Abstract
Large-scale affinity purification and mass spectrometry studies have played important roles in the assembly and analysis of comprehensive protein interaction networks for lower eukaryotes. However, the development of such networks for human proteins has been slowed by the high cost and significant technical challenges associated with systematic studies of protein interactions. To address this challenge, we have developed a method for building local and focused networks. This approach couples vector algebra and statistical methods with normalized spectral counting (NSAF) derived from the analysis of affinity purifications via chromatography-based proteomics. After mathematical removal of contaminant proteins, the core components of multiprotein complexes are determined by singular value decomposition analysis and clustering. The probability of interactions within and between complexes is computed solely based upon NSAFs using Bayes' approach. To demonstrate the application of this method to small-scale datasets, we analyzed an expanded human TIP49a and TIP49b dataset. This dataset contained proteins affinity-purified with 27 different epitope-tagged components of the chromatin remodeling SRCAP, hINO80, and TRRAP/TIP60 complexes, and the nutrient sensing complex Uri/Prefoldin. Within a core network of 65 unique proteins, we captured all known components of these complexes and novel protein associations, especially in the Uri/Prefoldin complex. Finally, we constructed a probabilistic human interaction network composed of 557 protein pairs.
Keywords: chromatin remodeling, normalized spectral abundance factor, multidimensional protein identification technology
The assembly of protein interaction networks provides critical insight into the interrelationships of multiprotein complexes and the interconnections of their respective functions. To date, the study of protein interaction networks has largely been derived from yeast two-hybrid analyses in model organisms (1, 2) and higher eukaryotes (3, 4) and from large-scale affinity purification and mass spectrometry (APMS) analyses in the model organisms Saccharomyces cerevisiae (5, 6) and in humans (7). Although all of these approaches and datasets have proven to be highly valuable sources of information, the large-scale APMS analyses in yeast and humans were designed to determine the confidence of protein complex membership (5–7). Binary interactions, based on the presence and absence of proteins in purifications, are typically reported (8). In particular in yeast, the mathematical approaches for assembling protein complexes relies on very large-scale datasets (9) and on the reciprocity of bait and prey interactions where as many preys as possible are also baits (5, 6). Collins et al. (9) reported that applying such methods to a relatively small dataset resulted in less successful identification of protein–protein interactions. This raises the question of whether a human protein interaction network can be assembled from focused studies if a systematic dataset, which would require thousands of costly APMS experiments to generate, is not available. To address this challenge, we have developed a method for building probabilistic local networks that will allow focused studies of smaller-scale networks.
The mammalian TIP49a (Rvb1) and TIP49b (Rvb2) proteins (hereafter refer to as TIP49a/b) belong to an evolutionary conserved family of AAA+ ATPases and are involved in multiple protein complexes. In S. cerevisiae, TIP49a/b are subunits of two distinct ATP-dependent chromatin remodeling complexes SWR1 (10, 11) and INO80 (12, 13). Protein complexes that share components are difficult to be computationally separated and analyzed. The complexity of such analysis was shown in yeast, where, for instance, the portion of the protein interaction network that includes the SWR1, INO80, and NuA4 complexes was grouped as one large module by using the Markov Clustering procedure (14), a key mathematical component used for the large-scale yeast APMS studies (5). In humans, TIP49a/b are components of at least four multiprotein complexes that play roles in chromatin remodeling [SRCAP (15), hINO80 (16), TRRAP/TIP60 (17), or nutrient sensing (Uri/Prefoldin (18)]. The complexity of the TIP49a/b local network in humans presents the analytical challenge of distinguishing these complexes from one another.
Previous protein interaction network analyses have not taken advantage of quantitative shotgun proteomics technologies like spectral counting. The total number of peptides identifying a protein correlates strongly with the abundance of the protein (19–22). We have shown that the relative abundance of proteins can be estimated by using normalized spectral abundance factors (NSAFs) (23, 24), which are calculated from the total number of spectra identified for each protein, normalized to the protein's length and the total number of identified spectra for all proteins in the sample. Here, we show that NSAFs provide a foundation for a systematic approach to remove nonspecific interactions, define core complexes, and build a probabilistic protein interaction network.
Results
A High-Quality Dataset of Human TIP49a/b-Associated Proteins.
A total of 27 different proteins were FLAG-tagged (hereafter referred to as “baits”), expressed in and purified by affinity purification from human tissue culture cells and analyzed by MudPIT [supporting information (SI) Fig. 5], leading to the identification of 1,278 nonredundant (NR) proteins (SI Table 1 A and B). Parallel analyses of 35 negative controls (extracts from untransformed parental cells passed through Flag affinity purification and analyzed by MudPIT) identified 812 NR proteins (SI Table 2 A and B). A crucial step in analyzing proteomics data is unraveling the subset of specific proteins from the nonspecific binders, i.e., contaminant proteins. To do so, we represented each detected protein (hereafter referred to as “prey”) as two vectors consisting of the NSAF values for each of the specific and the negative purifications, respectively. We calculated the vector ratio magnitude between the two sets (α) as a way to extract contaminants. A protein was considered a contaminant if α was >1, suggesting the protein was more abundant in the negative controls than in the specific experiments. After purging, the remaining 945 proteins were used for further analysis. Next, we constructed a matrix A (27 × 945), with the matrix element Aij representing the normalized spectral count, i.e., NSAF, for prey i and bait j, and applied singular value decomposition (SVD) (25) to extract the proteins enriched from the immunoprecipitations by using a rank estimated method. The resulting 125 proteins (SI Table 1C) included all previously reported members of the SRCAP (15), hINO80 (16), and TRRAP/TIP60 (17) multiprotein complexes and were subsequently used to determine the core complexes.
Determination of Protein Complexes.
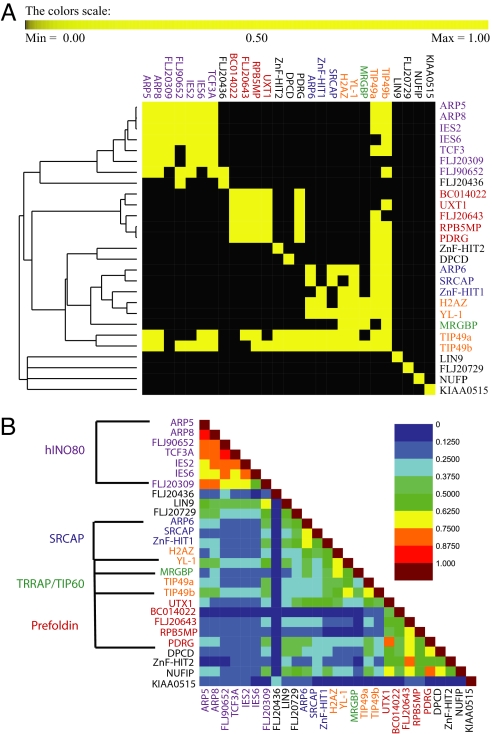
We first focused our analysis on a cluster procedure based on reciprocal pull-down of bait pairs. This resulted in five main groups corresponding to (i) baits for which there was no or little reciprocal pull-down with other purifications, (ii) hINO80, (iii) Uri/prefoldin, (iv) SRCAP/TRRAP/TIP60 complexes, and (v) a cluster containing TIP49a and TIP49b, which also belong to groups 2, 3, and 4 (Fig. 1A). To determine the similarity between purifications, we then calculated Jaccard indices between each of the bait pairs. Because the Jaccard index is proportional to the number of overlapping preys between two baits, it is expected that baits found in the same cluster have a high similarity index. In the symmetric matrix of Jaccard indices (Fig. 1B), 20 of the baits were partitioned in four different groups, three of which correspond to the well characterized hINO80 (16), SRCAP (15), and TRRAP/TIP60 (17) complexes, which function as chromatin-modifying and remodeling complexes. The fourth group corresponds to the recently described Uri/prefoldin complex, which has poorly understood roles in nutrient sensing and TOR signaling (18). The remaining seven baits, KIAA0515, FLJ20436, FLJ20729, NUFIP, DPCD, ZnF-HIT2, and LIN9, could not be considered core components of any of these four complexes according to the clustering procedure used in Fig. 1A. However, DPCD, NUFIP, and ZnF-HIT2 had high Jaccard indices with components of the Uri/prefoldin complex, whereas LIN9, FLJ20729, and FLJ20436 showed some prey overlap with elements of the hINO80 complex (Fig. 2).
Fig. 1.
Similarity and organization of bait-dependent analyses. (A) A symmetrical matrix was constructed based on the reciprocal pull-down for each baits pair as described in Materials and Methods. Each bait pair is labeled black (0) for no reciprocal pull-down and yellow (1) for reciprocal pull-down. These values were then hierarchical clustered by using the Euclidian distance metric and UPGMA as a method. A total of four clusters were identified corresponding to the SRCAP, hINO80, and TRRAP/TIP60 and Uri/Prefoldin complexes. (B) The Jaccard index value for each MudPIT analysis of a given bait is shown in a symmetrical 27 × 27 matrix. As the color progresses from black to maroon, the similarity between the two baits becomes greater.
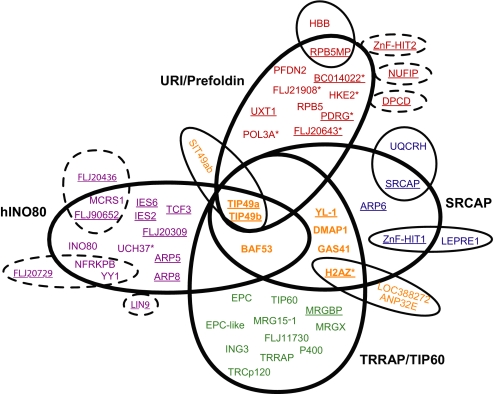
Fig. 2.
Assembly of protein complex cores and attachments. Using the rules described in Materials and Methods, four complexes were assembled and represented in the Venn diagram that included the SRCAP (blue), hINO80 (violet), TRRAP/TIP60 (green), and Uri/Prefoldin (red) complexes. Underlined proteins are the baits used in this study. Proteins with asterisks are previously undescribed core components of these complexes. The proteins that were shared between the four complexes, i.e., modules, are colored dark orange. The attachments have the same color as the protein they associate with and were detected only in single baits. The six baits that were not part of the core components but shared significant prey recovery with hINO80 or Uri/Prefoldin are circled with dashed lines.
Next, we predicted that all prey proteins overlapping between the baits within the same group and lying above a threshold form the actual complexes, where a prey protein had to appear in at least half of the baits used to define a given complex. The prey proteins that belonged to a single complex and were not shared by the other complexes are defined as the core components of the corresponding complex. Overall, the results obtained through this approach were consistent with reports from the literature (15–17). In addition to already-known components of the Uri/Prefoldin complex (18), we identified six additional subunits: HKE2, BC014022, POL3A, PDRG, FLJ21908, and FLJ20643. H2AZ was also assigned as a bona fide component of the SRCAP complex (15).
Several prey proteins in the dataset were part of more than one complex and were defined as modules (Fig. 2). A module can be two or more proteins. Examples of modules in this dataset include TIP49a/b, which were core components of the four complexes. BAF53 has been shown to form a complex with TIP49a/b (26); in the current study, BAF53 is present in SRCAP, hINO80, and TRRAP/TIP60, and in these complexes likely forms a module with TIP49a/b. In addition, DMAP1, GAS41, H2AZ, and YL-1 are present in SRCAP and TRRAP/TIP60 and may also form a module. Other proteins were strongly associated with only one bait and were defined as attachments (Fig. 2). Of particular interest to the local TIP49a/b protein interaction network is the uncharacterized protein FLJ21945 that we named as specific interactor with TIP49a/b (SIT49ab). Although TIP49a/b were detected in most purifications, SIT49ab was present only in the MudPIT analyses of TIP49a/b affinity purifications. The baits DPCD, NUFIP, and ZnF-HIT2 recovered most of the URI/Prefoldin complex, although none of them was ever detected in purifications by using bona fide core components as baits or in each other preparations. Similarly with hINO80, FLAG-LIN9 interacted with half of the complex, FLAG-FLJ20729 specifically pulled-down YY1 and NFRKPB, and FLAG-FLJ20436 reciprocally associated with FLJ90652 and MCRS1. This indicates that subassemblies could occur (Fig. 2).
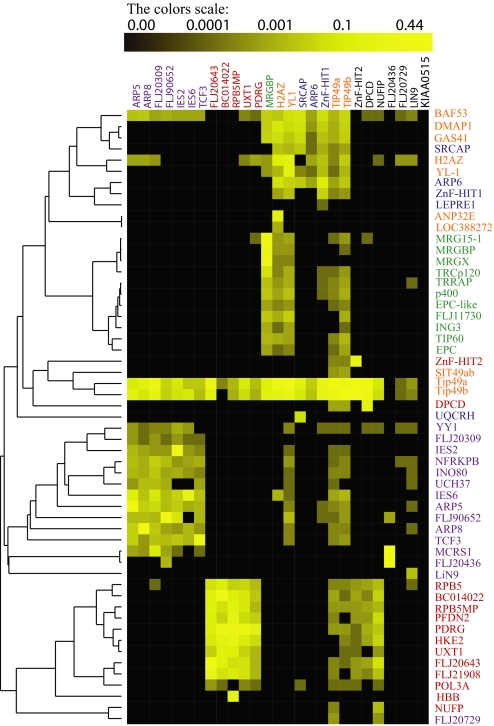
In the analysis of protein complexes, a clear distinction is sometimes made between core components and modules or attachments (6). Core components are normally stably associated with the complex and are experimentally recovered in reproducible stoichiometric yields. In contrast, modules and attachments, which may modulate the activity of the core complex, are often loosely or transiently associated with a specific protein or module and recovered in substoichiometric yields (27, 28). To assess these features, we performed hierarchical clustering analyses on the 27 immunoaffinity purifications (Fig. 3). The relative protein abundances expressed as NSAF values were clustered by using Pearson correlation as a distance metric and unweighted paired group average linkage (UPGMA) as a method (see SI Text). The results of the cluster analysis demonstrate that the core components of the complexes were well separated and partitioned at the major branches of the dendrogram (Fig. 3). Interestingly, all of the previously undescribed core components of the Uri/Prefoldin complex showed similar abundance levels as the known components of the complex, indicating strong interactions with the complex (Fig. 3). This analysis strongly suggests that quantitative proteomics values based on NSAFs can be used to group and order proteins across multiple experiments and to identify protein interactions.
Fig. 3.
Hierarchical clustering. A hierarchical cluster using the UPGMA algorithm and Pearson correlation as distance metric was performed on the relative proteins abundances expressed as NSAFs. Each column represents an isolated purification (bait), and each row represents an individual protein (prey). The color intensity represents protein abundance, with the brightest yellow indicating highest abundance and decreasing intensity indicating decreasing abundance. Black indicates that the protein was not detected in a particular sample. The protein complexes are hINO80 (violet), URI/Prefoldin (red), TRRAP/TIP60 (green), and SRCAP (blue). The modules are colored in orange.
Probabilistic Network Analysis with the Bayes Classifier.
During the partitioning of the proteins into complexes, 10 other proteins consistently copurified with only a subset of baits in a complex. Because these proteins were not contaminants, they could either be essential for the synthesis/folding/stability/function of one or more components of the four major complexes or, alternatively, could represent a physical association outside these complexes. For instance, both human TIP49a/b and NOP5/NOP58 are known to interact with U14 snoRNA (29). Likewise, SRCAP is capable of remodeling chromatin by catalyzing the incorporation of H2AZ/H2B dimers into nucleosomes, perhaps explaining the specific presence of H2B in the purifications. Therefore, these 10 proteins, along with the 43 deemed core components of the complexes (Fig. 2), the six baits that could not be stringently assigned to the complexes (Fig. 1), and the six proteins that were considered attachments (Fig. 2), were included in the final TIP49a/b interaction map containing 65 proteins. The bait KIAA0515 was clearly not part of the TIP49a/b network and was not considered any further (Fig. 3). The remaining 59 proteins that did not pass any of the criteria described above are considered highly frequent proteins, such as chaperones and ribosomal proteins, and were deliberately removed.
Although binary representation of APMS data has been successfully used to predict protein complexes and protein interactions, quantitative information based on NSAF could be a useful alternative to ascertain these predictions. Therefore, we used a probabilistic model for a protein interaction network that provides quantitative information for each interaction. In this model, each pair of proteins (bait–prey) received a probability computed only from the observed experimental NSAF values by using a Bayesian approach. For a bait–prey pair, the resulting probability quantifies the preference of the prey to associate with the bait.
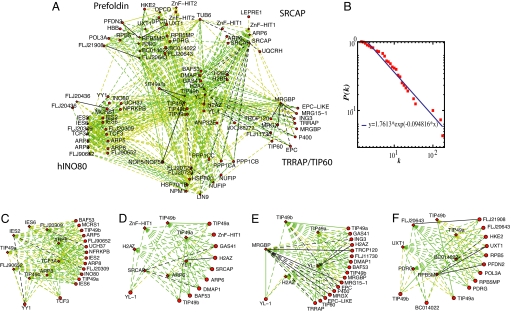
As suggested (30, 31), we used these probabilities to construct a probabilistic network of human TIP49a/b-containing complexes. To visualize our complexes, probabilistic networks were displayed in Fig. 4 by using the Cytoscape software environment (32). We used a posterior probability cutoff of 0.1 to define relatively high probability for two proteins to associate (black dashed lines), 0.01 for relatively moderate probability (green dashed lines), and 0.001 for relatively low probability (yellow dashed lines) (Fig. 4 A and C–F). The complete set of protein pairs and their corresponding probabilities is reported in SI Table 3. The plot of node degree distribution P(k) of human TIP49a/b-associated complexes generated by our core data of 557 protein interactions between 65 different human proteins followed an exponential decay (Fig. 4B).
Fig. 4.
Probabilistic network of TIP49a/TIP49b interactions. (A) The entire network is shown, in which nodes represent protein baits (depicted by diamonds) or protein preys (circles), and the weighted edges represent the calculated posterior probability. Black dashed lines represent the interactions with the highest probability, green dashed lines represent interactions with a moderate probability, and yellow dashed lines represent interaction with the lowest probability. Solid lines represent protein–protein interactions validated by the literature or the present study. (B) The node degree connectivity of the overall network. Focused probabilistic displays of the human INO80 (C), SRCAP (D), TRRAP/TIP60 (E), and the Uri/Prefoldin F complexes are shown with the same color coding as in A.
Validation of Predicted Interactions.
In vivo coimmunoprecipitation assays were conducted to test several interactions within the TRRAP/TIP60 complex (SI Fig. 6). This orthogonal analysis confirmed the interactions between MRGBP and TRCp120, DMAP, MRG15, TIP60, and YL1 (SI Fig. 6 A–E), which were all predicted with medium to high probabilities in our analysis (SI Table 4). The distinct interaction between SIT49ab (FLJ21945) and TIP49a/b was also confirmed in two separate experiments (SI Fig. 6 F–G). In addition, we systematically analyzed the human protein reference database (33) for additional confirmations of high-probability protein associations (SI Text). For instance, our analysis predicted strong association between YL-1 and H2AZ, which is supported by the demonstrated interaction between the S. cerevisiae orthologs of YL-1 (Swc2) and H2AZ (Htz1) (34). In TRRAP/TIP60, MRGBP has the highest probability of interaction with MRGX followed by MRG15–1, and these probabilities were the eighth and ninth highest in the entire dataset. Cai et al. (35) have shown that MRGBP-MRGX heterodimers, MRGBP-MRG15 heterodimers and MRGBP-MRG15-MRGX heterotrimers can be resolved by analytical superpose 6 gel filtration of a FLAG-MRGBP eluate.
Discussion
In this study, we have demonstrated the value of quantitative proteomics for organizing proteins into complexes and for generating probabilistic interaction networks. To begin, NSAF values are valuable for the extraction of contaminants. Many proteins known to be part of complexes can also be found in negative control purifications. By comparing the level of protein abundance between samples and an equal number of negative control purifications, we are able to separate those proteins that are quantitatively enriched in the samples over the negative controls. Indeed, all known components of each of the complexes were faithfully recovered among the putative true positive sets. Other large-scale studies, which did not use negative control runs, removed only those proteins that appear in more than a certain percentage of the purifications (5, 6, 36). If we were to take this approach with the current dataset, we would have to remove TIP49a and TIP49b from the datasets, even though these proteins are the foundation of this network.
We devised a strategy that uses normalized spectral counts to generate a probabilistic measure of the preference of proteins to interact with one another. The probability between two proteins is calculated from the bait-to-prey relationship alone, whereas other methods require reciprocal bait–prey interactions or copurification of preys by a third bait (5, 6). By using this approach, we assigned probabilities not only to the interactions inside the complexes but also to the interactions outside the complexes. For instance, for SIT49a/b, which is not part of the four complexes, we were able to assign a high probability to TIP49a/b that was experimentally verified. The same holds true for FLJ20436, which forms an external interaction with the MCRS1 component of hINO80 complex.
Thus far, there is no precise way to predict direct interaction based on APMS data. Nonetheless, there is a possibility that some pairs with high probability could form a direct contact. This information is particularly important when designing focused experimentation to disrupt particular interactions within a network. For example, the highest-probability pairs predicted for subunits of hINO80 were IES2/YY1 and IES2/FLJ20309 (Fig. 4C). The portion of the network containing the transcription factor YY1, a member of the hINO80 complex, is especially important, because overexpression of YY1 is strongly implicated in cancer development (37). In fact, although YY1 is clearly a member of hINO80, YY1 is linked to the Prefoldin complex via the protein DPCD and the SRCAP complex via ZnF-HIT1 (Fig. 4). As could be the case for therapeutic targeting of YY1 involvement in cancer (37), calculating probability-based interaction networks should result in superior model building. This is an advantageous starting point before chemical modulation of protein interaction networks when targeting specific protein–protein interactions for disruption, potentially improving treatments of human disease.
Materials and Methods
Identification of Proteins by MudPIT.
The cloning, expression, and purification of the human TIP49a, TIP49b, Arp8, PAPA-1 (hIES2), C18orf37 (hIes6), TCF3-Amida, and FLJ90652 full-length proteins and a fragment of FLJ20309 (residues 106–544) were reported by Jin et al. (16). N-terminally FLAG-tagged human MRGBP, YL-1, ZnF/HIT1, and H2AZ were obtained as described by Cai et al. (17). SCRAP-associated proteins were purified as described by Ruhl et al. (15).
Full-length cDNAs encoding the human ZnF/HIT2, ARP5, ARP6, PDRG, UXT1, BC014022, FLJ20643, FLJ20436, FLJ21908, NUFIP, LIN9, FLJ20729, and DPCD proteins were obtained from the American Type Culture Collection (ATCC), subcloned with FLAG tags into pcDNA5/FRT, and introduced into HEK293/FRT cells by using the Invitrogen Flp-in system as reported (17). Next, TIP49a- and TIP49b-associating proteins were purified by anti-FLAG agarose immunoaffinity chromatography as described by Jin et al. (16). As a control for the specificity of immunoaffinity purifications, extracts prepared from untransformed parental cells (23 independent preparations from HeLa and 12 from HEK/293 cells) were subjected to the same procedure (SI Fig. 5). Identification of proteins was accomplished by Multidimensional Protein Identification Technology (MudPIT) as described (38), and details are provided in SI Text. Protein spectral counts were converted to the NSAF for subsequent analysis (SI Text).
Contaminant Extraction.
In this study, we define contaminants as follows: for M purifications and N identified proteins, let xij be the NSAF value of ith identified protein and jth purification. The vector [xi1 xi2… xiM] represents the protein vector with 1 ≤ i ≤ N, and 1 ≤ j ≤ M. Similarly, let yij represent the NSAF value of ith identified protein in the negative controls and jth control purification. The vector [yi1 yi2… yiM] represent the negative control protein vector. For each protein with two vectors x and y, the vector magnitude (α) is calculated as:
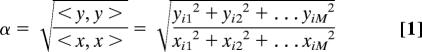 |
α > 1 indicates that the value expected as y is a “greater” vector. The symbols <, > represent the inner product or simply norm of a vector and are defined as the root-square of the sum of each term of the vector taken at square (39). A protein with a value of α > 1 was considered contaminant and was excluded from the data, leading to the removal of 336 proteins. A visual examination was performed to ensure that the removed proteins were nonspecific. The remaining 945 proteins were subjected to SVD analysis.
SVD.
SVD is an established method (25, 39–41), and a mathematical definition is provided in SI Text. Here, we used SVD to find a group of proteins in the dataset that contributes most to the matrix by using a ranking estimation method. SVD analysis revealed that the first singular value and associated singular vectors contribute the most to the matrix, restricting our subsequent analysis to the first left singular vector (lsv). The first lsv represents a weighted average and distinguishes proteins by their averaged overall expression. The coefficients of the first lsv were sorted based on their magnitudes. In this analysis, coefficients were retained if their magnitudes were larger than a cutoff ≈0.002. The significance of the cutoff is that it provides a scale-independent way to determine the proteins that were enriched from the immunoprecipitation experiments while reducing the excess noise. Using this cutoff, 125 proteins were found corresponding to the most essential proteins in the dataset. More importantly, these 125 proteins contained all reported members of the SRCAP (15), hINO80 (16), and TRRAP/TIP60 (17) multiprotein complexes.
Definition of Protein Complexes.
A symmetric binary matrix was constructed based on reciprocal pull down of the baits. For two baits, a value of 1 was assigned if they copurify in both direction (i.e., if one protein is prey in the purification by using the second protein as a bait and vice versa) and 0 otherwise. Hierarchical clustering was then applied to the binary matrix. Based on the resulting matrix (Fig. 1A), TIP49a and TIP49b copurify bidirectionally with the majority of the remaining baits and accordingly were assigned to all of the complex clusters; similarly H2AZ was assigned to the two clusters corresponding to SRCAP and TRRAP/TIP60 complexes.
Assuming that the baits belonging to the same cluster should generally pull-down common proteins, we verified this by calculating a similarity value defined by the Jaccard index to each of the bait pairs. Given two sets of purifications A and B, na and nb count the number of proteins in individual purifications A and B, and ni is the number of proteins present in both purifications. The Jaccard index is defined as the ratio between the number of proteins present in both sets of purifications and the number of proteins present in either one:
When two baits share a large number of proteins, the coefficient shows a value close to 1. By contrast, it has a value close to 0 if the two baits do not copurify with many common proteins. The pairs of baits with high indices are more likely to be part of the same cluster. The overlapping proteins between the baits found in the same group are sorted based on the number of times they are detected out of n bait purifications. In each complex, the bait that pulls down the lowest number of shared proteins determines the threshold below which prey proteins are not considered subunits of the complex.
Derivation of the Posterior Probabilities.
Our goal was to compute a probability for each bait–prey interaction pair based on the NSAF (SI Text).
To quantify the association preference between an affinity candidate protein i (i = 1,…, N) and a protein bait j (j = 1,…, M), we first estimated the conditional probability by:
where P(i,j) is the joint probability of association involving protein i and protein j and is defined as:
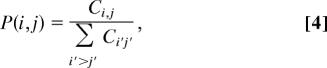 |
where Ci,j is the NSAF value of protein i in bait j, whereas Σi′≥j′ Ci′j′ sums the total NSAFs.
P(j) is the likelihood that protein j participates in an association and is estimated by:
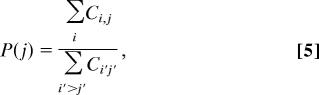 |
where Σi Cij sums the NSAF values of protein i in the bait j. When the conditional probability is known, we can calculate next the probability of protein i by using:
where the summation is over all possible values of j.
For a bait, l, j, and prey, i, the posterior probability P(j|i) defined by Bayes' rule:
quantifies the preference of a prey to associate with a bait. Because of the lack of previously published human protein interaction data for some of the proteins, no prior knowledge was incorporated in our analysis. Similarly to previous studies, in which external prior information is avoided (5, 42), we assumed that each of the proteins i in the dataset occurred with equal probability of 1/N. The posterior probability was calculated in house by using a computing C language. The program implementing the method described, and the source code is freely available from the authors upon request.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Arcady Mushegian, Matthias Wahl, and Timothy Doerr for valuable discussions during the preparation of this manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706983105/DC1.
References
- 1.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 4.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 6.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 7.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X, Gerstein M, Snyder M. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 9.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 12.Shen X, Mizuguchi G, Hamiche A, Wu C. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 13.Shen X, Ranallo R, Choi E, Wu C. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 14.Pu S, Vlasblom J, Emili A, Greenblatt J, Wodak SJ. Proteomics. 2007;7:944–960. doi: 10.1002/pmic.200600636. [DOI] [PubMed] [Google Scholar]
- 15.Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 16.Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, et al. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 18.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 19.Blondeau F, Ritter B, Allaire PD, Wasiak S, Girard M, Hussain NK, Angers A, Legendre-Guillemin V, Roy L, Boismenu D, et al. Proc Natl Acad Sci USA. 2004;101:3833–3838. doi: 10.1073/pnas.0308186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Sadygov RG, Yates JR., III Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 21.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Zybailov B, Coleman MK, Florens L, Washburn MP. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 23.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Weliky Conaway J, Florens L, Washburn MP. Proc Natl Acad Sci USA. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 25.Alter O, Brown PO, Botstein D. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, Wood MA, Cole MD. Mol Cell Biol. 2002;22:1307–1316. doi: 10.1128/mcb.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAfee KJ, Duncan DT, Assink M, Link AJ. Mol Cell Proteomics. 2006;5:1497–1513. doi: 10.1074/mcp.T500027-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 29.Watkins NJ, Dickmanns A, Luhrmann R. Mol Cell Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asthana S, King OD, Gibbons FD, Roth FP. Genome Res. 2004;14:1170–1175. doi: 10.1101/gr.2203804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Nat Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 32.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, Wu C. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 35.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 36.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 37.Gordon S, Akopyan G, Garban H, Bonavida B. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 38.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuruvilla FG, Park PJ, Schreiber SL. Genome Biol. 2002;3:RESEARCH0011. doi: 10.1186/gb-2002-3-3-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fogolari F, Tessari S, Molinari H. Proteins. 2002;46:161–170. doi: 10.1002/prot.10032. [DOI] [PubMed] [Google Scholar]
- 41.Wall ME, Dyck PA, Brettin TS. Bioinformatics. 2001;17:566–568. doi: 10.1093/bioinformatics/17.6.566. [DOI] [PubMed] [Google Scholar]
- 42.Slonim N, Atwal GS, Tkacik G, Bialek W. Proc Natl Acad Sci USA. 2005;102:18297–18302. doi: 10.1073/pnas.0507432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.