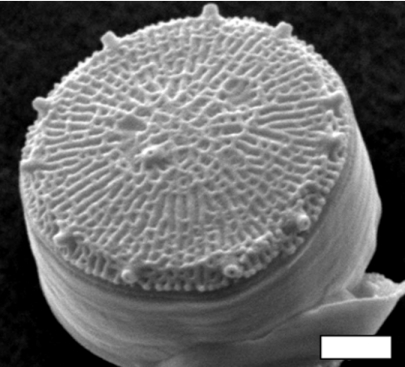
Ever since the invention of the microscope, scientists and laypeople alike have marveled at the intricate patterning of the silica within the diatom cell wall, termed the frustule (Fig. 1). There are well over 10,000 species of diatoms in marine and freshwaters, and each species has a unique wall morphology (1). Although organisms from multiple kingdoms of life produce siliceous structures (e.g., phytoliths in higher plants, spicules in sponges, frustules in diatom), those produced by diatoms are the most ornate and the most finely detailed. The ability of diatoms to manipulate silicon at the nanoscale exceeds that of human nanotechnology, making the genetic and biochemical underpinnings of biosilicification of great interest in material science. Diatoms are also important ecologically because they are major primary producers supporting the food webs in both marine and fresh waters. Despite the broad scientific interest in the silicification process, the biochemical pathways involved and the underlying genetics are largely a mystery. The sequencing of the first diatom genome (2) has enabled a new suite of approaches in the hunt for pathways involved in biosilicification. In this issue of PNAS, Mock et al. (3) exploit this potential by using whole genome expression profiling to elucidate candidate genes controlling silicification in the marine diatom Thalassiosira pseudonana.
Fig. 1.
The diatom Thalassiosira pseudonana. (Scale bar: 1 μm.) (Micrograph courtesy of Mark Hildebrand, Scripps Institution of Oceanography, San Diego, CA.)
Diatoms are microscopic unicellular protists and the vast majority are photoautotrophs. Their role in the ecology and biogeochemistry of our planet is impressive. They are responsible for 20% of the carbon fixed through photosynthesis on Earth (4), making diatoms major contributors to global carbon cycling and they collectively produce ≈10 km3 of hydrated amorphous silica each year (5). Diatoms are the only major group of phytoplankton that require silicon. Dissolved silicic acid concentrations in the surface ocean are often inadequate to meet the demand of the diatoms (6, 7), making knowledge of diatom silicification important for understanding the mechanisms governing the contribution of diatoms to ecological and biogeochemical processes. Diatoms dominate the biogeochemical cycling of silicon through the sea. Each atom of silicon entering the oceans from weathering is incorporated into the cell wall of a diatom and dissolved back into the sea an average of 40 times before being buried in the sea bed (8). The slow burial of diatoms in the sea bed over geologic time has produced a significant fraction of the world's petroleum, and diatom deposits that have been uplifted onto the land are mined worldwide as a source of diatomaceous earth, which is used as a filtration medium and in the manufacturing of explosives and other products.
The diatom cell wall is mulipartate consisting of two valves with interconnecting segments called girdle bands. Each component of the frustule is formed in a tightly timed sequence within the cell cycle (9, 10). In the case of T. pseudonana, its girdle bands are formed sequentially at a nearly constant rate during G1, silicification ceases in S phase, and valves are deposited in late in the cell cycle G2 and M (10). The process of biosilicification in diatoms begins with the active transport of dissolved silicic acid from the aqueous environment into the cell mediated by silicon transporters, or SITS, whose expression and activity also show tight regulation during the cell cycle (11, 12). Intracellular concentrations of dissolved silicon exceed the solubility of silicic acid (13), suggesting the presence of an organic silicon complex that may maintain silicon in solution and shepherd it to the silicon deposition vesicle (SDV). Silicon enters the SDV and is deposited as amorphous hydrated silica under precise genetic control to form the ornate patterns of the frustule (14). The process of frustule morphogenesis has been studied in great detail since the advent of the electron microscope, but the molecular underpinnings of the process are only now being discovered.
Our knowledge of the molecular basis of biosilicification in diatoms stems mainly from analysis of proteins isolated from the mature diatom cell wall. Several key molecules involved in silicon processes have been discovered by using this approach, including silicon transporters (11, 12) and proteins, named silaffins, which together with polyamines are able to produce silica nanospheres in vitro that bear a strong resemblance to those formed during frustule formation in vivo in some species (15, 16). Mock et al.'s (3) approach is different in that they attempt to identify all genes involved in the silicification process within the diatom genome. This poses a great challenge, because there is little homology among the proteins involved in silicification from different taxa, thwarting efforts to identify genes involved in biosilicificiation through in silico homology-based approaches. To circumvent this issue, they first employ whole genome tiling array-based expression profiling to identify novel genes. More than 34,470 transcriptional units not predicted by gene models were identified, suggesting that the T. pseudonana genome contains 30% more genes than previously thought (2). To examine which of these gene products may be involved in silicification pathways, the authors constructed impressively comprehensive gene expression arrays that included probes to all in silico predicted genes, all available EST sequences, the newly discovered unpredicted transcriptional units, as well as 16,000 additional tiling array probe clusters. To target silicon-related genes, Mock et al. (3) looked for differences in the hybridization of RNA isolated from silicon-limited versus silicon-replete cells. Then they took the additional step of hybridizing the same arrays against RNA expressed under a variety of other environmental stresses, including nitrogen and iron limitation.
Silicon limitation either up- or down-regulated 75 genes that had no known function pointing to their possible role in silicification. Potential regulatory mechanisms were revealed including a vertebrate-like consensus motif upstream of the initiator codon for the 75 transcripts, reduction of antisense expression of 10 genes under silicon limitation, and the up-regulation of a noncoding transcript, which may be the first noncoding RNA to be discovered that is involved in silicon processing.
A unique aspect of the work is the discovery of a possible direct coupling of silicon and iron pathways. Silicon and iron limitation each induced a common set of 84 genes, suggesting that iron and silicon metabolisms share common pathways or that iron is involved as a cofactor in silicon biochemistry. Iron limitation also resulted in the up-regulation of genes known to be involved in the silicification process. The frustules of the iron-limited cells contained significantly more silica than those of control cells suggesting a link between the up-regulation of these genes and the extent of silicification. The observation that iron stress produces more heavily silicified frustules is well established (17, 18); however, previous hypotheses to explain why iron-limited diatoms are more heavily silicified emphasized the lengthening of the duration of the cell cycle stages associated with silicification under low iron, which would presumably allow more time for additional silicon uptake and incorporation (13). The work of Mock et al. (3) suggests that a more complicated suite of regulatory processes are involved.
Diatoms dominate the biogeochemical cycling of silicon through the sea.
A direct link between silicon and iron metabolism would have profound ecological significance. Phytoplankton primary productivity is limited by iron in ≈30% of the global ocean in what are known as High-Nutrient Low-Chlorophyll (HNLC) regions. That acronym denotes the inability of phytoplankton in these areas to proliferate and deplete major nutrients such as nitrogen and phosphorus for lack of iron. Large-scale iron fertilization experiments in HNLC regions have consistently shown diatoms to be the group of phytoplankton that respond most strongly to iron addition (19). These areas are also low in dissolved silicon (20), suggesting that both silicon and iron may play a role in regulating diatom productivity in what are currently considered to be iron-limited waters.
By exploiting the entire diatom genome, Mock et al. (3) have increased the number of candidate genes and gene products involved in diatom silicification from <10 to 100 or more. This information should speed the advancement of our knowledge regarding the molecular basis of diatom silicification. The expression libraries obtained under other environmental stressors may prove equally valuable in revealing the linkages between silicon metabolism and that of other key nutrient elements affecting diatom growth in nature, such as iron and nitrogen. Interest in diatom silicification continues to grow, and now both material scientists and oceanographers have a refined roadmap to guide future research.
Footnotes
The authors declare no conflict of interest.
See companion article on page 1579.
References
- 1.Werner D. The Biology of Diatoms. Berkeley: Univ of California Press; 1977. [Google Scholar]
- 2.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 3.Mock T, Samanta MP, Iverson V, Berthiaume C, Robison M, Holtermann K, Durkin C, BonDurant SS, Richmond K, Rodesch M, et al. Proc Natl Acad Sci USA. 2008;105:1579–1584. doi: 10.1073/pnas.0707946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field CB, Behrenfeld MJ, Falkowski P. Science. 1998;281:237. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 5.Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Global Biogeochem Cycles. 1995;9:359–372. [Google Scholar]
- 6.Brzezinski MA, Nelson DM. Deep-Sea Res II. 1996;43:437–453. [Google Scholar]
- 7.Leynaert A, Tréguer P, Lancelot C, Rodier M. Deep-Sea Res I. 2001;48:639–660. [Google Scholar]
- 8.Tréguer P, Nelson DM, Bennekom AJV, DeMaster DJ, Leynaert A, Quéguiner B. Science. 1995;268:375–379. doi: 10.1126/science.268.5209.375. [DOI] [PubMed] [Google Scholar]
- 9.Chiappino ML, Volcani BE. Protoplasma. 1977;93:205–221. [Google Scholar]
- 10.Hildebrand M, Frigeri LG, Davis AK. J Phycol. 2007;43:730–740. [Google Scholar]
- 11.Hildebrand M, Volcani BE, Gassmann W, Schroeder JI. Nature. 1997;385:688–689. doi: 10.1038/385688b0. [DOI] [PubMed] [Google Scholar]
- 12.Thamatrakoln K, Hildebrand M. Eukaryotic Cell. 2007;6:271–279. doi: 10.1128/EC.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Jézéquel V, Hildebrand M, Brzezinski MA. J Phycol. 2000;36:821–840. [Google Scholar]
- 14.Li C-W, Volcani BE. Philos Trans R Soc London Ser B. 1984;304:519–528. [Google Scholar]
- 15.Kröger N, Deutzmann R, Sumper M. Science. 1999;286:1129. doi: 10.1126/science.286.5442.1129. [DOI] [PubMed] [Google Scholar]
- 16.Kröger N, Deutzmann R, Bergsdorf C, Sumper M. Proc Natl Acad Sci USA. 2000;97:14133–14138. doi: 10.1073/pnas.260496497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchins DA, Bruland KW. Nature. 1998;393:561–564. [Google Scholar]
- 18.Takeda S. Nature. 1998;393:774–777. [Google Scholar]
- 19.de Baar H, Boyd PW, Coale KH, Landry MR, Tsuda A, Assmy P, Bakker DCE, Bozec Y, Barber RT, Brzezinski MA, et al. J Geophys Res. 2005;110 doi: 10.1029/2004JC002601. [DOI] [Google Scholar]
- 20.Dugdale RC, Wilkerson FP, Minas HJ. Deep-Sea Res I. 1995;42:697–719. [Google Scholar]