The heat shock proteins (HSPs) comprise a family of highly conserved molecular chaperones that are central to protein structure homeostasis. HSPs prevent aggregation of cellular proteins, assist in protein folding, and act as “biochemical buffers” to guard proteins against diverse types of cellular stress including heat, changes in cellular pH, and hypoxia. The major functions of the HSPs in normal cells include coordination of higher-order interactions between “client” proteins and macromolecular machines, organelles, trafficking and metabolic vesicles, and structural proteins such as microtubules and actin filaments. The HSPs, and HSP90 in particular, facilitate signal transduction and gene transcription by stabilizing interactions between client proteins, proteins and nucleic acids, and proteins and their ligands (1, 2). A unique aspect of HSP90 biochemistry is that this protein stabilizes rather than modifies its client proteins; and this, too, seems to be how it is involved in malignant transformation (3). Mutations and/or amplifications in oncogenic receptor tyrosine kinases, such as EGFR and HER2, or oncogenic signal transducers, such as RAF and SRC, frequently lead to constitutively active proteins. In the normal course of events, intact negative feedback loops result in higher levels of protein degradation, thus maintaining relatively normal levels of growth promoting signal transduction. However, tumor cells harbor extensive genetic damage and exist in physiologically stressful conditions (hypoxia, pH, cytokine storm) and thus also tend to have elevated levels of activated HSP90. These higher levels of activated HSP90 can lead to stabilization of constitutively activated client proteins, in essence establishing a positive feedback loop that perpetuates growth signaling and survival in opposition to the normal regulatory pathways that induce growth arrest and programmed cell death (4, 5). In a recent issue of PNAS, Chakraborty et al. (6) extend the purview of HSP90-dependent tumor promoting activity by demonstrating that it acts as a bona fide physiologic inhibitor of a new client protein, the proapoptotic protein kinase, IP6K2.
To date, nearly all cancer-promoting HSP90-client interactions, including those that block programmed cell death, stem from HSP90-stabilized, aberrant signal transduction events (Fig. 1; important exceptions include FKBP38 and p53). In most cases, HSP90-mediated stabilization of protein–protein interactions requires an N-terminal ATPase activity (amino acids 1–210) and a core client binding motif (amino acids 272–629). Client proteins associate in transient low-affinity complexes with HSP90 dimers and affiliated proteins, whereupon bound ATP is hydrolyzed at the N terminus of HSP90, which leads to a conformational change in the HSP90–client complex. This conformational change in the complex activates and stabilizes client proteins. Because tumor cells have significantly higher levels of activated HSP90 than normal cells, small molecules that inhibit the N-terminal ATPase activity show a high degree of tumor specificity (7, 8). Several of these inhibitors are in early-stage cancer therapeutic clinical trials.
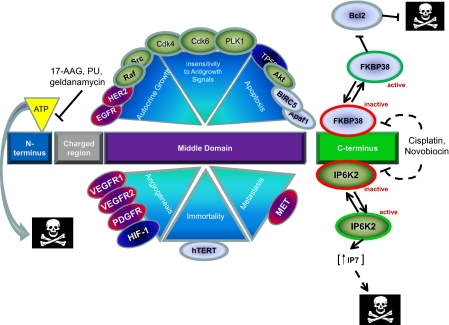
Fig. 1.
Schematic representation of HSP90 binding to client proteins, how these proteins affect six hallmarks of cancer, and drug target sites within HSP90. HSP90 interacts with proteins that contribute to all six hallmarks of cancer (for a complete list of HSP90 binding partners see ref. 16). HSP90 binds to many of the expressed kinase domains within the human genome. HSP90 stabilizes the active conformations of both WT and mutant tyrosine kinase receptors (red-purple), cytosolic serine-threonine and tyrosine kinases (green), transcription factors (blue), structural proteins and other enzymes (gray). Note: no specific sequence within the middle domain as a binding site is implied by the cartoon. Client binding occurs through the middle domain of HSP90 (purple rectangle), which leads to HSP90 dimerization, cochaperone binding (HSP70, HIP, HOP, cdc37), and ATP binding and hydrolysis. Many of these interactions are inhibited by small molecules that compete for the N-terminal ATP binding pocket such as the benzoquinone ansamycins (geldanamycin, 17-AAG), radicicols, their derivatives, and purine analogues (PU) (see refs. 4 and 8). Thus, many signal transduction pathways require HSP90 to perpetuate growth promoting signals, and attenuation of these signals by inhibition of HSP90 ATPase activity leads indirectly to cell death. On the other hand, proteins that bind to the C terminus (green rectangle) of HSP90, such as FKBP38 and IP6K2, are maintained in a constitutively inactive state by the interaction (red border). Drugs such as cisplatin and novobiocin (which interact with the C terminus of HSP90 at high concentrations) appear to disrupt these interactions, leading to the release and activation of the now cytosolic c-terminal binding partners (green border) and subsequent apoptosis, either through inhibition of Bcl2 (FKBP38) or increased cytosolic concentrations of IP7 (IP6K2). Dashed arrows indicate interactions that have yet to be demonstrated biochemically.
More than a decade of work has established that HSP90 affects all of the “hallmarks of cancer” (Fig. 1) (9). HSP90 interacts with several components of the apoptosis machinery to promote survival, and the best-described interactions feed through the AKT signaling pathway. Dysregulation of the phosphoinositol 3-kinase/AKT pathway is a common event in human malignancies and usually occurs through overactivation of AKT by mutation/amplification of genes in the pathway, or inactivation of PTEN by mutation or promoter hypermethylation (10). Constitutively active AKT is stabilized by its interaction with active HSP90-cdc37, which facilitates phosphorylation of AKT by pyruvate dehydrogenase kinase (11). HSP90 also seems to protect phospho-AKT from dephosphorylation and deactivation by PP2A. Activated AKT perpetuates survival signaling in several ways: AKT phosphorylates (and inhibits) several members of the proapoptotic Bcl2 family, inhibits caspase-9 activity, and promotes survival indirectly by stimulating NF-κβ-dependent transcription by phosphorylating IKK (10).
Chakraborty et al. (6) and others (12) have shown that IP6K2 catalyzes the production of diphosphoinositol pentakisphosphate (IP7) and has a role as a proapoptotic gene sensitizing cancer cells to apoptosis by cell stressors and anticancer drugs all of which depend on IP6K2 catalytic activity (6, 12). In addition, IP6K2 is located in the 3p21.31 chromosomal region, which often undergoes allele loss in a variety of human cancers. IP6K2 is located close to a series of other 3p21 genes including NPRL2, FUS1, RASSF1A, SEMA3B, and SEMA3F, all of which have some tumor suppressor activity (13). Thus, many human tumors will have lost a copy of IP6K2, and although it may ultimately turn out to be a tumor suppressor, there are no reports of genetic or epigenetic inactivation of IP6K2 as yet. Chakraboty et al. (6) appear to have identified a new method for inactivation that allows for the possibility of targeted small-molecule reactivation.
In their article, Chakraboty et al. (6) elegantly demonstrate that HSP90 regulates apoptosis through a normal physiologic interaction with IP6K2, an enzyme involved in the formation of inositol pyrophosphates, NF-κβ signaling, and cisplatin-mediated cell death (6, 14). They show that: IP6K2 selectively binds to HSP90, whereas a closely related family member IP6K1 (also located nearby on chromosome region 3p21) does not; binding of cellular IP6K2 occurs through the HSP90 C terminus and a 12-aa sequence in IP6K2 with Arg-133 and Arg-136 of IP6K2 being critical for this interaction; binding to HSP90 decreases IP6K2 catalytic activity; and HSP90 binds to a significant portion of cellular IP6K2, all of which leads to inhibiting IP6K2 activity.
It is of interest that the interaction between HSP90 and IP6K2 is inhibited by cisplatin and novobiocin, compounds that are known to bind to the C terminus of HSP90, but not by the geldanamycin derivative 17-AAG, which targets the N-terminal ATP binding pocket. Cisplatin, a drug commonly used in the treatment of many human cancers (and another drug, novobiocin, which is an antistaphylococcal agent), acts by disrupting the inhibitory effect of HSP90 on IP6K2, leading to increased IP7 formation (5). Thus, another implication of their findings is that IP6K2 expression or activity in tumors may correlate clinically with response to platinum-containing chemotherapy regimens, which, if confirmed, would be important for rational selection of chemotherapy. As Zhang and Burrows (5) discuss, the molecular mechanisms whereby IP6K2 induce cell death are unclear, but at the very least seem to involve IP7 production. In yeast, IP6K plays a role in modulating telomere length, so HSP90:IP6K2 interaction may also be a new target for controlling telomere length in cancer cells. The ability of HSP90 to inhibit the normal growth regulatory activity of IP6K2 may be a segue into the discovery of several similar interactions. For example, the peptidylprolyl cis-trans isomerase, FKBP38, also binds to the C terminus of HSP90 where it is prevented from blocking various Bcl-2 interactions (15).
The findings in the article by Chakraboty et al. (6) strengthen the case for HSP90 having oncogenic properties: its overactivation and promiscuous interaction with a range of oncogenic signal transducers leads to increased cell growth, and its overexpression appears to lead to increased cell survival through constitutive inactivation of several proapoptotic and growth inhibitory pathways. So far, the recent flurry of genomewide copy number and sequencing studies have not uncovered any genetic alterations in HSP90; however, there is an abundance of evidence demonstrating that its regulation by physiologic stresses could play a role in cancer pathogenesis (3).
Perhaps the most exciting outcome of the article by Chakraboty et al. (6) is the potential therapeutic application of finding new C-terminal HSP90 inhibitors, which might be active against cancer and other diseases. Their findings can be used to design a strategy for screening small-molecule libraries for compounds that can selectively disrupt the specific HSP90:IP6K2 12-aa interaction. There are also specific predictions that can be made in terms of which tumors should respond to such targeted therapy (e.g., those with increased HSP90 activity and decreased IP6K2 activity) and assays to show whether the drugs get to the target site and do their job. However, there are a number of elements that still need to be worked out, including exactly how IP6K2 mediates apoptosis, whether IP6K2 binds to activated HSP90, which is required for other client proteins, and whether targeted disruption of HSP90:IP6K2 has selectivity for cancer cells. In addition, any new HSP90:IP6K2 targeted drugs will have to be evaluated for additive, synergistic, or antagonistic tumor effects and normal tissue toxicity with standard chemotherapy agents. Nonetheless, the findings by Chakraboty et al. (6) provide an exciting new entry into the regulation of apoptosis and the targeted therapy of cancer and other diseases.
ACKNOWLEDGMENTS.
This work was supported by the International Association for the Study of Lung Cancer, National Cancer Institute Grant P50CA70907, and Department of Defense grants.
Footnotes
The authors declare no conflict of interest.
See companion article on page 1134 in issue 4 of volume 105.
References
- 1.Morimoto RI. Cell. 2002;110:281–284. doi: 10.1016/s0092-8674(02)00860-7. [DOI] [PubMed] [Google Scholar]
- 2.Smith DF, Whitesell L, Katsanis E. Pharmacol Rev. 1998;50:493–514. [PubMed] [Google Scholar]
- 3.Whitesell L, Lindquist SL. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 4.Burrows F, Zhang H, Kamal A. Cell Cycle. 2004;3:1530–1536. doi: 10.4161/cc.3.12.1277. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Burrows F. J Mol Med. 2004;82:488–499. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty AM, Koldobskly MA, Sixt KM, Juluri KR, Mustafa AK, Snowman AM, van Rossum DB, Patterson RL, Snyder SH. Proc Natl Acad Sci USA. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 8.Rodina A, Vilenchik M, Moulick K, Aguirre J, Kim J, Chiang A, Litz J, Clement CC, Kang Y, She Y, et al. Nat Chem Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Vivanco I, Sawyers CL. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 11.Basso AD, et al. J Biol Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 12.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zabarovsky ER, Lerman MI, Minna JD. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 14.Morrison BH, Bauer JA, Lupica JA, Tang Z, Schmidt H, DiDonato JA, Lindner DJ. J Biol Chem. 2007;282:15349–15356. doi: 10.1074/jbc.M700156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edlich F, Erdmann F, Jarczowski F, Moutty M-C, Weiwad M, Fischer G. J Biol Chem. 2007;282:15341–15348. doi: 10.1074/jbc.M611594200. [DOI] [PubMed] [Google Scholar]
- 16.Picard D. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]