Abstract
Emerging and re-emerging infectious diseases, especially those caused by drug-resistant bacteria, are a major problem worldwide. Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) appeared rapidly and unexpectedly in the United States, resulting in an epidemic caused primarily by isolates classified as USA300. The evolutionary and molecular underpinnings of this epidemic are poorly understood. Specifically, it is unclear whether there has been clonal emergence of USA300 isolates or evolutionary convergence toward a hypervirulent phenotype resulting in the independent appearance of similar organisms. To definitively resolve this issue and understand the phylogeny of USA300 isolates, we used comparative whole-genome sequencing to analyze 10 USA300 patient isolates from eight states in diverse geographic regions of the United States and multiple types of human infection. Eight of 10 isolates analyzed had very few single nucleotide polymorphisms (SNPs) and thus were closely related, indicating recent diversification rather than convergence. Unexpectedly, 2 of the clonal isolates had significantly reduced mortality in a mouse sepsis model compared with the reference isolate (P = 0.0002), providing strong support to the idea that minimal genetic change in the bacterial genome can have profound effects on virulence. Taken together, our results demonstrate that there has been recent clonal expansion and diversification of a subset of isolates classified as USA300. The findings add an evolutionary dimension to the epidemiology and emergence of USA300 and suggest a similar mechanism for the pandemic occurrence and spread of penicillin-resistant S. aureus (known as phage-type 80/81 S. aureus) in the 1950s.
Keywords: genomics, phylogeny, virulence, single-nucleotide polymorphism
Study of bacterial genome diversification and pathogen emergence has historically relied on strategies that index genomic variation at a relatively small number of nucleotides. Although these strategies are powerful, the fact that members of a bacterial species can differ dramatically in gene content and allelic variation means that analysis of strains at the whole genome level is required to understand important biomedically relevant processes such as epidemics. Whole genome sequencing approaches represent a next generation of studies to address bacterial genome diversification.
One of the most unexpected events in bacterial infectious diseases in recent years has been the rapid emergence of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections, now a leading cause of disease in otherwise healthy individuals (1–3). For example, the majority of all skin and soft tissue infections in patients presenting to emergency departments in the United States are caused by CA-MRSA, primarily by isolates classified as pulsed-field gel electrophoresis (PFGE) type USA300 (4–6). In addition, a recent study by Klevens et al. (7) indicates that USA300 isolates account for the majority (67%) of invasive community MRSA infections, some of which result in death. These and other studies clearly indicate there is an ongoing epidemic of CA-MRSA in the United States (8). The origin of isolates classified as USA300 is not known nor do we understand the extent of genetic diversity or the basis of enhanced virulence in these organisms.
To address this deficiency in knowledge and gain understanding about molecular evolutionary processes contributing to temporal variation in disease frequency, we compared the genome sequence of 10 geographically and clinically diverse isolates classified as USA300, and evaluated strain virulence in a mouse infection model.
Results and Discussion
Analysis of Geographically Diverse S. aureus Isolates Classified as USA300 by PFGE.
As a first step toward understanding the extent of genetic diversity of epidemic community-MRSA, we analyzed 11 isolates classified as USA300 that were isolated from patients with diverse infections and hospitalized at several regions of the U.S. between 2002 and 2005 (Table 1). These strains included community- and healthcare-associated MRSA recovered from patients with diverse clinical manifestations, including skin and soft tissue infections, bacteremia, endocarditis, and necrotizing pneumonia. Organisms were selected largely from a repository of 864 MRSA isolates collected over a 1-year period by the Active Bacterial Core Surveillance system (ABCs) at the Centers for Disease Control and Prevention (CDC) and are representative of major geographic regions of the United States (7). The strain sampling strategy is described in detail in Materials and Methods. Five of the 11 isolates had PFGE profiles that were indistinguishable from one another, suggesting that these organisms are related (Fig. 1). The other 6 isolates differed from the common PFGE type by 1–4 bands (Fig. 1).
Table 1.
USA300 isolates analyzed by comparative genome sequencing
| Isolate | Spa | PFGE* | SCCmec | ACME† | State | Syndrome | Origin‡ | Date | Age§ |
|---|---|---|---|---|---|---|---|---|---|
| FPR3757¶ | 1 | 0114 | IVa | + | California | Abscess | CA | 2003 | 36 |
| LAC‖ | 1 | 0114 | IVa | + | California | Abscess | CA | 2002 | n/a |
| 18805 | 168 | 0114 | IVa | + | Oregon | Endocarditis | CA | 2005 | 54 |
| 18806 | 1 | 0180 | IVa | – | Georgia | Bacteremia | HA | 2005 | 49 |
| 18807 | 1 | 0114 | IVa | + | Colorado | Bacteremia | CA | 2005 | 39 |
| 18808 | 1 | 0047 | IVa | + | New York | Endocarditis | CA | 2005 | 43 |
| 18809 | 1 | 0114 | IVa | + | Minnesota | Bacteremia | CA | 2005 | 63 |
| 18810 | 1 | 0114 | IVa | + | Pennsylvania | Necrotizing pneumonia | CA | 2005 | 21 |
| 18811 | 1 | 0247 | IVa | + | California | Osteomyelitis | HA | 2005 | 94 |
| 18813 | 1 | 0045 | IVb | – | Tennessee | Bacteremia | HA | 2005 | 3 |
| 19321 | 1 | 0120 | IVb | – | Oregon | Pneumonia / bacteremia | HA | 2005 | 50 |
* PFGE subtype.
†ACME, arginine catabolic mobile element.
‡CA, community-associated; HA, healthcare-associated.
§n/a, not available.
¶Strain FPR3757 is the reference strain to which all other isolates were compared.
‖Los Angeles County clone, LAC.
Fig. 1.
Analysis of USA300 isolates by pulsed-field gel electrophoresis (PFGE). PFGE was performed as described in Materials and Methods. Although there is variation among the strains, the PFGE patterns are typical of USA300. FPR3757 is the reference strain. The white line indicates a merge of two separate images.
Comparative Genome Sequencing of USA300 Isolates Indicates Recent Clonal Descent of Epidemic CA-MRSA.
PFGE profiling indexes variation at a relatively small number of base pairs and thus does not determine the extent of single nucleotide polymorphisms (SNPs) and many other molecular events contributing to genetic variation. In principle, similarity in PFGE profile can be caused by different molecular processes, including evolutionary convergence or recent clonal descent from a common ancestor. To differentiate between these two possibilities, we performed comparative genome sequencing (CGS), a process that identifies the full extent of SNPs and non-SNP regions of difference (RDs) in genomic and plasmid DNA. [Fig. 2, and supporting information (SI) Tables 2–7]. Strain FPR3757 (USA300-0114) was used as the reference organism because its genome was recently sequenced by conventional methods (9). Non-SNP insertions, deletions, and transpositions were defined as RDs. All putative SNPs and RDs identified by CGS were evaluated by conventional capillary DNA sequencing, and 91% of the SNPs (526 of 578 SNPs) were confirmed to be true polymorphisms. Compared with the reference strain FPR3757, there was a total of 578 unique SNPs in the core genome (≈2,753 kbp excluding foreign elements) of the 10 USA300 isolates. This frequency (1 SNP in every 4,763 bp) is similar to the SNP frequency of 1 in 6,707 bp for the core metagenome of clonally related serotype M3 strains of group A Streptococcus reported recently (10). Eighteen percent (n = 106) of SNPs in these USA300 isolates were located in intergenic regions and 29% (n = 165) and 53% (n = 307) were synonymous and nonsynonymous nucleotide changes, respectively (SI Table 2 and SI Fig. 6).
Fig. 2.
SNPs in the core genome of USA300 isolates. SNPs were identified by using comparative genome sequencing. Each bar represents a SNP. Green and dark blue vertical lines indicate synonymous and nonsynonymous nucleotide changes, respectively. Light blue indicates a SNP in an intergenic region. The gray shaded regions represent notable genetic elements. Forward (above the line) and reverse (below the line) strand ORFs of the FPR3757 reference strain (Ref.) are illustrated in red at the top of the aligned sequences.
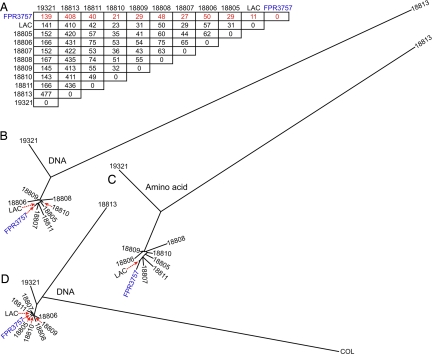
Compared with the reference genome, the number of SNPs in each of the 10 clinical isolates ranged from 11 to 408 SNPs, although 8 of these organisms had a more limited range of SNPs (11 to 48 SNPs) (Fig. 3A). The 8 most closely related isolates differed on average from the reference strain by 32 SNPs and by 50 SNPs from one another. The 2 most divergent isolates (18813 and 19321, Table 1) differed from each other by 477 SNPs, and differed from the reference strain by 408 and 139 SNPs, respectively (Fig. 3A).
Fig. 3.
SNP matrix and SNP-based phylogenetic analysis. (A) SNP matrix. Red font indicates number of SNPs compared with reference strain FPR3757. (B) Phylogenetic analysis of the 11 USA300 isolates is based upon 578 concatenated core genomic SNPs and includes SNPs present in intergenic regions. (C) Phylogenetic analysis using 472 concatenated amino acid residues containing SNPs, synonymous, and nonsynonymous. (D) Phylogenetic analysis of USA300 isolates and strain COL is based upon 1,063 concatenated core genomic SNPs.
To determine overall genetic relationships among the core chromosome of these 11 isolates, we next conducted SNP-based phylogenetic analysis. As implied by the SNP data, 9 of the 11 isolates are closely related, whereas 18813 and 19321 are more divergent (Fig. 3 B and C). Moreover, these 2 isolates were more similar to S. aureus strain COL, another reference strain whose genome also has been sequenced (11). These results suggest that COL, 18813, and 19321 diverged from a common ancestor before the genetic diversification events characterizing the cluster of USA300 isolates (Fig. 3D).
Importantly we found a relatively high overall ratio of nonsynonymous-to-synonymous SNPs (1.9:1) in the 10 query genomes relative to FPR3757, indicating that the SNP diversity in these strains has accumulated recently (12, 13). Notably, the ratio of nonsynonymous to synonymous SNPs among the 8 most closely related isolates was 2.6:1, a value far greater than the ratio of 1.8:1 between divergent isolates 18813 and 19321. The only reasonable interpretation of these findings is that there has been very recent clonal expansion and geographic dissemination of the nine most closely related isolates (8 query organisms plus FPR3757). These results rule out the formal hypothesis of convergent evolution.
Frequency of Core Genome SNPs Present in Genes Encoding Molecules Associated with Virulence.
When considering only the core genome, we found 472 SNPs in 350 unique ORFs (i.e., excluding intergenic regions) of the 10 isolates. These SNPs were distributed widely among ORFs assigned to many functional categories based upon Clusters of Orthologous Groups of proteins (COGs). Many of these 472 SNPs were present in more than one isolate, consistent with divergence from a common ancestor (SI Table 2). However, only 16 nonsynonymous SNPs were present in genes encoding proven or putative S. aureus virulence determinants, including exotoxin 7, clumping factor A and B (clfA and clfB), alpha-hemolysin (hla), gamma-hemolysin (hlgC), serine and cysteine proteases (splF, sspA and sspB), and fibronectin binding protein A and B (fnbA and fnbB) (SI Table 2). These genes encode proteins that participate in important pathogenesis processes such as adhesion, host cell lysis, and proteolysis. Isolate 18811 contained a missense mutation in agrA that lies within the putative DNA binding domain of the encoded protein, thereby potentially altering function of the accessory gene regulator (Agr) (SI Table 2). Agr is a major regulator of S. aureus virulence and controls production and secretion of virulence factors, including hemolysins, adhesions, and leukotoxins (14).
The occurrence of multiple nonsynonymous SNPs present in a single ORF can be a signature of elevated selective pressure or reflect a premium on diversification. Among all strains studied, 57 ORFs had more than one SNP, including cap5E (encoding capsular polysaccharide biosynthesis protein 5E), fnbB, sdrC and sdrD (encoding adhesins), secF, tagX (encoding teichoic acid biosynthesis protein X), and several cell surface or membrane proteins of unknown function (SI Table 2). Most of these SNPs occurred in isolate 18813 (SI Table 2).
SNPs Present in Foreign Mobile Genetic Elements Support Recent Clonal Expansion of a Subset of Isolates Classified as USA300.
Many genes contributing to S. aureus virulence or antibiotic resistance are encoded by mobile elements, such as insertion sequences, transposons, plasmids, prophages, and genomic islands (9, 11, 15–17). In addition to SNPs in the core genome, we identified 365 SNPs in apparently mobile elements, including the SCCmec IV cassette (encoding the methicillin resistance genes), arginine catabolic mobile element (ACME), staphylococcal pathogenicity island 5 (SaPI5), prophages φSa2usa and φSa3usa, and plasmids pUSA01 and pUSA03 (SI Table 3). The two most genetically divergent isolates (19321 and 18813) had the greatest number of SNPs in mobile elements (83 and 262 of the 365 SNPs, respectively), compared with only 5–20 SNPs in all other organisms (some SNPs are present in more than one isolate) (SI Table 3). The ratio of nonsynonymous to synonymous SNPs in mobile elements was 1:1.7 for isolates 18813 and 19321 (relative to the reference strain), and this ratio was much different for the other isolates (1:0.9). Collectively, these findings are in accordance with the analysis of SNPs present in the core genome, which indicate earlier divergence of 18813 and 19321 followed by subsequent clonal expansion and diversification of isolates 18805–18811, LAC, and FPR3757.
Identification of Non-SNP Polymorphisms in USA300 Isolates.
Compared with FPR3757, there were 17 unique RDs (excluding nonsense mutations, SI Table 4) among the 10 USA300 query isolates, mapping to seven loci and two intergenic regions within the core genome and 8 chromosomal RDs in foreign mobile elements (SI Tables 5 and 6 and SI Fig 7). There were 36 additional RDs identified in plasmids by CGS, but these RDs could not be confirmed by targeted conventional capillary DNA sequencing (SI Table 7). Consistent with data reported for group A Streptococcus (10), there was a high level of false-positive calls among the RDs (≈63%). This phenomenon may be due to segments of repetitive DNA, low DNA melting temperature, or DNA secondary structure. Three RDs were deletions within or encompassing ACME in isolates 18806, 18813, and 19321 (SI Table 6). Among S. aureus, ACME is primarily associated with isolates classified as USA300 and may promote survival within the host (9, 11, 15–17). Although 18806 lacks ACME, its core genome clearly places it among organisms involved in the recent clonal expansion of USA300 strains (Fig. 3B). In addition, 18806 has a SCCmec subtype IVa mecA gene versus subtype IVb in 18813 and 19321 (Table 1). Isolate 18813 had the greatest number of non-SNP RDs (6 of the 17 total) and PCR analysis confirmed that the ACME and φSA2usa were absent in this isolate. Isolate 18805 had a 227-bp deletion that would produce a frameshift in SAUSA300_2589, a gene encoding a secreted protein that has an LPXTG-cell wall anchor peptide (SI Table 5). It is also notable that this gene is juxtaposed to secY, which regulates secretion of exported proteins.
Allelic Variation among USA300 Isolates Significantly Affects Virulence.
The restricted allelic variation identified in the core genome of USA300 isolates suggested there would be little or no difference in virulence among these organisms. However, there is an emerging literature that modest genetic changes such as SNPs can dramatically alter pathogen-host interaction and strain virulence (10, 18, 19). Understanding whether differences exist in virulence among clonally related organisms is a critical issue as the population size of USA300 isolates expands and genetic diversification increases. We used a mouse sepsis/bacteremia model to test the hypothesis that the 11 isolates we analyzed differed in virulence (Fig. 4). Consistent with this hypothesis, our analysis revealed significant variance in virulence among these 11 isolates (Fig. 4). Survival was significantly greater for animals infected with isolates 18805 and 18811 compared with the other 9 isolates (Fig. 4). Remarkably, the core genome of 18805 and 18811 was relatively similar to those of the most virulent isolates, differing from the reference strain (FPR3757) by only 29 and 40 SNPs, respectively (Fig. 3A). There were a limited number of nonsynonymous SNPs or other polymorphisms unique to these two isolates, including those in the core genome and mobile elements excluding plasmids (7 and 14 SNPs in 18805 and 18811, respectively; and 2–3 non-SNP deletions in each isolate) (SI Tables 2, 3, and 5). None of the polymorphisms in 18805 were within putative or proven virulence factors, whereas 18811 had nonsynonymous SNPs in genes encoding AgrA and VraF (SAUSA300_0647), each of which could negatively impact virulence (SI Table 2).
Fig. 4.
Virulence of S. aureus isolates in a mouse sepsis model. Strain-to-strain variation in virulence. Survival after infection (5 × 107 CFU, tail vein) was determined as described in Materials and Methods using 14–15 mice per group. Each isolate is compared with reference strain FPR3757 (gray circle).
Correlation of SNPs and Virulence with Patterns of Exoprotein Expression.
S. aureus produces many secreted exoproteins that contribute significantly to virulence, such as enterotoxins, cytolytic toxins, and protein A (Spa). Given the significant variance in mouse virulence among the highly related isolates in our sample, we tested the hypothesis that one or more of the SNPs we identified resulted in differences in exoprotein profiles (Fig. 5 A–D). Nine of the 11 isolates had a very similar exoprotein profile, but this profile was remarkably different from those observed for 18805 and 18811, the two organisms of low virulence in the mouse model (Fig. 5A). The culture supernatant of these two isolates lacked α-hemolysin (Hla) and had reduced levels of LukF-PV (a subunit of Panton-Valentine leukocidin, PVL) compared with the reference strain (Fig. 5C). There was also more Spa in the culture supernatant of 18811 and 73-fold more spa transcript relative to the reference strain (P < 0.01) (Fig. 5B and SI Table 8). Of the 11 isolates tested, all except 18811 expressed delta-toxin, a result consistent with the occurrence of a missense mutation in agrA revealed by the SNP data for this isolate (Fig. 5D). Inasmuch as Agr is a primary regulator of exoprotein gene expression and virulence in S. aureus (14, 20), the amino acid polymorphism in agrA and associated alterations in Agr-regulated transcripts and global exoprotein profile may explain the reduced virulence of this isolate in the mouse bacteremia model (SI Tables 2 and 8 and Fig. 4). Elucidation of the mechanism underlying altered virulence and exoprotein profile in isolate 18805 requires further investigation. Lack of LukF-PV in culture supernatant of 18813 can be explained by the absence of φSA2usa (Fig. 5C and SI Table 6). The observation that this mutation had no significant/apparent impact on virulence in the mouse bacteremia model is in accordance with our previous study using isogenic lukS/F-PV deletion strains (PVL-negative) in mice (21).
Fig. 5.
Variation in S. aureus exoprotein profile. (A) Exoproteins present in the supernatant obtained from overnight cultures were analyzed by 10–20% gradient SDS/PAGE. (B and C) Production of protein A (Spa) (B) and of α-hemolysin (Hla) and LukF-PV (C) in the culture media. Aliquots of media from overnight cultures were analyzed by SDS/PAGE, and the indicated proteins were detected by Western immunoblot analysis. (D) Production of delta-toxin. Aliquots of the indicated media were subjected to LC/MS to quantitate delta-toxin. Data are the mean ± SEM of three experiments.
Concluding Comment.
Although not commonly appreciated, S. aureus has long been known to cause epidemics (2, 22–24). In addition, S. aureus is typically multidrug resistant and therefore infections can be difficult to treat. The first penicillin-resistant strains were reported in the mid-1940s and became widespread causes of infections in hospitalized patients (25, 26). A penicillin-resistant S. aureus genotype designated phage type 80/81 caused pandemic disease in the 1950s and 1960s in hospitals and the community (24). Although strains of phage type 80/81 disappeared after methicillin was introduced in 1959, methicillin-resistant S. aureus (MRSA) strains emerged almost immediately and are now epidemic in hospitals (3, 22, 27).
The molecular genetic basis of the current epidemic of CA-MRSA has remained obscure. Here, we used comparative whole-genome sequencing to understand the extent of genetic diversity in clinical isolates classified as USA300. Recent whole genome sequencing approaches have provided unprecedented detail about mechanisms of bacterial pathogen success or multidrug resistance. For example, the molecular basis for the origin and emergence of a highly successful clone of serotype M1 group A Streptococcus was made possible only by full genome resequencing (18), and studies by Mwangi et al. (19) used whole genome sequencing to identify steps in the evolution of antibiotic resistance in S. aureus.
Our studies reveal there has been recent clonal expansion (emergence) of a subset of USA300 isolates followed by diversification and thus provide insight into the molecular genetic basis of the current CA-MRSA epidemic. Previous work suggested CA-MRSA infections are caused by clonal groups of strains, but the strain typing methods used index genetic variation at a relatively few nucleotides and were not sufficient to demonstrate clonal emergence (5, 6).
Although our data unambiguously show that a number of USA300 isolates had a common ancestor, this finding should not be construed to mean that they are genetically identical nor have similar virulence characteristics. In fact, two of the clonal isolates analyzed in this study had a different virulence and exoprotein profile compared with the reference strain (Figs. 4 and 5). We acknowledge that relatively small changes in growth conditions in the laboratory or during processing may have a substantial impact on virulence. However, all strains tested had similar growth kinetics in vitro (SI Fig. 8) and phenotypic differences noted with the two isolates of low mouse virulence (e.g., absence of Hla) indicate the phenomenon is not likely related to variances in day-to-day growth in vitro. Taken together, these results are consistent with the idea that modest genetic changes in bacterial pathogens can have a dramatic impact on host-pathogen interaction, virulence and/or antibiotic resistance phenotype (10, 18, 19, 28). Therefore, it is reasonable to conclude that within a short period there will be an increasing pool of derivative USA300 isolates that differ in virulence potential and/or pathogenicity. A comprehensive understanding of genomic variation at the single nucleotide level is critical to elucidating the events that define bacterial virulence, multidrug resistance, and epidemics.
Materials and Methods
For full details, see SI Materials and Methods.
Bacteria and Culture Conditions.
Nine MRSA isolates (Table 1) classified as USA300 by PFGE were selected from among 864 MRSA isolates from invasive infections collected over an ≈1-year period through the Active Bacterial Core Surveillance system (ABCs) at the Centers for Disease Control and Prevention (CDC) in Atlanta (7). Of these 864 isolates, 252 (29.5%) were classified as USA300. Nine study isolates were selected from among 252 USA300 isolates to yield broad geographic representation from across the U.S., a variety of clinical syndromes, and a range of patient ages. The isolates were drawn from eight surveillance sites (California, Colorado, Georgia, Minnesota, New York, Oregon, Pennsylvania, and Tennessee). Five isolates met criteria for community-associated MRSA infections (1, 5) and 4 were healthcare-associated infections. Strain FRP3757 was kindly provided by Binh An Diep (University of California, San Francisco) and is the sequenced reference strain used for these studies. LAC was selected for this study because it was recovered early during the CA-MRSA epidemic (2002), has been studied in virulence models, and was recovered from a skin and soft tissue infection, which represents the majority of CA-MRSA infections.
Comparative Genome Sequencing (CGS).
The protocol used for comparative genome sequencing (NimbleGen, Inc.) is available at www.nimblegen.com.
Confirmation of SNPs and Analysis of RDs.
SNPs and RDs identified by CGS were verified by conventional capillary DNA sequencing using oligonucleotide primers (Sigma Genosys, The Woodlands, TX) designed with Vector NTI software (Invitrogen Corp., Carlsbad, CA).
Phylogenetic Analyses.
Evolutionary relationships were reconstructed by using IQPNNI software (29). The DNA sequences analyzed were generated by concatenating nucleotides at all of the core genomic DNA SNP loci identified in sequential order relative to the FPR3757 reference genome sequence.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the Rocky Mountain Laboratories (RML) Genomics Unit, Research Technologies Section, Research Technologies Branch, National Institute of Allergy and Infectious Diseases (NIAID) for assistance with DNA sequencing and bioinformatics analyses; and the ABCs system of the Centers for Disease Control and Prevention (CDC, Atlanta, GA). The findings and conclusions in this publication are those of the authors and do not necessarily represent the views of the CDC. This work was supported by the Intramural Program of the NIAID, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710217105/DC1.
References
- 1.Zetola N, Francis JS, Nuermberger EL, Bishai WR. Lancet Infect Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 2.Chambers HF. N Engl J Med. 2005;352:1485–1487. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- 3.Chambers HF. Emerg Infect Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 5.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F. J Clin Microbiol. 2004;42:2080–2084. doi: 10.1128/JCM.42.5.2080-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, et al. J Am Med Assoc. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 8.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. Ann Intern Med. 2006;144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 9.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, et al. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 10.Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, et al. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, Cousins DV, Graviss EA, Shashkina E, Kreiswirth BN, Musser JM. Genetics. 2002;162:1533–1543. doi: 10.1093/genetics/162.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocha EP, Smith JM, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 14.Novick RP. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 15.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, et al. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 16.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, et al. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 18.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, et al. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 19.Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de LH, Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, et al. Proc Natl Acad Sci USA. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 21.Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, et al. J Infect Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 22.Stewart GT, Holt RJ. Br Med J. 1963;1:308–311. doi: 10.1136/bmj.1.5326.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. Proc Natl Acad Sci USA. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson DA, Kearns AM, Holmes A, Morrison D, Grundmann H, Edwards G, O'Brien FG, Tenover FC, McDougal LK, Monk AB, et al. Lancet. 2005;365:1256–1258. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- 25.Kirby WM. Science. 1944;99:452–453. doi: 10.1126/science.99.2579.452. [DOI] [PubMed] [Google Scholar]
- 26.Barber M, Hayhoe FG, Whitehead JE. Lancet. 1949;2:1120–1125. doi: 10.1016/s0140-6736(49)91144-7. [DOI] [PubMed] [Google Scholar]
- 27.Jevons MP. Br Med J. 1961;1:124–125. [Google Scholar]
- 28.Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Shang Y, Williams TM, Fortunov RM, Liu Y, et al. BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinh LS, von Haeseler A. Mol Biol Evol. 2004;21:1565–1571. doi: 10.1093/molbev/msh176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.