Abstract
The spread of sexually transmitted diseases (e.g., chlamydia, syphilis, gonorrhea, HIV, etc.) across populations is a major concern for scientists and health agencies. In this context, both the data collection on sexual contact networks and the modeling of disease spreading are intensive contributions to the search for effective immunization policies. Here, the spreading of sexually transmitted diseases on bipartite scale-free graphs, representing heterosexual contact networks, is considered. We analytically derive the expression for the epidemic threshold and its dependence with the system size in finite populations. We show that the epidemic outbreak in bipartite populations, with number of sexual partners distributed as in empirical observations from national sex surveys, takes place for larger spreading rates than for the case in which the bipartite nature of the network is not taken into account. Numerical simulations confirm the validity of the theoretical results. Our findings indicate that the restriction to crossed infections between the two classes of individuals (males and females) has to be taken into account in the design of efficient immunization strategies for sexually transmitted diseases.
Keywords: bipartite graphs, epidemic threshold, sexual contact networks
Disease spreading has been the subject of intense research for a long time (1–3). On the one hand, epidemiologists have developed mathematical models that can be used as guides to understanding how an epidemic spreads and to design immunization and vaccination policies (1–3). On the other hand, data collections have provided information on the local patterns of relationships in a population. In particular, persons who may have come into contact with an infectious individual are identified and diagnosed, making it possible to contact-trace the way the epidemic spreads and to validate the mathematical models. However, up to a few years ago, some of the assumptions at the basis of the theoretical models were difficult to test. This is the case, for instance, for the complete network of contacts, the backbone through which the diseases are transmitted. With the advent of modern society, fast transportation systems have changed human habits, and some diseases that just a few years ago would have produced local outbreaks now are a global threat for public health systems. A recent example is severe acute respiratory syndrome (SARS), which spread very fast from Asia to North America a few years ago (4–6). Therefore, it is of utmost importance to carefully take into account as many details as possible of the structural properties of the network in which the infection dynamics occurs.
Strikingly, a large number of statistical properties have been found to be common in the topology of real-world social, biological, and technological networks (7–9). Of particular relevance, because of its ubiquity in nature, is the class of complex networks referred to as scale-free (SF) networks. In SF networks, the number of contacts or connections of a node with other nodes in the system, the degree (or connectivity) k, follows a power-law distribution, Pk ∼ k−γ. Recent studies have shown the importance of the SF topology on the dynamics and function of the system under study (7–9). For instance, SF networks are very robust to random failures but at the same time extremely fragile to targeted attacks of the highly connected nodes (10, 11). In the context of disease spreading, SF contact networks lead to a vanishing epidemic threshold in the limit of infinite population when γ ≤ 3 (12–15), which is because the exponent γ is directly related to the first and second moment of the degree distribution, 〈k〉 and 〈k2〉, and the ratio 〈k〉/〈k2〉 determines the epidemic threshold above which the outbreak occurs. When 2 < γ ≤ 3, 〈k〉 is finite whereas 〈k2〉 goes to infinity; that is, the transmission probability required for the infection to spread goes to zero. Conversely, when γ > 3, there is a finite threshold and the epidemic survives only when the spreading rate is above a certain critical value. The concept of a critical epidemic threshold is central in epidemiology. Its absence in SF networks with 2 < γ ≤ 3 has a number of important implications in terms of prevention policies: If diseases can spread and persist even in the case of vanishingly small transmission probabilities, then prevention campaigns in which individuals are randomly chosen for vaccination are not very effective (12–15).
Our knowledge of the mechanisms involved in disease spreading and the relation between the network structure and the dynamical patterns of the spreading process has improved in the last several years (16–19). Current approaches are either individual-based simulations (18) or metapopulation models in which network simulations are carried out through a detailed stratification of the population and infection dynamics (20). In the particular case of sexually transmitted diseases (STDs), infections occur within the unique context of sexual encounters, and the network of contacts (19, 21–26) is a critical ingredient of any theoretical framework. Unfortunately, ascertaining complete sexual contact networks in significantly large populations is extremely difficult. However, here we show that it is indeed possible to make use of known global statistical features to generate more accurate predictions of the critical epidemic threshold for STDs.
Networks of Sexual Contacts.
Data from national sex surveys (21–25) provide quantitative information on the number of sexual partners, the degree k, of an individual. Usually, surveys involve a random sample of the population stratified by age, economical and cultural level, occupation, marital status, etc. The respondents are asked to provide information on sexual attitudes such as the number of sex partners they have had in the last 12 months or in their entire life. Although in most cases the response rate is relatively small, the information gathered is statistically significant, and global features of sexual contact patterns can be extracted. In particular, it turns out that the number of heterosexual partners reported from different populations is well described by power-law SF distributions. Table 1 summarizes the main results of surveys conducted in Sweden, the U.K., Zimbabwe, and Burkina Faso (21–24).
Table 1.
Statistical properties of sexual contact networks from national sex surveys conducted in four different countries: Sweden, the U.K., Zimbabwe, and Burkina Faso
| Survey | Ref. | 12 months |
Lifetime |
Respondents |
||||
|---|---|---|---|---|---|---|---|---|
| γF | γM | γF | γM | Total | Female | Male | ||
| Sweden | 21 | 3.54 ± 0.20 | 3.31 ± 0.20 | 3.1 ± 0.30 | 2.6 ± 0.30 | 2,810 | — | — |
| U.K. | 22, 23 | 3.10 ± 0.08 | 2.48 ± 0.05 | 3.09 ± 0.20 | 2.46 ± 0.10 | 11,161 | 6,399 | 4,762 |
| Zimbabwe | 23 | 2.51 ± 0.40 | 3.07 ± 0.20 | 2.48 ± 0.15 | 2.67 ± 0.18 | 9,843 | 5,424 | 4,419 |
| Burkina Faso | 24 | 3.9 ± 0.2 | 2.9 ± 0.1 | — | — | 466 | 179 | 287 |
The exponents γF and γM are referred to the distribution of number of sexual partners cumulated in 12 months and over the respondent's lifetime. The number of respondents also is reported.
The first thing to notice is the gender-specific difference in the number of sexual acquaintances (21–24). This difference is manifested by the existence of two different exponents in the SF degree distributions, one for males (γM) and one for females (γF). Interestingly enough, the predominant case in Table 1 (no matter whether data refer to time frames of 12 months or to entire life span) consists of one exponent being smaller and the other >3. This is certainly a borderline case that requires further investigation on the value of the epidemic threshold.
The differences found in the two exponents γF and γM have a further implication for real data and mathematical modeling. In an exhaustive survey, able to reproduce the whole network of sexual contacts, the total number of female partners reported by men should equal the total number of male sexual partners reported by women. Mathematically, this means that the number of links ending at population M (of size NM) equals the number of links ending at population F (of size NF), which translates into the following closure relation:
Assuming that the degree distributions for the two sets are truly SF, then PkG = (γG − 1) × k−γG/k01−γG, with the symbol G standing for the gender (G = F, M), and k0 being the minimum degree. Moreover, if NG ≫ 1 and γG > 2 for any G, Eq. 1 gives the relation between the two population sizes as
which implies that the less heterogeneous (in degree) population must be larger than the other one.
In conclusion, the empirical observation of two different exponents demands for a more accurate description of the network of heterosexual contacts as bipartite SF graphs, i.e., graphs with two set of nodes and links connecting nodes from different sets only. In the following section, we will consider a graph with NM nodes, representing males and characterized by the exponent γM, and NM nodes, representing females and characterized by γF. Concerning the choice of the couple of exponents from those reported in Table 1, one must be careful that different STDs have different associated (recovery) time scales, and that the spreading is based on the assumption that the links are concurrent on the time scale of the disease. In this sense, the exponents extracted from 1-year data seem better suited to most of the STDs, with HIV being an important exception. However, during the lifetime of sexually active individuals, sexual behavior is likely to change because of changes in residence, marital status, age-linked sexual attitudes, etc. (27). We thus prefer to use lifecycle data collections that integrate all these patterns and can consequently be regarded as better statistical indicators. After all, the values reported in Table 1 indicate that both 1-year and cumulative data produce exponents in the same range.
Theoretical Modeling.
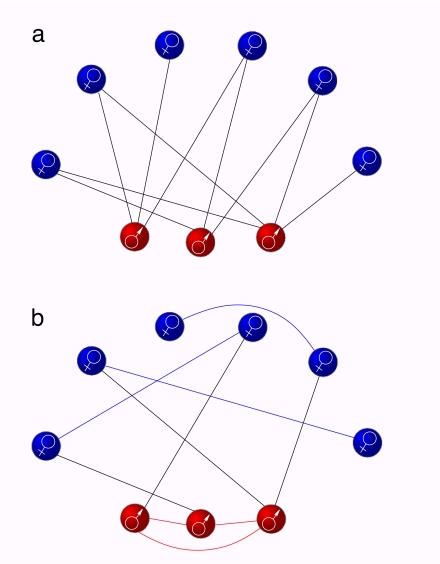
The problem of how a disease spreads in a population consisting of two classes of individuals can be tackled by invoking the so-called criss-cross epidemiological model (3). As illustrated in the bipartite network of Fig. 1a, in the criss-cross model, the two populations of individuals (NM males and NF females) interact so that the infection can only pass from one population to the other by crossed encounters between the individuals of the two populations, incorporating in this way one of the basic elements of the heterosexual spreading of STDs. We adopted here the indexes M and F to denote quantities relative to male and female populations in networks of heterosexual contacts. However, the present approach is more general and applies to any spreading of diseases in which crossed infections between two populations occur. In particular, we consider a susceptible–infected–susceptible (SIS) dynamics, in which individuals can be in one of two different states, namely, susceptible (𝒮) and infectious (ℐ). If 𝒮M and ℐM (𝒮F, ℐF) stand for a male (female) in the susceptible and infectious states, respectively, the epidemic in the SIS criss-cross model propagates by the following mechanisms:
being νM, νF, μM, and μF the infection and recovery probabilities for males and females. In the case of heterogeneous contact networks, there is a further compartmentalization of the population into classes of individuals with the same degree k, i.e., the same number of sexual partners. Denoting the fraction of males (females) with degree k in the susceptible or infectious state by skM and ikM (skF and ikF), respectively, and adopting a mean-field approach (3, 12, 14), the differential equations describing the time evolution of the densities of susceptible and infected individuals in each population are
and
where, λG = νG/μG (G = F, M) are the effective transmission probabilities. The quantities ϴkM(t), ϴkF(t) stand for the probabilities that a susceptible node of degree k of one population encounters an infectious individual of the other set. Eqs. 3 and 4 have the same functional form of the equation derived in ref. 12 for unipartite networks. Neglecting degree–degree correlations, the critical condition for the occurrence of an endemic state reduces to
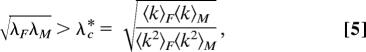 |
yielding that a necessary condition for the absence of the epidemic threshold is the divergence of at least one of the second moments of the degree distributions, 〈k2〉M and 〈k2〉F. Eq. 5 can be compared with the condition obtained without taking into account that, in heterosexual networks, the infection can occur only between male–female couples (12, 13). In fact, working with a unipartite representation of a sexual network, such as that shown in Fig. 1b, with NM + NF nodes and a degree distribution Pk = (NM PkM + NF PkF)/(NM + NF), one can express the epidemic threshold as a function of the first and second moments of the male and female degree distributions as
Eqs. 5 and 6 are clearly different. For real SF networks of sexual contacts, the two thresholds are finite (in the infinity size limit) only when the two exponents γM and γF are both >3, for example, for the 1-year number of partners in Sweden (see Table 1). In such a case, the two expressions read as follows:
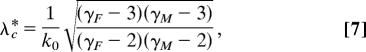 |
These two thresholds are equal only in the case γM = γF (studied in ref. 15). More importantly, when γF ≠ γM, we have λ∗c ≥ λc, namely the epidemic state occurs in bipartite networks for larger transmission probabilities than in unipartite networks. This result is good news and highlights the importance of incorporating the crossed infections scheme in the propagation of STDs. However, as shown in Table 1, most of the real networks have at least one exponent γG (G = F, M) < 3, which means that, in most of the practical cases, the two epidemic thresholds vanish as the system size goes to infinity, no matter the formulation used to model the disease propagation. However, real populations are finite, and thus the degree distributions have a finite variance regardless of the exponents. Consequently, an epidemic threshold does always exist and, to compare unipartite with bipartite networks, one must then pay attention to the scaling of the threshold with the size of the population.
Fig. 1.
Bipartite and unipartite networks. (a) Representation of a bipartite network accounting for heterosexual contact networks. In such a network, we have NM (NF) nodes representing males (females), and only male–female links are allowed. (b) A rewired version of the network in a in which the bipartite nature is lost and the degree of the nodes is preserved. The two graphs have the same couple of degree distributions (one for males and one for females), although only the graph in a reflects the bipartite character of heterosexual contact networks.
Finite Populations.
We now analyze in more details the differences between λc and λ∗c when one of the two exponents (say, γM without loss of generality) is in the range 2 < γM < 3, whereas γF > 2. First, we derive the size scaling of the critical threshold in unipartite graphs, λc. Eq. 6 yields
Manipulating this expression by considering again the limit of large (but finite) population sizes, NG ≫ 1 (G = F, M), we obtain
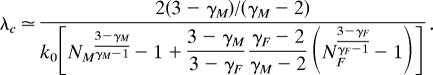 |
The final expression for λc now can be obtained by using the closure relation of Eq. 2 for the M and F population sizes, yielding
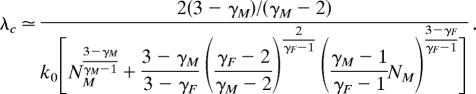 |
In this formula, only one population size NM appears. Finally, if, for example, γF > γM, the above equation reduces to
which contains simultaneously the cases when 2 < γF < 3 and γF > 3.
Now we calculate the scaling of the epidemic threshold in bipartite (heterosexual) networks, λ∗c. Manipulating Eq. 5, λ∗c can also be expressed as a function of the two exponents γM and γF and one population size:
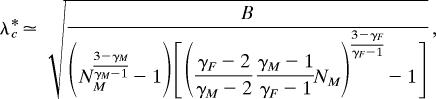 |
with B = (3 − γM)(3 − γF)/[k02(2 − γM)(2 − γF)]. The above expression, when evaluated for 2 < γG < 3 (G = F, M) and, for example, γF > γM, yields
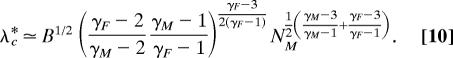 |
However, when, for example, γF > 3, the expression reduces to
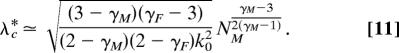 |
Comparing the Scalings.
Although both epidemic thresholds, λc and λ∗c, tend to zero as the population goes to infinity, the scaling relations, λc(NM, γM, γF) ∼ NM−α and λ∗c(NM, γM, γF) ∼ NM−α*, are characterized by two different exponents, α and α*. Table 2 reports the expression of these two exponents as a function of γF and γM, showing that α* is always smaller than α. In particular, for the most common case (see Table 1), i.e., when one degree distribution exponent is in the range ]2,3] and the other one is >3, the value of α* found for bipartite networks is two times smaller than α. As a consequence, the results show that in finite bipartite populations the onset of the epidemic takes place at larger values of the spreading rate. In other words, it could be the case that for a given transmission probability, in the unipartite representation shown in Fig. 1b, the epidemic would have survived, infecting a fraction of the population, whereas when only crossed infections are allowed, as in Fig. 1a, the same disease would not have produced an endemic state.
Table 2.
Scaling exponents of the epidemic thresholds
| Network | α* | α |
|---|---|---|
| 2 < γF < 3 | (3 − γM)/(γM − 1) | |
| γF > 3 | (3 − γM)/(γM − 1) |
Scaling exponents, α and α*, of the epidemic thresholds, λc ∼ NMα and λc ∼ NMα*, obtained for the SIS model on unipartite networks and when a bipartite network is considered, respectively. The two situations considered (2 < γF < 3 and γF > 3) correspond to 2 < γM < 3.
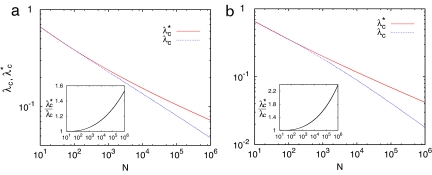
Moreover, the difference between the epidemic thresholds predicted by the two approaches increases with the system size. This dependency is shown in Fig. 2, where we have reported, as a function of the system size, the critical thresholds obtained by numerically solving Eqs. 5 and 6 with the values of γM and γF found for the lifetime distribution of sexual partners in Sweden (21) and the U.K. (22, 23).
Fig. 2.
Epidemic thresholds as a function of the total population size. The thresholds λ∗c (Eq. 5) and λc (Eq. 6) obtained for bipartite and unipartite networks are plotted as a function of the population size N for two networks with degree distributions as those found for the sexual networks of Sweden (a) (21) and the U.K. (b) (22). Insets show that the ratio λ∗c/λc grows as N increases, so that for a typical population size of N = 106, the thresholds for heterosexual networks are 53% (Sweden) and 130% (U.K.), respectively, larger than the values expected for the unipartite networks.
Numerical Simulations.
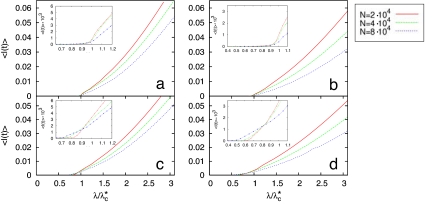
To check the validity of the analytical arguments and also to explore the dynamics of the disease above the epidemic threshold, we have conducted extensive numerical simulations of the SIS model in bipartite and unipartite computer-generated networks. Bipartite and unipartite graphs of a given size are built up (see Methods) having the same degree distributions, PkM and PkF, and thus they only differ in the way the nodes are linked. A fraction of infected individuals initially is randomly placed on the network, and the SIS dynamics is evolved: At each time step, susceptible individuals get infected with probability ν if they are connected to an infectious one and get recovered with probability μ = 1 (hence, the effective transmission probability is λ = ν). After a transient time, the system reaches a stationary state in which the total prevalence of the disease, 〈I(t)〉, is measured (see Methods). Finally, the results are averaged over different initial conditions and network realizations. Fig. 3 shows the fraction of infected individuals as a function of λ/λ∗c for several system sizes and for the bipartite (Fig. 3 a and b) and unipartite (Fig. 3 c and d) graphs. Here, the infection probability λ has been rescaled by the theoretical value λ∗c given by Eq. 5. The purpose of the rescaling is twofold. First, it allows us to check the validity of the theoretical predictions and, at the same time, provides a clear comparison of the results obtained for bipartite networks with those obtained for the unipartite case. Again we have used the values of γM and γF extracted from the lifetime number of sexual partners reported for Sweden and the U.K. (21–23). Fig. 3 indicates that the analytical solution, Eq. 5, is in good agreement with the simulation results for the two-gender model formulation. Conversely, when the bipartite nature of the underlying graph is not taken into account, the epidemic threshold is underestimated, being λc/λ∗c < 1. In addition, the error in the estimation grows as the population size increases, in agreement with our theoretical predictions.
Fig. 3.
Monte Carlo simulations for the U.K. and Sweden lifetime networks. The SIS phase diagram, 〈I(t)〉 versus λ/λ∗c, is reported for synthetic networks of different sizes. Note that for each curve, the x axis has been rescaled by the theoretical value (Eq. 5) for the critical point in bipartite networks, λ∗c, of the corresponding network size and exponents γF and γM. These exponents used in the network construction (see Methods) are those extracted from the lifetime degree distributions (see Table 1) of Sweden (21) (a and c) and the U.K. (22, 23) (b and d). The results plotted in a and b correspond to the SIS model on a bipartite network, whereas those shown in c and d correspond to a unipartite substrate. The numerical results clearly indicate (see Insets) the validity of the analytical predictions for the epidemic threshold in heterosexual (bipartite) contact networks and the underestimation of the epidemic threshold when a unipartite substrate is used.
Conclusions
The inclusion of the bipartite nature of contact networks to describe crossed infections in the spread of STDs in heterosexual populations is seen to strongly affect the epidemic outbreak and leads to an increase of the epidemic threshold. Our results show that, even in the cases when the epidemic threshold vanishes in the infinite network size limit, the epidemic incidence in finite populations is less dramatic than actually expected for unipartite SF networks. The results also point out that the larger the population, the greater the gap between the epidemic thresholds predicted by the two models, therefore highlighting the need to accurately take into account all of the available information on what heterosexual contact networks look like. Our results also have important consequences for the design and refinement of efficient degree-based immunization strategies aimed at reducing the spread of STDs. In particular, they pose new questions on how such strategies have to be modified when the interactions are further compartmentalized by gender and only crossed infections are allowed. We finally stress that the present approach is generalizable to other models for disease spreading (e.g., the “susceptible–infected–removed” model) and other processes in which crossed infection in bipartite networks is the mechanism at work.
Methods
Bipartite Network Construction.
Synthetic bipartite networks construction starts by fixing the number of males, NM, and the two exponents γM and γF of the power-law degree distributions corresponding to males and females, respectively. The first stage consists of assigning the connectivity kiM (i = 1, …, NM) to each member of the male population by generating NM random numbers with probability distribution PkM = AM k−γM (Σk0∞ AM k−γM = 1, with k0 = 3). The sum of these NM random numbers fixes the number of links Nl of the network. The next step is to construct the female population by means of an iterative process. For this purpose, we progressively add female individuals with a randomly assigned degree following the distribution PkF = AF k−γF (Σk0∞ AF k−γF = 1, with k0 = 3). Female nodes are incorporated until the total female connectivity reaches the number of male edges, Σi kiF ≤ Nl. In this way, one sets the total number of females NF. Once the two sets of NM males and NF females with their corresponding connectivities are constructed, each one of the Nl male edges is randomly linked to one of the available female edges, avoiding multiple connections. Finally, those few female edges that did not receive a male link in the last stage are removed, and the connectedness of the resulting network is checked.
Unipartite Network Construction.
Synthetic unipartite networks have been constructed in two ways. The simplest way consists of taking the two sets of NM males and NF females constructed for the bipartite network and apply a rewiring process to the entire population, i.e., allowing links between individuals of the same sex. In the second method, a set of N = NM + NF individuals whose connectivities are randomly assigned following the degree distribution P(k) = (NM/N) PkM + (NF/N) PkF is generated before applying a wiring process between all pairs of edges. In both methods, the wiring process avoids multiple and self connections, and those isolated edges that remain at the end of the network construction are removed. The connectedness of the networks also is checked.
Numerical Simulations of SIS Dynamics.
Monte Carlo simulations of SIS dynamics are performed by using networks of sizes ranging from N = 2 × 104 to N = 8 × 104. The initial fraction of infected nodes is set to 1% of the network size. The SIS dynamics is initially evolved for a time of typically 104 time steps, and after this transient the system is further evolved over consecutive time windows of 2 × 103 steps. In these time windows, we monitor the mean value of the number of infected individuals, 〈I(t)〉. The steady state is reached if the absolute difference between the average number of infected individuals of two consecutive time windows is .
ACKNOWLEDGMENTS.
We thank K. T. D. Eames and J. M. Read for their useful suggestions. Y.M. was supported by Ministerio de Educación y Ciencia through the Ramón y Cajal program. This work was partially supported by Spanish DGICYT Projects FIS2006-12781-C02-01 and FIS2005-00337 and by the Italian TO61 Istituto Nazionale di Fisica Nucleare project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Anderson RM, May RM, Anderson B. Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford Univ Press; 1992. [Google Scholar]
- 2.Daley DJ, Gani J. Epidemic Modeling. Cambridge, UK: Cambridge Univ Press; 1999. [Google Scholar]
- 3.Murray JD. Mathematical Biology. Berlin: Springer; 2002. [Google Scholar]
- 4.Hufnagel L, Brockmann D, Geisel T. Proc Natl Acad Sci USA. 2004;101:15124–15129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guimera R, Mossa S, Turtschi A, Amaral LAN. Proc Natl Acad Sci USA. 2005;102:7794–7799. doi: 10.1073/pnas.0407994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colizza V, Barrat A, Barthélemy M, Vespignani A. Proc Natl Acad Sci USA. 2006;103:2015–2020. doi: 10.1073/pnas.0510525103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert R, Barabási A-L. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 8.Newman MEJ. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 9.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU. Phys Rep. 2006;424:175–308. [Google Scholar]
- 10.Callaway DS, Newman MEJ, Strogatz SH, Watts DJ. Phys Rev Lett. 2000;85:5468–5471. doi: 10.1103/PhysRevLett.85.5468. [DOI] [PubMed] [Google Scholar]
- 11.Cohen R, Erez K, ben Avraham D, Havlin S. Phys Rev Lett. 2001;86:3682–3685. doi: 10.1103/PhysRevLett.86.3682. [DOI] [PubMed] [Google Scholar]
- 12.Pastor-Satorras R, Vespignani A. Phys Rev Lett. 2001;86:3200–3203. doi: 10.1103/PhysRevLett.86.3200. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd AL, May RM. Science. 2001;292:1316–1317. doi: 10.1126/science.1061076. [DOI] [PubMed] [Google Scholar]
- 14.Moreno Y, Pastor-Satorras R, Vespignani A. Eur Phys J B. 2002;26:521–529. [Google Scholar]
- 15.Newman MEJ. Phys Rev E. 2002;66 016128. [Google Scholar]
- 16.Read JM, Keeling MJ. Proc R Soc London B. 2003;270:699–708. doi: 10.1098/rspb.2002.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Read JM, Keeling MJ. Theo Pop Biol. 2006;70:201–213. doi: 10.1016/j.tpb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Eubank S, Guclu H, Anil-Kumar VS, Marathe MV, Srinivasan A, Toroczkai Z, Wang N. Nature. 2004;429:180–184. doi: 10.1038/nature02541. [DOI] [PubMed] [Google Scholar]
- 19.Eames KTD, Keeling MJ. Proc Natl Acad Sci USA. 2002;99:13330–13335. doi: 10.1073/pnas.202244299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colizza V, Pastor-Satorras R, Vespignani A. Nat Phys. 2007;3:276–282. [Google Scholar]
- 21.Liljeros F, Edling CR, Amaral LAN, Stanley HE, Aberg Y. Nature. 2001;411:907–908. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- 22.Fenton KA, Korovessis C, Johnson AM, McCadden A, McManus S, Wellings K, Mercer CH, Carder C, Copas AJ, Nanchahal K, et al. The Lancet. 2001;358:1851–1854. doi: 10.1016/S0140-6736(01)06886-6. [DOI] [PubMed] [Google Scholar]
- 23.Schneeberger A, Mercer CH, Gregson SA, Ferguson NM, Nyamukapa CA, Anderson RM, Johnson AM, Garnett GP. Sex Transm Dis. 2004;31:380–387. doi: 10.1097/00007435-200406000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Latora V, Nyamba A, Simpore J, Sylvette B, Diane S, Sylvere B, Musumeci S. J Med Vir. 2006;78:724–729. doi: 10.1002/jmv.20614. [DOI] [PubMed] [Google Scholar]
- 25.De P, Singh AE, Wong T, Yacoub W, Jolly AM. Sex Transm Inf. 2004;80:280–285. doi: 10.1136/sti.2003.007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freiesleben de Blasio B, Svensson A, Liljeros F. Proc Natl Acad Sci USA. 2007;104:10762–10767. doi: 10.1073/pnas.0611337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris M. Nature. 1993;365:437–440. doi: 10.1038/365437a0. [DOI] [PubMed] [Google Scholar]