enigmatic. It is not thought that the 1918 virus was more easily transmitted than other influenza viruses. In the United States, it has been shown that the transmissibility (R) of the 1918 virus, was 2.0, with a median extreme R of 2.6 (8). This indicates that, on average, there were two to three secondary cases for each primary case, which is not particularly high. According to our calculations based on both morbidity and mortality data, the R of the influenza virus in Iceland was 1.9–2.0, with an extreme R of 3.5, suggesting that the epidemic had similar characteristics in Iceland as in the United States and England (15). These similarities also suggest that our study on transmission within families may have implications beyond Iceland.
Until recently, no isolates of the 1918 virus were available for direct analysis. Studies by Taubenberger and colleagues suggest that the virus was more closely related to avian influenza viruses than influenza from any other species, indicating that humans became infected by an avian virus that subsequently gained the ability to cause human-to-human transmission (3). The reconstructed 1918 virus has been shown to be unusually virulent in animal models of infection, through synergy among all viral genes (4). High viral burden in both upper and lower respiratory tracts with corresponding dysregulation of the antiviral responses are correlated with fatal outcome (16). Host range restriction is mediated in part through the hemagglutinin (HA) protein that mediates binding of the virus to sialic acid-containing cell surface molecules (17). Human influenza viruses prefer sialic acid linked to galactose by α-2,6 linkage, whereas avian viruses prefer α-2,3 linkage. The α-2,3 receptors for H5N1 are found deep in the respiratory tract of humans (18), which may explain the current low level of transmission. However, variation in both expression and distribution of these receptors could play a role in determining susceptibility of humans to these pathogens. In subsequent steps, the viruses must be able to use human enzyme systems to replicate efficiently. Recent data suggest that pathogenicity of both the 1918 H1N1 virus and the H5N1 virus is mediated in part by their ability to replicate to extremely high numbers in respiratory epithelia and even blood, with an accompanying “cytokine storm,” which correlates well with fatal outcome (19, 20). Evasion from the immune system may also be an important virulence determinant, because the H5N1 virus seems most likely to evade or suppress the immune responses, most likely through the NS1 protein, which renders it resistant to the antiviral effects of interferons and TNFα (21). In addition, recent epidemiological data on H5H1 infections in Indonesia have shown a high percentage of clusters among blood relatives within families, which is compatible with a possible genetic susceptibility to H5N1 influenza (22).
The ability of the 1918 pandemic virus to induce severe hemorrhagic pneumonia, particularly in otherwise healthy young adults, thus raises the question of whether host genetic factors may have determined the severity of the Spanish flu. We attempted to address this question by studying familial aggregation of fatal infections by using the unique information sources in Iceland from the epidemic of 1918. Our data allowed us to examine the clustering of fatalities within families and calculate the risk of death for relatives of the victims and their mates. When the datasets were adjusted for age, sex, and residence, the risk of dying from the Spanish flu clearly depended on relatedness within families. However, the importance of the environmental factor is underlined by the high relative risk for mates of influenza victims. As a result, we attempted to correct for this factor by performing identical calculations for the families of unaffected mates, which yielded results that were similar to those from relatives of patients. The effect for parents and fathers shown in Table 2 is unlikely to be significant when taking the number of comparisons into account. Thus, our study suggests that, under the circumstances prevailing in 1918, genetic diversity did not contribute much to differences in the clinical course of the Spanish flu.
One potential explanation for our results is that crowding or greater proximity to highly infectious patients was associated with influenza of greater severity, leading to death. We propose that these effects could be mediated through higher inocula or infectious burden of the virus at the onset. Similarly, it has been suggested that, in the case of measles, crowding and the exposure to a high dose of a pathogen may increase the case fatality proportion and is more important than the nutritional status and genetically determined susceptibility (23). A recent study from two parishes in Norway supports this hypothesis, because it shows that residence in smaller apartments was associated with greater risk of dying of the 1918 influenza (24). We are not aware of any studies in humans on the association between varying doses of influenza viruses and disease severity.
Although it could be argued that the high mortality in the 1918 pandemic may be explained by the low or even nonexistent level of immunity within the population, it would not explain the unusually high severity in young adults, which remains enigmatic. It can be speculated that exposure to a prior pandemic, possibly in 1890, could have conferred immunity against the Spanish flu, which could explain in part the unusual W-shaped mortality curve. We corrected for this potential confounder by using age-matched categories for calculations of risk ratios. Another limitation of our study could be due to the uneven spread of the epidemic through the country, in part because of regional differences in public health measures, as has been shown in the United States (25, 26). Reykjavik, the capital, was hardest hit, but the entire northern and eastern parts of the country were spared, most likely because of aggressive quarantine measures, which were remarkably effective. It has been shown that the population structure in Reykjavik in 1918 was a mixture of people from all parts of the country (27). In addition, we corrected for residence in the familial risk calculations by matching cases and population controls within geographical regions defined by fatality proportions and thus corrected for this confounder. Last, the impact of an early wave of influenza, such as the one described for New York City (10) could, in theory, confer immunity to a fraction of the population and confound our results. Health records report that an influenza-like illness outbreak hit Iceland in July 1918, causing mild symptoms and no excess mortality (11). According to the same sources, infection by this agent did not systematically confer immunity against the subsequent Spanish flu and is therefore unlikely to have an impact on our results.
In summary, we have demonstrated that the Spanish influenza of 1918 carried an elevated risk of fatalities within families of Spanish flu victims and a similar risk pattern was seen among relatives of unaffected mates. Therefore, our data do not provide conclusive evidence for the role of genetic factors in susceptibility to the Spanish flu. It can be speculated that during early stages of the infection, greater proximity or repeated exposure to infectious patients was the single most important determinant of fatal outcome, possibly through greater infective dose of the virus, resulting in higher viral burden with “cytokine storm” and death.
Materials and Methods
Setting.
Reykjavik, the capital of Iceland, was hit by the second wave of the Spanish influenza on October 19 and 20, respectively, in 1918. The epidemic subsequently spread through the capital and nearby villages through the southern and western part of the island, where ≈64% of the population resided. The northern and eastern parts were spared, in part due to aggressive quarantine. According to official reports, 484 fatalities were registered (11).
The deCODE Genetics Genealogical Database.
Iceland was settled ≈870 AD by a few thousand individuals who originated from Scandinavia and the British Isles (28). Building on a series of national and regional censuses (including the oldest preserved national census in the world from 1703), parish records and additional sources, a population-wide genealogical database has been compiled, which contains information about genealogical links between >95% of all Icelanders who have been born since 1703. More than 715,000 individuals are registered in the database. This tool can therefore be used to look for aggregation of diseases within families, including historical outbreaks.
Definition of Fatal Influenza Cases.
Three sources of information were used to identify fatal cases of Spanish flu in Iceland. First, the deCODE genealogical database was used to identify fatalities occurring from October 26 to December 6,1918 (12). Identical periods in 1917 and 1919 were used as controls to derive baseline mortality in the absence of influenza pandemic. Second, cases were also found by searching through the Icelandic burial registry (accessible at www.gardur.is) and, third, by searching manually through a newspaper “Morgunblaðið” (archives available at http://timarit.is/mbl/), that published a list of fatal Spanish flu victims. The following individuals were excluded from the patient cohort: (i) foreign-born persons, (ii) Icelanders who verifiably died of causes other than influenza, (iii) Icelanders who died in regions of the country not hit by the pandemic, and (iv) Icelanders who died abroad. The case list was encrypted through a process approved by the Data Protection Authority of Iceland (29).
Age-Specific and Excess Mortality.
The Icelandic national population registry was used to calculate age-specific mortality by 5-year intervals, using the age composition of the Icelandic population on December 31, 1917 (total population, 91,368) as a denominator (data available from www.statice.is). Excess mortality for the 1918 epidemic was calculated by subtracting the weekly average mortality during a 42-day period (October 26 to December 6) in 1917 and 1919 from 1918 (identical period) and presented as the number of deaths/week/100,000 inhabitants.
Transmissibility of Influenza.
Estimates of transmissibility (R) based on increasing phase of the epidemic for both morbidity and mortality were performed by using methods described in ref. 15 and a case fatality proportion of 2.8%. Sensitivity analysis using the entire morbidity curve was performed by using the method of Ferrari et al. (30).
Familial Aggregation Analysis.
To evaluate familial aggregation of fatal influenza cases we calculated a relative risk (RR) for close and distant relatives by using methods described in ref. 14. The RR for relatives of patients with fatal influenza in the 1918 pandemic was defined as the risk of fatal influenza among the relatives of individuals who died of influenza, divided by the prevalence of fatal influenza in the general population of Iceland, in the 6-week study period. If PA denotes the event that the proband is affected, RA denotes the event that a relative for a given relation is affected, and P is a probability, then RR for that relation is defined as the ratio P(RA | PA)/P(RA). We estimate this ratio directly in subpopulations defined by region of residency, sex, and year of birth in 5-year intervals, and the overall estimate is derived from the original estimates by using a geometric weighted average, where the weights are defined by the probability that an individual belongs to a given subpopulation (14). In this way, the regional- and age-dependency of mortality from the 1918 pandemic is addressed. To assess the significance of the RR obtained for a given group of patients, we compared their observed values with the RR computed for 1,000 independently drawn and matched groups of control individuals. Each influenza fatality was matched to a single control individual in each control group. The control individuals were drawn at random and matched with patients who died of influenza on year of birth (rounded to 5 years), residency, sex, and number of ancestors in the database five generations back. By using these 1,000 matched controls groups one can get an estimate of the distribution of RR for each relative type under the null hypothesis of there being no increase in RR of influenza deaths. This distribution is then used to report an empirical P value based on how many RR values for the control groups exceed the RR value for the proband. By using a variance-stabilizing square-root transform, an approximate confidence interval for the RR of the relatives may be constructed based on the distribution of RR for the control groups. The RR for relatives of unaffected mates was calculated in the same way. We also report the weighted average RR for individuals belonging to multiple-degree relationship. Here, the RR is computed for each degree relationship independently and then the values are combined by taking a weighted average where the weights are determined by the expected relative size of the degree relationship classes (14). A P value for the null hypothesis that the relative risks in relatives of affected victims and relatives of mates was equal was computed by comparing the difference in variance-stabilized square root transform relative risk values with differences in relative risks computed from RR values of the randomly drawn cohorts.
Mates were defined as individuals that have at least one offspring in common. In instances where individuals had more than one mate, the mate with the youngest offspring was used in the analyses. The geographic location of individuals at the time of the pandemic was obtained by using two major sources, a national census from 1910 and information about residency that is linked to the genealogical database. When information about the residency of individuals before 1918 was not available, the place of burial was used as a proxy; in the absence of this information, the place of birth was used. This approach is justified by the knowledge that migration was limited at this point in Icelandic history (27). Children born after 1910, with no direct information about residency before 1918 or place of birth, were assumed to have the same residency as their mothers.
ACKNOWLEDGMENTS.
We thank Valgeir Thorvaldsson at Vesturfararsetrid, Hofsos, Iceland, for providing information on emigration of Icelanders to North America and Dr. Haraldur Briem, State Epidemiologist, for valuable discussions. This work was supported, in part, by National Institutes of Health/National Institute of Allergy and Infectious Diseases HHSN266200400064C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1109.
References
- 1.Potter CW. In: Textbook of Influenza. Nicholson KG, Webster RG, Hay AJ, editors. Oxford: Blackwell Scientific; 1998. pp. 3–18. [Google Scholar]
- 2.Johnson NP, Mueller J. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 4.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, et al. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 5.Oner A, Bay A, Arslan S, Akdeniz H, Sahin H, Cesur Y, Epcacan S, Yilmaz N, Deger I, Kizilyildiz B, et al. N Engl J Med. 2006;355:2179–2185. doi: 10.1056/NEJMoa060601. [DOI] [PubMed] [Google Scholar]
- 6.Kandun I, Wibisono H, Sedyaningsih E, Yusharmen, Hadisoedarsuno W, Purba W, Santoso H, Septiawati C, Tresnaningsih E, Heriyanto B, et al. N Engl J Med. 2006;355:2186–2194. doi: 10.1056/NEJMoa060930. [DOI] [PubMed] [Google Scholar]
- 7.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, et al. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 8.Mills CE, Robins JM, Lipsitch M. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous. London: His Majesty's Stationery Office; 1920. Supplement to the 81st Annual report of the registrar general: Mortality from influenza in England and Wales during the epidemic of 1918–19; pp. 1–47. [Google Scholar]
- 10.Olson DR, Simonsen L, Edelson PJ, Morse SS. Proc Natl Acad Sci USA. 2005;102:11059–11063. doi: 10.1073/pnas.0408290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anonymous. Heilbrigðisskýrslur, 51–56. Icelandic: 1918. [Google Scholar]
- 12.Thoroddsen TJ. Laeknabladid (Icelandic Med J) Vol. 5. Icelandic: 1919. pp. 13–23.pp. 74–79. [Google Scholar]
- 13.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, Stefansson K. Eur Heart J. 2006;27:708–712. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 14.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, et al. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viboud C, Tam T, Fleming D, Handel A, Miller MA, Simonsen L. Vaccine. 2006;24:6701–6707. doi: 10.1016/j.vaccine.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 16.Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, et al. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 17.Neumann G, Kawaoka Y. Emerg Infect Dis. 2006;12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 19.de Jong MD, Simmons CP, Thanah TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Van Vinh Chau N, Khanh TH, Dong VC, et al. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, Garcia-Sastre A, et al. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo SH, Hoffmann E, Webster RG. Nat Med. 2004;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 22.Sedyaningsih ER, Isfandari S, Setiawaty V, Rifati L, Harun S, Purba W, Imari S, Giriputra S, Blair PJ, Putnam SD, et al. J Infect Dis. 2007;196:522–527. doi: 10.1086/519692. [DOI] [PubMed] [Google Scholar]
- 23.Aaby P, Bukh J, Lisse IM, Smits AJ. J Infect Dis. 1983;147:693–701. doi: 10.1093/infdis/147.4.693. [DOI] [PubMed] [Google Scholar]
- 24.Mamelund SE. Soc Sci Med. 2006;62:923–940. doi: 10.1016/j.socscimed.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 25.Bootsma MCJ, Ferguson N. Proc Natl Acad Sci USA. 2007;104:7588–7593. doi: 10.1073/pnas.0611071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatchett RJ, Mecher CE, Lipsitch M. Proc Natl Acad Sci USA. 2007;104:7582–7587. doi: 10.1073/pnas.0610941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. Nat Genet. 2005;37:90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- 28.Helgason A, Hickey E, Goodacre S, Bosnes V, Stefansson K, Ward R, Sykes B. Am J Hum Genet. 2001;68:723–737. doi: 10.1086/318785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulcher JR, Kristjansson K, Gudbjartsson H, Stefansson K. Eur J Hum Genet. 2000;8:739–742. doi: 10.1038/sj.ejhg.5200530. [DOI] [PubMed] [Google Scholar]
- 30.Ferrari MJ, Bjornstad ON, Dobson AP. Math Biosci. 2005;198:14–26. doi: 10.1016/j.mbs.2005.08.002. [DOI] [PubMed] [Google Scholar]