Abstract
Bcl-2 inhibitor of transcription (Bit1) is a mitochondrial protein that functions as a peptidyl-tRNA hydrolase, but, when released into the cytoplasm, it elicits apoptosis. The proapoptotic function is uniquely counteracted by integrin-mediated cell attachment. We generated a conditional KO mouse of the Bit1 gene by using the Cre-LoxP recombination system. Bit1-null mice were born alive but with some developmental abnormalities. They developed a runting syndrome after birth and died within the first 2 weeks. Cultured fibroblasts from the Bit1-null embryos [mouse embryo fibroblasts (MEFs)] were more resistant to cell death induced by loss of attachment to extracellular matrix (anoikis) than cells from the wild-type or heterozygous littermates. MEFs and tissues from Bit1 KO mice displayed a marked increase in Erk phosphorylation. Knocking down Bit1 expression in cultured cells resulted in increased Erk activation, and partially knocking down Erk reversed the increased anoikis resistance of Bit1 knockdown. The enhanced Erk activation was associated with decreased Erk phosphatase activity. These studies establish the physiological significance of Bit1 activity and begin to delineate a Bit1 signaling pathway that acts through Erk regulation.
Keywords: apoptosis, cell adhesion, integrins, kinases, mitogen-activated
Bcl2-inhibitor of transcription 1 (Bit1) is a mitochondrial protein evolutionarily conserved from bacteria to humans (1–3). The 22-kDa Bit1 polypeptide of archaea and eukaryotes contains an N-terminal mitochondrial localization domain (2). This extension is absent in bacterial Bit1. A known function of Bit1 is to serve as one of the two enzymes that release tRNA from a peptidyl-tRNA complex. Hence, the protein also is known as peptidyl-tRNA hydrolase 2. The crystal structure of the human Bit1 hydrolase domain has been solved (3).
Like many mitochondrial proteins, Bit1 “moonlights” in another function: It acts as a proapoptotic factor when released into the cytosol or when experimentally expressed there (2). The proapoptotic activity of Bit1 requires the small Groucho family transcriptional regulator amino-terminal enhancer of split (AES), with which Bit1 forms a proapoptotic complex.
Bit1-induced apoptosis is uniquely regulated by integrin-mediated cell attachment. Plating cells onto fibronectin or vitronectin protects against death induced by cytoplasmic Bit1 by α5β1 and αvβ3 integrins and inhibits the formation of Bit1–AES complexes. TLE1, a member of the Groucho family that is negatively regulated by AES (4, 5), also protects against Bit1-induced apoptosis. Various antiapoptotic signaling molecules, such Bcl-2, Bcl-xL, PI-3K, and Akt, can block the release of Bit1 from mitochondria, but are unable to inhibit apoptosis induced by cytosolic Bit1. Only integrin-mediated cell attachment counteracts cytosolic Bit1. Thus, this apoptosis pathway, which functions independently of caspases by an unknown mechanism, seems to be particularly important in anoikis, which is cell death caused by the loss of cell attachment.
To examine the role of Bit1 in vivo, we generated Bit1 KO mice. In this study, we demonstrate that these Bit1-null mice die soon after birth of a runting syndrome with muscle weakness, ataxia, and decreased weight. The null tissues exhibited abnormal Erk activation that was associated with an increased resistance to anoikis by fibroblasts cultured from null embryos. Cell culture experiments further showed that Bit1 is a regulator of Erk activation.
Results
Bit1-Null Mice.
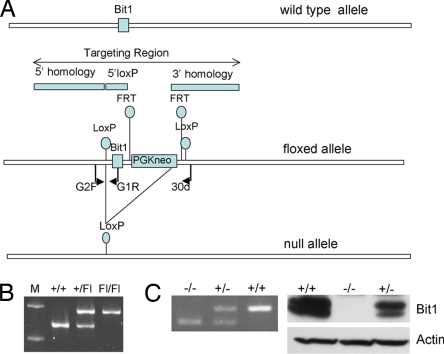
A mouse strain where the Bit1 gene was flanked by loxP sites was generated for us commercially (Fig. 1A). We intercrossed heterozygous Bit1flxNeo/+ mice to obtain homozygous floxed mice (Bit1flxNeo/flxNeo) and confirmed the genotype by using RT-PCR (Fig. 1B). Thus, it was unnecessary to remove the Neo-selectable gene used in the targeting procedure. To ensure that the floxed allele did not adversely affect Bit1 expression, we determined by using RT-PCR that the Bit1 mRNA is expressed in several tissues isolated from Bit1flxNeo/flxNeo mice, including the liver, kidney, skin, lung, gut, spleen, heart, and skeletal muscle (data not shown). Also, to determine whether the Bit1flxNeo allele affected viability or fertility, we bred homozygous Bit1flxNeo/flxNeo mice and found them to be fertile with viable pups.
Fig. 1.
Bit1 KO mice. (A) Schematic of targeting strategy. The wild-type, floxed, and null alleles are shown. (B) PCR genotyping assay to detect the floxed allele using primers G2F and G1R. M, molecular weight standard. (C) Genotyping MEFs from Bit1 wild-type (+/+), heterozygotes (+/−), and KO (−/−) mice using PCR primers G2F, G1R, and 30d (Left) and 14.5-day-old embryos using immunoblot analysis (Right). The Bit1 protein typically appears as a double band; the reason for this heterogeneity is not known.
To obtain Bit1-null mice, we crossed Bit1flxNeo/flxNeo mice with MoreCre mice and selected offspring with the Bit1flxNeo/+, Cre+ genotype. These mice were labeled as Bit1+/−, resulting from the deletion of Bit1 by the Cre recombinase in the germline. To obtain a pure line of Bit1 heterozygous mice without the Cre recombinase, we backcrossed Bit1+/−, Cre+ mice to wild-type BL/6 mice. The pure Bit1+/− were intercrossed to generate KO mice, expected as 25% of the offspring. RT-PCR analyses allowed us to identify Bit1 heterozygous and homozygous mice (Fig. 1C Left). Immunoblotting demonstrated reduced Bit1 protein expression in heterozygous mice and the absence of detectable Bit1 in null mice (Fig. 1C Right).
Bit1-Null Mice Are Postnatal-Lethal with a Runting Syndrome.
The Bit1 heterozygous mice displayed no obvious phenotypic differences relative to age-matched, wild-type littermates and were fertile. Analysis of the offspring revealed no viable Bit1-null pups at weaning, but we were able to detect Bit1-null embryos at gestation days 12.5, 14.5, and 18.5 at close to the expected Mendelian frequency (data not shown). This finding led us to believe that the null mice were dying after birth, but before weaning. We transferred all pups to ICR/FVBN strain foster mothers at birth. Using this strategy, we were able to obtain viable Bit1-null mice.
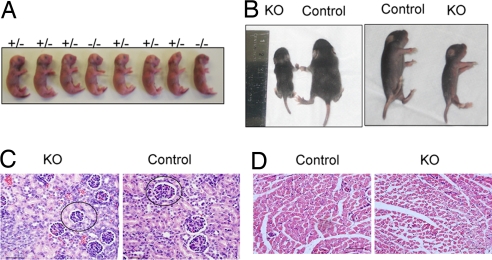
The null mice appeared normal at birth (Fig. 2A). Initial feeding behavior appeared normal because milk was present in the newborns' stomachs. Further, histological examination at embryonic day 18.5 (E18.5), just before birth, did not reveal obvious differences between null and control pups. However, when the mice were examined several days after birth, it was evident that the Bit1 nulls were smaller and weaker than age-matched littermates (Fig. 2B). The KO pups showed no signs of respiratory distress. The weakness became progressively worse, characterized by a runting syndrome with muscle wasting, ataxia, and death between postnatal day 8.5 (P8.5) and P12.5. Bit1 nulls weighed 50–60% of controls just before death (P < 0.01).
Fig. 2.
Bit1 KO mice exhibit stunted growth after birth. (A) Wild-type (+/+), heterozygous (+/−), and KO embryos delivered by C-section are shown. (B) Representative KO and control littermate mice are depicted in dorsal and side shots at approximately P10. (C and D) Bit1 KO mice exhibit delayed development. H&E-stained sections of the kidney (C) and the muscle (D). The glomeruli are smaller in the kidney than in wild-type control. A representative glomerulus is encircled. The fibers are smaller in epaxial muscle in the KO mice. This size difference was particularly striking for the older mice (P < 0.001; one-way ANOVA and Tukey test). (Size bars: 50 μm.)
Bit1-Null Mice Exhibit Delayed Development and Neutropenia.
We proceeded to examine tissues from various organs of Bit1 wild-type, heterozygous, and KO mice. We found a trend for delayed development in the KO mice relative to age-matched control littermates. In particular, the subcapsular glomeruli of the kidney were smaller in Bit1-null mice than controls (Fig. 2C), which may be due to delayed kidney maturation. In addition, there was evidence of retarded muscle development in the epaxial muscle of KOs, which had myofibers with a smaller diameter (Fig. 2D). These differences were statistically highly significant (P < 0.001; one-way ANOVA). No lung pathologies were observed. Blood analyses revealed neutropenia (neutrophilic leukopenia) in Bit1 KO mice, which had an 80% reduction in neutrophil counts relative to controls (P = 0.01). Further studies of plasma did not identify any abnormality in the liver or kidney function, and RBC parameters also were normal.
Bit1 KO Cells Are Resistant to Anoikis.
Because Bit1 protein released from the mitochondria plays a particular role in anoikis (2), we investigated the response of Bit1 KO MEFs to the loss of cell attachment. After 24 h in suspension culture, KO MEFs displayed 25–30% more viable cells than wild-type cells (P = 0.04) (Fig. 3A). At 72 h, there were almost twice as many surviving KO cells as wild-type cells (P = 0.002) (Fig. 3A). In contrast, the KO cells were more sensitive than the wild-type cells to treatment with the apoptosis-inducing kinase inhibitor staurosporine (Fig. 3B), suggesting that the Bit1 KO MEFs are selectively resistant to anoikis.
Fig. 3.
Bit1 KO MEFs are resistant to anoikis. (A) MEFs from wild-type (WT) and KO mice were grown in suspension, harvested, stained with annexin/propidium iodide, and analyzed by flow cytometry at the indicated time points. The percentage of viable cells is shown. The graphs represent MEFs isolated from at least two different WT and KO mice. (B) MEFs from WT and KO mice were treated with 0.2 μM staurosporine for 24 h. Cell death was analyzed as in A by flow cytometry (Left). Cell proliferation (Right) was assessed by using the MTS cell proliferation assay. (C and D) Bit1 KO mice display increased Erk activation. (C) Lysates were made from cultured MEFs of WT, heterozygous (HET), and KO mice. The lysates were analyzed by SDS/PAGE and immunoblotted for total and phosphorylated (active) Erk 1/2 (Thr-202/Tyr-204) and for Bit1. GAPDH immunoblotting was used as a loading control. (D) Lysates from mouse tissues were analyzed as in C.
Erk Activation Is Increased in Bit1 KO MEFs and Tissues.
Erk activation promotes cell survival (6–8), and cell attachment has been reported to affect Erk activation by suppressing a phophatase that inactivates Erk (9, 10). Hence, we investigated Erk activation (phosphorylation) in MEFs and tissues from Bit1 KO mice. We found the Erk protein levels to be equal in the KO and control samples, but the levels of phospho Erk were significantly higher in the KO MEFs (Fig. 3C). No activation of the upstream signaling molecules Akt and Mek was seen in the MEFs (data not shown). KO tissues also tended to show increased Erk phosphorylation, particularly in the kidney and the liver (Fig. 3D).
Bit1 Regulates Erk Activation.
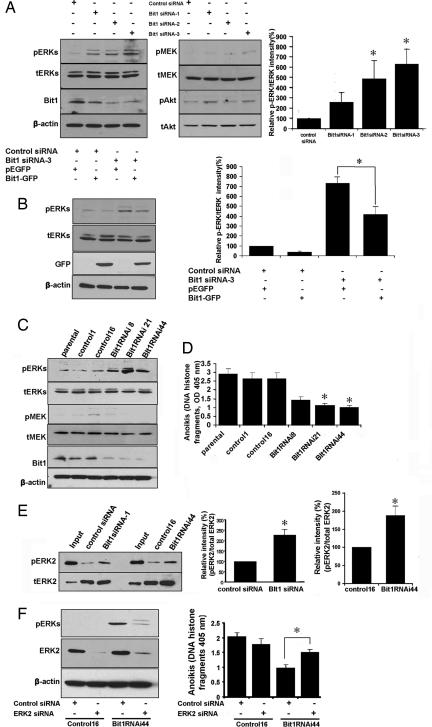
The results shown in Fig. 3 suggest that Bit1 may be involved in Erk regulation. We studied this question in the cultured human HeLa cancer cell line, which can be more easily transfected than MEFs. Transient knockdown of Bit1 in HeLa cells by using three different Bit1-specific siRNAs resulted in a robust Erk activation (phospho-Erk/total Erk ratio) compared with the control siRNA-transfected cells (Fig. 4A). Conversely, enforced overexpression of Bit1 in the Bit1 siRNA-treated cells attenuated Erk activation (Fig. 4B). We also tested the activation level of two signaling molecules upstream of Erk: Mek and Akt. There were no consistent differences in Mek or Akt activation between the knockdown and control cells, and the minor variations seen did not correlate with the level of Bit1 suppression (Fig. 4A).
Fig. 4.
Bit1 regulates Erk activation through Erk phosphatase activity, and Erk down-regulation partially restores the anoikis resistance of Bit1 KO cells. (A) HeLa cells were transfected with control siRNAs or with three different specific Bit1 siRNAs. (Left) Three days after transfection, cells were lysed, and the total lysate was subjected to immunoblotting (Left) to detect the phosphorylated Erk (pErk), total Erk (tErk), active Mek (pMek), total Mek, active Akt (pAkt), total Akt (tAkt), Bit1, and β-actin. (Right) The relative p-Erk/t-Erk ratio with the level in the control siRNA was taken as 100%. *, P < 0.05, compared with control siRNA (Student's t test). The values represent the average of four independent experimental results. (B) Three days after siRNA transfection, control siRNA- and Bit1 siRNA-treated HeLa cells were transfected with the empty vector or C-terminally GFP-tagged Bit1. (Left) After 48 h, cells were harvested, and p-Erk, t-Erk, GFP-tagged Bit1, and β-actin were detected by immunoblotting (Left). The normalized pErk/tErk intensity from three independent experiments. *, P < 0.01 (Student's t test) (Right). (C and D) Stable Bit1 knockdown and control clones were established in the HeLa cancer cell line as described in Materials and Methods. Exponentially growing, stable Bit1 knockdown cells (clones nos. 8, 21, and 44), control clones (nos. 1 and 16), and the parental cell line were harvested and subjected to immunoblotting (C) and anoikis assay; *, P < 0.001, compared with control clones (Student's t test) (D). (E) Total cell lysates from HeLa cells transfected with control siRNA or Bit1 siRNA-1, as well as lysates from stable control16 and Bit1 RNAi44 clones, were subjected to an Erk phosphatase assay as described in Materials and Methods. A representative immunoblot of isolated His-6-tagged Erk2 is shown to reveal pErk2 or total Erk2 levels. The relative intensity of pErk2/tErk2 was determined, and the values represent the average of at least three independent experiments. *, P < 0.01, compared with respective control (Student's t test). (F) Stable control 16 and Bit1 RNAi44 clones were transfected with control- or Erk2-specific siRNAs; 72 h after transfection, cells were harvested and subjected to immunoblotting with antibodies against total Erk2 and phosphorylated Erk1/2 (pErks), and to an anoikis assay. Results are representative of three independent experiments. *, P < 0.001 (Student's t test).
To further evaluate the effect of Bit1 down-regulation on Erk activity, we also established stable Bit1 knockdown clones in the HeLa cells (Fig. 4C). All three Bit1 knockdown clones exhibited decreased sensitivity to anoikis (Fig. 4D) and increased levels of phospho-Erk (Fig. 4C). The differences to the parental cells and control clones in these assays were statistically significant. Phosphorylation of the upstream kinase, Mek, was not significantly altered by the changes in Bit1 expression (Fig. 4 A and C). Instead, we found Bit1 knockdown cells to contain less Erk-directed phosphatase activity (Fig. 4E). Finally, siRNA-mediated down-regulation of endogenous Erk2, which is the Erk kinase isoform primarily affected by changes in Bit1 expression (Fig. 4 A and C), partially restored the sensitivity to anoikis in a Bit1 knockdown clone (Fig. 4F). These results show that Bit1 regulates Erk activation.
Discussion
In this study, we show that deleting Bit1 from mice results in a runting syndrome that culminates in early postnatal death. Mechanistic studies showed that MEFs from the KO mice were less susceptible to anoikis and displayed more Erk activation than wild-type MEFs. Bit1 expression and Erk activation in cultured tumor cells were inversely correlated, apparently because Bit1 regulates a phosphatase that inactivates Erk. Thus, this mouse KO study has yielded the beginnings of a molecular pathway through which Bit1 regulates anoikis.
The lethal phenotype of the Bit1 KO mice is not surprising given the constitutive expression profile of Bit1 in the adult mouse (data not shown), signifying that Bit1 may be required for general tissue homeostasis. The histological evidence of delayed development in epaxial muscle and the kidney may reflect a more important function of Bit1 in the development of these specific tissues, although blood analyses revealed no evidence of alteration in kidney function. It is not clear whether these changes, the marked neutropenia, and the general wasting relate to the proapoptotic function of Bit1 or its mitochondrial role. In addition to its proapoptotic function, Bit1 is a peptidyl-tRNA hydrolase (3), and that is its mitochondrial function at least in yeast (11). Rescue experiments with Bit1 that lacks either the proapoptotic or enzymatic function, but not both, are needed to answer these questions.
The most interesting findings from this study relate to anoikis and Erk activation. The increased anoikis resistance as a result of loss of Bit1 is in keeping with previous findings using siRNA (2), whereas the striking increase in Erk1/2 activation in Bit1 KO cells has not been noted before. The elevated Erk activity may account for the increased survival of the KO MEFs. The KO MEFs were more sensitive to the general apoptosis inducer, staurosporine (12, 13), indicating that these cells are selectively resistant to anoikis, not apoptosis in general. The target for Bit1 activity in the Erk regulation appears to be an Erk phosphatase. Erk is activated by phosphorylation on threonine and tyrosine residues and is deactivated by the removal of the phosphate (14, 15). We found no significant changes in the activity of Mek, the upstream kinase that phosphorylates Erk, and Akt activation also was not altered. Moreover, two chemical inhibitors of Mek had no effect on the anoikis sensitivity of the KO MEFs (data not shown). Rather, the Bit1 target seems to be Erk dephosphorylation because we found significantly decreased Erk phosphatase activity in Bit1 knockdown cells. Erk is a substrate of at least two protein phosphatases: MAPK phosphatase-1 and MAPK phosphatase-3 (16, 17). It remains to be determined whether these phosphatases are involved in the Bit1 regulation of Erk. Bit1 is a hydrolase, but it is unlikely to be a phosphatase, even when in complex with its obligate partner, the small Groucho protein AES (2), because there is no structural similarity between Bit1 and known phosphatases (3, 18). Hence, Bit1 may be a phosphatase activator or it could regulate one.
The proapoptotic function of cytoplasmic Bit1 is uniquely regulated by cell adhesion. Integrin-mediated adhesion to fibronectin or vitronectin was the only treatment among many antiapoptotic regimens that was capable of suppressing apoptosis initiated by cytoplasmic Bit1 (2). Interestingly, cell adhesion has been shown to regulate an Erk phosphatase (9), and a recent article indicates that this regulatory pathway likely involves vinexin β. This focal adhesion-associated scaffold protein can induce anchorage-independent activation of Erk2, through a mechanism that involves the suppression of Erk phosphatase activity (10). Our present results on Bit1 regulation of Erk activity through an Erk phosphatase are similar, and we have previously shown that Bit1, like vinexin β, is regulated by cell adhesion (2). These findings suggest that vinexin β and Bit1 may be on the same pathway. Moreover, cell cycle arrest caused by overexpression of inhibitors of cyclin-dependent kinases leads to anoikis resistance that is, at least in part, due to Erk-mediated suppression of the activity of the proapoptotic BH3-only protein Bim (19). Erk regulation in this system is independent of Ras and Mek and thought to take place at the level of Erk dephosphorylation. There may be a link between this system and Bit1 because Bit1 blocks Erk activation through a mechanism that appears to involve Erk dephosphorylation (this study), and Bit1 also inhibits the expression of the antiapoptotic protein Bcl-2 (2). Thus, Bit1 may occupy a central position in anoikis control that involves multiple mechanisms, one of which is the Erk regulation demonstrated here.
Finally, the increased Erk activation observed in Bit1 KOs may have implications in cancer. Sustained Erk activation is associated with enhanced cell survival and motility (6–8, 20, 21). Further, the cytoskeletal protein vinculin controls cell survival by regulating paxillin–focal adhesion kinase (FAK) interactions and altering Erk1/2 activation as a key step, resulting in highly metastatic and motile cells (22). These observations suggest that the loss of Bit1 in cancer cells could provide a survival advantage, making it important to study a potential role of Bit1 as a tumor-suppressor gene.
Materials and Methods
Bit1 Mice.
Bit1-floxed mice were made by Ozgene by using standard molecular cloning procedures and transfected in C57BL/6 ES cells. The floxed colony was expanded and intercrossed to obtain Bit1 +/Fl and Fl/Fl genotypes. The latter were crossed with MoreCre mice (The Jackson Laboratory) to obtain Bit1 heterozygotes, which were subsequently backcrossed to wild-type BL/6 mice to obtain a pure Bit1 +/− line. These mice were intercrossed to generate Bit1 KO mice on a BL/6 genetic background.
DNA for genotyping was isolated from embryos, mouse toes, or tails by using a Qiagen DNA purification kit. Bit1-floxed mice were genotyped by using primers G2F (5′-TGG GTC TTT GAA TCA ACT AG-3′) and G1R (5′-ACA TGC CAC AAG CAA CTC CA-3′). Bit1 wild-type, heterozygous, and KO mice were detected by using the primer combination G2F/G1R/30d (30d: 5′-TTT GAG ACC CTA TCA CTC CAC ACG-3′). PCR was performed with an initial denaturation of 95°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min, with a final extension at 72°C for 5 min. Quantitative real-time PCR was performed by using a Transcriptor First Strand cDNA synthesis kit (Roche) and a Power SYBR PCR Master Mix (ABI) with Bit1 forward and reverse primers 5′-TGGTTCATCCTGGTACACTC-3′ and 5′-GCCTCACTTGTTGAGTTCTG-3′. Cycling conditions were 95°C for 10 min, followed by 40 cycles of 56°C for 1 min and 72°C for 30 s.
Histological Examination.
Embryos were collected by C-section and fixed in 10% formalin for 24 h at 4°C. After a PBS wash, the embryos were preserved in 70% ethanol before histological sectioning, staining, and examination at the University of California, Davis, CPL facility. KO pups were anesthetized, a cardiac puncture was performed to collect blood for hematological examination, and an incision was made in the abdominal cavity. Pups were fixed in formalin for 2 h at 4°C, washed with PBS, and transferred to 70% ethanol before histological processing and analysis (University of Missouri, College of Veterinary Medicine, Columbia, MO). All animal procedures were undertaken in accordance with the guidelines and approval of the Burnham Institute Animal Research Committee.
MEFs and Cell Viability Assays.
Multiple independent MEF strains were isolated from the carcasses of E12.5 embryos and cultured in DMEM containing 10% FBS supplemented with glutamine, penicillin, and streptomycin. MEFs that had been passaged <10 times were used. For anoikis assays, 5 × 105 cells were seeded in 10-cm plates coated with 10−1 polyHEMA (Sigma-Aldrich) in regular medium containing either 5% BSA or 0.5% methylcellulose. Cell clumps in the BSA cultures were dissociated by passing through a syringe before flow-cytometry analysis. Samples in 0.5% methylcellulose were digested in trypsin to disperse the cells. Staurosporine assays used 0.2 μM of the drug on 2 × 105 cells per well seeded onto six-well plates. Cell viability was assessed by staining cells with annexin-FITC and propidium iodide (BioVision), followed by analysis with flow cytometry.
RNA Interference.
Three Bit1-specific siRNAs (Bit1 siRNA-1, [CGUACUCAGAUUGCACCAGtt], Bit1 siRNA-2, [GCCUGUCAGAUUCUAACAAtt], and Bit1 siRNA-3, [GGACCAGCAGACCUAAUUtt]) and control siRNAs were obtained from Ambion. The Erk2 siRNAs were purchased from Santa Cruz Biotechnology. For transient transfection experiments, 2 × 105 HeLa cells were transfected with 25 μM of each siRNA by using the Lipofectamine 2000 transfection reagent (Invitrogen). Three days after transfection, cells were harvested, and the resulting total cell lysate was subjected to immunoblotting or in vitro Erk-directed phosphatase assay. To overexpress Bit1 after siRNA transfection, control or Bit1 siRNA-treated cells were transfected with 2 μg of the empty or C-terminally GFP-tagged Bit1 vector 3 days after siRNA transfection; 24 h later, cells were harvested and analyzed by immunoblotting. For stable Bit1 knockdown, synthetic double-stranded oligonucleotides derived from the Bit1 siRNA-3 (5′-GATCCGGACCAGCAGACCTAATTGTTCAAGAGACAATTAGGTCTGGTCCTGA-3′) was cloned into a pSilencer 4.1-CMV neo expression vector (pSilencer 4.1-CMV Bit1RNAi). The negative control pSilencer 4.1-CMV neo vector (Ambion) that expresses a hairpin siRNA with limited homology to human coding cDNAs was used to generate control clones. To generate stable cell lines, HeLa cells was transfected with pSilencer 4.1-CMV Bit1RNAi or the negative control vector and subsequently selected with 500 μg/ml G418, and clones were isolated by serial dilution. Three Bit1 RNAi clones (8, 21, 44) and two control clones (1, 16) were chosen for the subsequent experiments.
Anoikis Assay After RNA Interference.
Stable control and Bit1 knockdown clones were plated in PolyHEMA-coated 96-well plates in complete growth medium containing 0.5% methylcellulose at a density of 1.0 × 104 per well for 48 h. Cells were collected and subjected to an apoptosis assay by using the cell death detection ELISA kit (Roche Molecular Biochemicals) to detect for the presence of cytosolic nucleosomal fragments according to the manufacturer's instructions. To examine the effect of Erk2 knockdown on the anoikis resistance of Bit1 knockdown cells, stable control 16 and Bit1 RNAi44 clones were transfected with control or Erk2-specific siRNAs; 48 h after siRNA transfection, attached cells were resuspended in PolyHEMA-coated wells for another 48 h and subjected to apoptosis assay cell death detection ELISA.
Immunoblotting.
Mouse organs were homogenized in RIPA buffer [1% Triton X-100, 50 mM Tris·HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% deoxycholic acid (sodium salt), 10% glycerol, and 0.1% SDS] supplemented with protease and phosphatase inhibitor cocktails (Sigma-Aldrich). Lysates were spun at maximum speed in a table-top centrifuge to remove debris. Cultured cells were lysed in ice-cold Nonidet P-40 lysis buffer [1% Nonidet P-40, 20 mM Tris·HCl (pH 7.4), 150 mM NaCl, and 10% glycerol] with inhibitor cocktails added. Lysis was performed at 4°C for 15 min, and immunoblotting was undertaken according to standard procedures. Bit1 antisera were produced by immunizing rabbits with the Bit 1 hydrolase domain, and antibodies were on the immunogen-insolubilized onto agarose. Antibodies against total Erk1/2, phosphorylated Erk1/2 (Thr-202/Tyr-204), Erk2, phosphorylated Mek, and total Mek, and activated and total Akt were from Cell Signaling Technology.
Erk Phosphatase Assay.
The in vitro Erk phosphatase assay was performed as previously reported (9). This method relies on detecting dephosphorylation of a purified, phosphorylated, His6-tagged Erk upon incubation with total cell lysate. Briefly, 150 μg of cell lysate prepared without phosphatase inhibitors was diluted 1:4 in phosphatase assay buffer [10 mM MgCl2, 10 mM Hepes (pH 7.4), and 10 μM Mek inhibitor UO126] and incubated with 30 ng of recombinant phosphorylated His6-tagged Erk (Biomol) at room temperature for 30 min. The reaction was terminated by adding 8 M urea (pH 8.6) containing 10 mM imidazole, and the samples were placed on ice. The His-Erk was subsequently captured by adding nickel-conjugated agarose and incubating at 4°C for 90 min. The samples were washed three times with 8 M urea (pH 8.6) and 10 mM imidazole and two times in 300 mM NaCl2 and 25 mM Tris (pH 7.5). The isolated His-Erk was subjected to immunoblotting to determine the level of remaining phosphorylated Erk and total Erk to control for protein loading
ACKNOWLEDGMENTS.
We thank Dr. Eva Engvall for comments on the manuscript, Ms. Grace Cecena for the isolation of MEFs, the Burnham Institute animal facility for assistance with mice, and Mr. Albert Wahbe for assistance with photography. This work was supported by National Cancer Institute Grants CA102583 and CA098162.
Footnotes
The authors declare no conflict of interest.
References
- 1.Lai C, Chou C, Chang L, Liu C, Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jan Y, et al. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell. 2004;116:751–762. doi: 10.1016/s0092-8674(04)00204-1. [DOI] [PubMed] [Google Scholar]
- 3.de Pereda JM, et al. Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity. J Biol Chem. 2004;279:8111–8115. doi: 10.1074/jbc.M311449200. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 5.Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 6.Yujiri T, Sather S, Fanger GR, Johnson GL. Role of MEKK1 in cell survival and activation of JNK, ERK pathways defined by targeted gene disruption. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- 7.Zugasti O, et al. Raf-MEK-Erk cascade in anoikis is controlled by Rac1 and Cdc42 via Akt. Mol Cell Biol. 2001;21:6706–6717. doi: 10.1128/MCB.21.19.6706-6717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavria G, et al. ERK-MAPK signaling opposes Rho-kinase to promote endothelial cell survival and sprouting during angiogenesis. Cancer Cell. 2006;9:33–44. doi: 10.1016/j.ccr.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Laakko T, Juliano RL. Adhesion regulation of stromal cell-derived factor-1 activation of ERK in lymphocytes by phosphatases. J Biol Chem. 2003;278:31621–31628. doi: 10.1074/jbc.M304700200. [DOI] [PubMed] [Google Scholar]
- 10.Mitsushima M, Ueda K, Kioka N. Involvement of phosphatases in the anchorage-dependent regulation of ERK2 activation. Exp Cell Res. 2007;313:1830–1838. doi: 10.1016/j.yexcr.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Rosas-Sandoval G, et al. Orthologs of a novel archaeal and of the bacterial peptidyl-tRNA hydrolase are nonessential in yeast. Proc Natl Acad Sci USA. 2002;99:16707–16712. doi: 10.1073/pnas.222659199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugimoto K, et al. A serine/threonine kinase, Cot/Tpl2, modulates bacterial DNA-induced IL-12 production and Th cell differentiation. J Clin Invest. 2004;114:857–866. doi: 10.1172/JCI20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moshal KS, et al. Regulation of homocysteine-induced MMP-9 by ERK1/2 pathway. Am J Physiol. 2005;290:C883–C891. doi: 10.1152/ajpcell.00359.2005. [DOI] [PubMed] [Google Scholar]
- 14.Saxena M, Williams S, Brockdorff J, Gilman J, Mustelin T. Inhibition of T cell signaling by mitogen-activated protein kinase-targeted hematopoietic tyrosine phosphatase (HePTP). J Biol Chem. 1999;274:11693–11700. doi: 10.1074/jbc.274.17.11693. [DOI] [PubMed] [Google Scholar]
- 15.Pettiford SM, Herbst R. The MAP-kinase ERK2 is a specific substrate of the protein tyrosine phosphatase HePTP. Oncogene. 2000;19:858–869. doi: 10.1038/sj.onc.1203408. [DOI] [PubMed] [Google Scholar]
- 16.Keyse SM. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr Opin Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 17.Camps M, Nichols S, Arknstall S. Dual specificity phosphatases: A gene family for control of MAP kinase function. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- 18.Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Collins NL, et al. G1/S cell cycle arrest provides anoikis resistance through Erk-mediated Bim suppression. Mol Cell Biol. 2005;25:5282–5291. doi: 10.1128/MCB.25.12.5282-5291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glading A, Chang P, Lauffenburger DA, Wells A. Epidermal growth factor receptor activation of calpain is required for fibroblast motility and occurs via an ERK/MAP kinase signaling pathway. J Biol Chem. 2000;275:2390–2398. doi: 10.1074/jbc.275.4.2390. [DOI] [PubMed] [Google Scholar]
- 21.Vial E, Sahai E, Marshall CJ. ERK-MAPK signalling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 22.Subauste MC, et al. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. Cell Biol. 2004;165:371–381. doi: 10.1083/jcb.200308011. [DOI] [PMC free article] [PubMed] [Google Scholar]