Abstract
Mechanisms by which the extracellular domain of Notch1 controls Notch1 signaling are not well defined. Here, we show that the O-fucose glycan in the Notch1 ligand-binding domain regulates the strength of Notch1 signaling during embryogenesis, postweaning growth, and T cell development in the mouse. Heterozygotes carrying a Notch112f allele and an inactive Notch1 allele die at approximately embryonic day (E)12 with a typical Notch1 null phenotype. Homozygous Notch112f/12f mice are viable and fertile but grow somewhat more slowly than littermates after weaning. Notch112f/12f thymocytes bind less Delta1 and exhibit reduced Notch1 signaling. The number of double-positive (DP) and single-positive (SP) T cells are decreased in Notch112f/12f thymus, and DP T cells are more apoptotic. By contrast, proportionately more SP cells have matured, and SP-to-DP ratios are increased in mutant thymus. Thus, the O-fucose glycan in EGF12 of mouse Notch1 is required for optimal Notch1 signaling and T cell development in mammals.
Keywords: hypomorphic mutation, Notch signaling, O-fucosylation mutant
Notch receptors in Drosophila and mammals are covalently modified with O-linked fucose glycans at EGF repeats in the extracellular domain (1–3). An evolutionarily conserved O-fucose site resides in epidermal growth factor-like repeat 12 (EGF12), the only O-fucose site in the ligand-binding domain (EGF11+EGF12) of Drosophila Notch (4, 5). Mutation of this site results in enhanced binding of both Delta and Serrate Notch ligands and a hyperactive Notch that is refractory to Fringe (6). Thus, O-fucose in Drosophila Notch EGF12 plays an inhibitory role. By contrast, mouse Notch1 with this mutation (Notch112f) does not signal when overexpressed in cultured cells (7, 8), predicting a Notch1 null phenotype in vivo. Here, we show that homozygous Notch112f/12f mice are unexpectedly viable and fertile. On the other hand, heterozygotes with a single copy of Notch112f and an inactive Notch1 allele are not born. Thus, the Notch112f mutation is hypomorphic in mammals.
Of the four mammalian Notch receptors, Notch1 is the only one absolutely required for the development of T cells (9, 10). Deletion of Notch1 in bone marrow prevents T cell development (11) and deletion in thymus at the DN2 to DN3 stage blocks the generation of double-positive (DP) and single-positive (SP) T cells (12, 13). Stimulation of Notch1 signaling in the thymus occurs through the interaction of Notch1 on T cells with Delta1 and Delta4 Notch ligands on stromal cells (14, 15). The Lunatic Fringe glycosyltransferase (Lfng), which adds N-acetylglucosamine to O-fucose on Notch EGF repeats (1, 3, 16) greatly facilitates the binding of Delta1 to T cells and T cell development (17). However, of the 23 consensus sites for the addition of an O-fucose glycan on Notch1 (18), it is not known which, if any, are important for Notch1 functions in the thymus. We report here that Notch112f/12f mice that lack the single O-fucose site in the Notch1 ligand-binding domain have defects in T cell development that provide insights into Notch1 signaling in the thymus.
Results
The O-Fucose Glycan in EGF12 of Notch1 Promotes Notch1 Signaling.
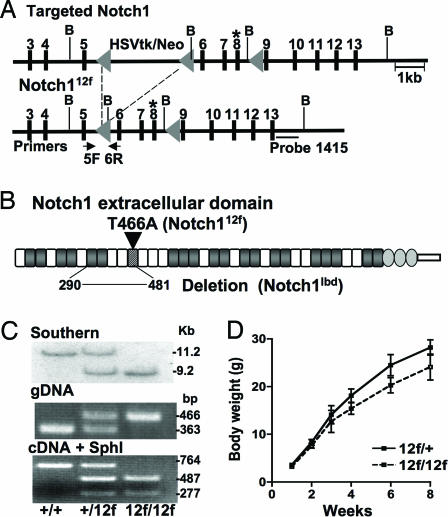
To generate mice with Notch1 lacking the O-fucose glycan in EGF12, Thr-466 was replaced with Ala, and the mutant allele Notch112f was knocked into the Notch1 locus (Fig. 1 A and B). Chimeric mice were crossed with mice carrying the MeuCre40 recombinase transgene (19) to obtain the floxed Notch112f allele. Southern blot analysis and PCR of genomic DNA were used to genotype progeny of Notch1+/12f crosses (Fig. 1C). The Notch112f allele was expressed equivalently to wild type based on RT-PCR of RNA from thymocytes (Fig. 1C). Only Notch112f transcripts were digested by SphI, a restriction site introduced during vector construction.
Fig. 1.
Targeting of the Notch1 gene. (A) The O-fucosylation site Thr-466 in Notch1 EGF12 in exon 8 (*) was replaced by Ala and introduced into the Notch1 locus. Notch112f and Notch1lbd alleles were obtained by crossing targeted mice with MeuCre40 transgenic mice. B, BamHI. (B) Notch1 extracellular domain showing the Notch112f and Notch1lbd ligand-binding domain (lbd) mutations. EGF repeats with an O-fucose site are shaded; EGF12 is striped. (C Top) Southern blot analysis after BamHI digestion and hybridization to probe 1415. (Middle) PCR genotyping with primers 5F and 6R. (Bottom) Notch1 RT-PCR products digested by SphI. (D) Body weight of Notch112f/12f and Notch1+/12f males (mean ± SD; n ≥ 11).
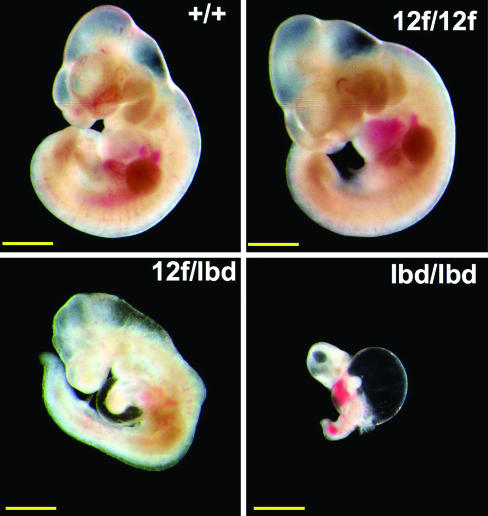
Homozygous Notch112f/12f mice were viable, and the ratio of pups from heterozygous crosses was 1:2:1. However, both male and female (data not shown) Notch112f/12f mice grew more slowly than littermates after weaning (Fig. 1D). To determine whether Notch112f is a hypomorphic allele, homozygous Notch112f/12f mice were crossed with heterozygotes carrying the Notch1lbd allele lacking the Notch1 ligand-binding domain (Fig. 1B). Notch1lbd/lbd homozygotes die at approximately embryonic day (E)10 with the typical phenotype of Notch1 null embryos (20) (C.G., T. Liu, and P.S., unpublished work). When Notch1+/lbd heterozygotes were crossed with Notch112f/12f mice, there were no Notch112f/lbd pups born (Table 1). However, Notch112f/lbd embryos survived longer than Notch1lbd/lbd embryos (Table 1). At E9.5, control (n = 8) and Notch112f/12f (n = 3) embryos had 23–26 somites, Notch112f/lbd embryos (n = 5) had 17–21 somites, and Notch1lbd/lbd embryos had only 13–17 (n = 3). At E10.5 Notch112f/lbd embryos were noticeably smaller than controls, exhibited an undeveloped heart loop, aberrant neuroepithelium, and an anemic appearance due to defective vasculogenesis (Fig. 2), similar to Notch1 null embryos (21, 22). Therefore, although a single copy of Notch1 supports development to adulthood in Notch1+/lbd heterozygotes, a single copy of Notch112f lacking O-fucose in the ligand-binding domain supports development for only ≈1.5 days longer than Notch1lbd/lbd embryos (Fig. 2), most of which are resorbed by E10.5.
Table 1.
Notch112f is a hypomorphic allele
| Stage | Litters | Progeny | Genotype |
|
|---|---|---|---|---|
| 12f/+ | 12f/lbd | |||
| E9.5 | 5 | 38 | 18 | 20 |
| E10.5 | 8 | 70 | 37 | 33 |
| E11.5 | 7 | 48 | 28 | 20 |
| E12.5 | 8 | 33 | 28 | 5* |
| E13.5 | 5 | 23 | 23 | 0 |
| Birth | 8 | 30 | 30 | 0 |
Notch112f/12f and Notch1+/lbd mice were mated and progeny genotyped.
*Heart not beating.
Fig. 2.
Notch112f is a hypomorphic allele. Embryos at E10.5 with the genotypes shown. Notch112f/lbd embryos die at approximately E12 (Table 1). Some bleeding occurred during photography. (Scale bars, 1 mm.)
The O-Fucose Glycan in EGF12 of Notch1 Promotes T Cell Development.
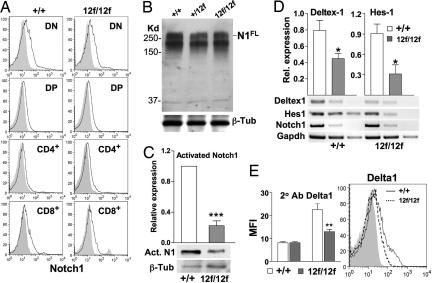
Notch112f function was investigated in T cell development. Flow cytometry with antibodies to CD4, CD8, and Notch1 extracellular domain (23) revealed similar patterns of cell surface Notch1 expression in T cell subsets from Notch112f/12f and Notch1+/+ littermates (Fig. 3A). Western blot analysis showed equivalent levels of full-length Notch1 in Notch112f/12f and Notch1+/+ thymocytes (Fig. 3B). However, activated Notch1 generated by γ-secretase cleavage and detected by antibody Val-1744 (24) was markedly reduced in Notch112f/12f thymocytes (Fig. 3C). The expression of the Notch1 target genes Deltex1 and Hes1 was also reduced, although Notch1 expression was not changed (Fig. 3D). Notch112f/12f thymocytes also bound less of the Notch ligands Delta1 (Fig. 3E) and Jagged1 (data not shown). Therefore, the O-fucose glycan in the Notch1 ligand-binding domain plays a positive role in the interaction of Notch1 with Delta1 and is required for optimal Notch1 signaling in the thymus.
Fig. 3.
Notch112f cell surface expression, signaling, and ligand binding. (A) Flow cytometry analysis of thymocyte subsets using Notch1 antibody 8G10. Shaded profiles represent secondary antibody alone. (B) Western blot analysis of thymocyte lysates using Notch1 antibody 8G10, followed by anti-β-tubulin III. Results are representative of four experiments. (C) Western blot analysis of thymocyte lysates using antiactivated Notch1 Val-1744 antibody. The histogram shows normalized expression (mean ± SEM; ***, P < 0.001; n = 4). (D) cDNA was prepared from thymocytes of control and mutant mice (n = 3 pairs), serially diluted 5-fold, and subjected to PCR twice. Histograms show normalized average expression (mean ± SEM; *, P < 0.05). (E) Binding of soluble Delta1 to Notch1+/+ (solid line) and Notch112f/12f (dashed line) thymocytes. Secondary antibody profile is shaded. Histograms show mean fluorescence intensity (MFI) ± SEM (**, P < 0.01; n = 4).
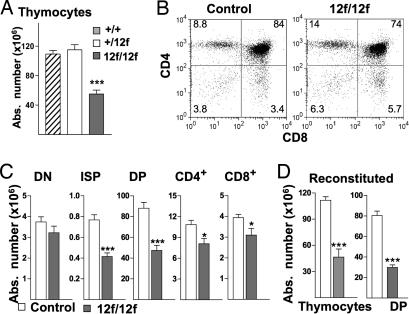
There were no morphological abnormalities in Notch112f/12f thymi based on hematoxylin and eosin or peanut agglutinin staining of sections (data not shown). However, total thymocytes (hereafter termed thymocytes) in Notch112f/12f thymus were reduced ≈50% compared with Notch1+/12f and Notch1+/+ thymi (Fig. 4A). Flow cytometry revealed no differences in the distribution of T cell subsets in Notch1+/12f versus Notch1+/+ mice [supporting information (SI) Fig. 7A]. However, all of the more mature T cell subsets were significantly reduced in Notch112f/12f mice (Fig. 4 B and C), although the expression of genes required for T cell development including pre-Tα, Rag1, and Rag2 was not altered (data not shown). In the DN subset, the numbers of B cells, natural killer (NK) cells, dendritic cells, and γδ T cells were similar in Notch112f/12f and controls (SI Fig. 7B). There was no apparent effect of the Notch112f mutation on T cell emigration to the spleen or on the activation of splenic T cells by anti-CD3ε/anti-CD28 antibodies (SI Fig. 8).
Fig. 4.
T cell development in Notch112f/12f mice. (A) Total thymocytes in Notch1+/+ (n = 5), Notch1+/12f (n = 11), and Notch112f/12f mice (n = 11); mean ± SEM (***, P < 0.001). (B) Representative flow cytometric analysis of thymocytes from 8-week-old Notch112f/12f and control littermates using anti-CD4 and anti-CD8α. Percentages of T cell subsets are shown. (C) Absolute numbers of DN, ISP (CD4−CD8+CD3−), DP, CD4+, and CD8+ in thymocytes from control and Notch112f/12f mice; mean ± SEM; ISP, n = 8 pairs; others, n = 11 pairs (*, P < 0.05; ***, P < 0.001). (D) Reconstituted thymi. Numbers of thymocytes and DP T cells expressing CD45.2 after 6-week repopulation of irradiated hosts (CD45.1+) by bone marrow from CD45.2+ Notch1+/12f (n = 6) or Notch112f/12f (n = 6) mice.
To determine whether the partial block in DN-to-DP transition is a cell-autonomous property of Notch112f/12f T cells, bone marrow transfer and ex vivo coculture experiments were performed. Notch112f/12f or Notch1+/12f bone marrow cells were transferred to irradiated C57BL/6 mice that express CD45.1 on T cells. After 6 weeks, thymi from mice that received Notch112f/12f bone marrow were small and contained ≈60% fewer thymocytes than those that received control bone marrow (Fig. 4D). Although the average number of Notch112f/12f donor SP T cells was reduced, differences from control did not reach significance at n = 6. However, the number of CD45.2+ donor DP T cells was markedly reduced, ≈50% in Notch112f/12f bone marrow recipients (Fig. 4D). Furthermore, the proportion of DP T cells obtained by coculture of lineage-depleted Notch112f/12f fetal liver cells with OP9-DL1 stromal cells (15) was markedly reduced compared with controls (SI Fig. 9). Therefore, the Notch112f/12f T cell developmental phenotype derives from defective Notch1 signaling in mutant T cells.
T Cell Development Is Partially Blocked at the DN3–DN4 Stage in Notch112f/12f Thymus.
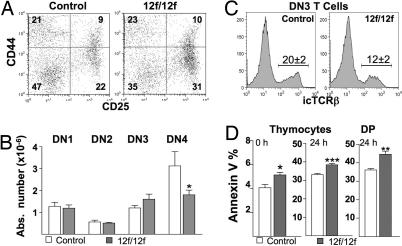
Flow cytometry of the DN T cell subset (Fig. 5A) showed that DN1 and DN2 cell numbers were similar in Notch112f/12f and controls, but CD44−CD25− DN4 T cells were decreased by ≈40% (Fig. 5B). By contrast, there was a slight increase in the DN3 subset (Fig. 5B). Consistent with retarded development at the DN3-to-DN4 stage, intracellular TCRβ (icTCRβ) was reduced ≈60% in Notch112f/12f DN3 T cells (Fig. 5C). However, icTCRβ expression was unaffected in Notch112f/12f DN4 T cells, and icCD3ε expression was similar in control and mutant DN3 and DN4 T cells (data not shown). Therefore, lack of an O-fucose glycan in the Notch1 ligand-binding domain partially blocks T cell development at the DN3-to-DN4 transition.
Fig. 5.
DN3 and DN4 T cells in Notch112f/12f mice. (A) Representative flow cytometric analysis using antibodies to CD44 and CD25 after gating on DN thymocytes from Notch112f/12f and control mice. Percentages of CD44+CD25− (DN1), CD44+CD25+ (DN2), CD44−CD25+ (DN3), and CD44−CD25− (DN4) thymocytes are indicated. (B) Absolute numbers of DN T cell subsets in Notch112f/12f mice and controls (mean ± SEM; n = 6; *, P < 0.05, one-tailed t test). (C) Percentage of cells expressing intracellular TCRβ (icTCRβ) in DN3 T cells of control and Notch112f/12f mice (mean ± SEM, n = 4, P < 0.001). (D) Annexin V binding to freshly isolated thymocytes (0 h) or thymocytes or DP T cells isolated by flow cytometry and cultured for 24 h in DMEM/10% FBS (mean ± SEM) *, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 5–6.
Notch112f/12f Thymocytes Are More Sensitive to Apoptosis.
Overexpression of activated Notch1 protects T lineage cells from apoptosis (25–28). To examine apoptosis in Notch112f/12f thymus, annexin V binding and TUNEL assays were performed. Freshly isolated Notch112f/12f thymocytes bound more annexin V than controls and, after culturing for 24 h, this difference was enhanced (Fig. 5D). Similar results were obtained by using the TUNEL assay (data not shown). When T cell subsets were examined, only DP T cells exhibited enhanced apoptosis (Fig. 5D and data not shown). Consistent with this, the expression of the antiapoptosis genes Bcl-2 and Bcl-xL was decreased in Notch112f/12f thymocytes, whereas expression of the proapoptosis gene Bax was increased (SI Fig. 10). Therefore, the reduction in thymocytes in Notch112f/12f mice appears to be caused in part by increased apoptosis of the DP T cell subset.
The DP-to-SP Transition Is Enhanced in the Notch112f/12f Thymus.
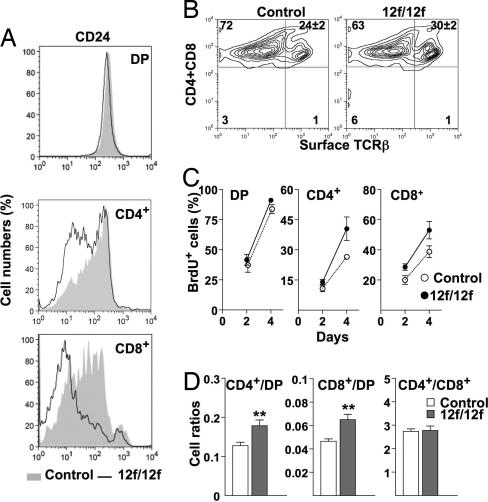
The expression of heat-stable antigen CD24 is reduced with T cell maturation (29). The Notch112f/12f SP T cell population expressed less CD24 than controls (Fig. 6A) indicating enhanced maturation of the mutant SP population. Cell surface TCRβ expression, another marker of T cell maturation, was increased in Notch112f/12f CD4+ and/or CD8+ T cells (Fig. 6B), although two early SP maturation markers, CD5 and CD69, were expressed similarly in mutant and control SP T cells (data not shown). Bromodeoxyuridine (BrdU) content of SP T cells is a measure of DP-to-SP transition, and BrdU+ SP T cells were proportionately increased in Notch112f/12f thymus (Fig. 6C). Furthermore, the ratios of both CD4+ to DP and CD8+ to DP T cells were increased in Notch112f/12f thymus, whereas the CD4+-to-CD8+ T cell ratio was unaltered (Fig. 6D). Therefore, reduced Notch1 signaling in Notch112f/12f thymocytes alters the DP-to-SP transition, favoring the generation of proportionately more SP T cells.
Fig. 6.
Enhanced transition of DP-to-SP T cells in Notch112f/12f thymus. (A) Surface expression of CD24 on DP, CD4+, and CD8+ T cells. Representative profiles for control (shaded) and Notch112f/12f (solid line) mice; n = 4. (B) Surface expression of TCRβ on CD4+ and CD8+ thymocytes. Mean ± SEM (P < 0.01, n = 4). (C) DP, CD4+, and CD8+ T cells that incorporated BrdU (BrdU+) were calculated as a percentage of the total DP, CD4+, or CD8+ cells, respectively (12). Mean ± SEM (n = 4). Average numbers of DP/BrdU+ T cells on day 2 were 2.89 × 107 (control), 1.89 × 107 (mutant); average numbers of CD4+BrdU+ were 1.20 × 106 (control), 1.33 × 106 (mutant); and average numbers of CD8+BrdU+ were 8.54 × 105 (control), 9.32 × 105 (mutant). (D) Ratios of SP/DP and CD4/CD8 subsets were calculated. Mean ± SEM; **, P < 0.01, n = 11.
Discussion
Here, we identify the O-fucose glycan in the mouse Notch1 ligand-binding domain as a regulator of the interaction between Notch1 on T cells and Delta1 and of Notch1 signaling during embryogenesis and T cell development. We showed that it is the loss of the O-fucose glycan at Thr-466 and not the change of Thr to Ala that causes reduced Notch112f signaling (8). We show here that precluding the addition of O-fucose to EGF12 in Notch1 results in a hypomorphic allele that cannot support embryogenesis beyond E11.5 when expressed with the inactive Notch1lbd allele. Notch112f/lbd heterozygous embryos develop ≈1.5 days longer than Notch1 null embryos but have a similar range of developmental defects. However, the Notch112f mutation has comparatively mild effects in homozygotes. Thus, Notch112f/12f pups appear normal until after weaning, when their growth slows. They have no apparent increase in morbidity for up to 16 months of age. However, they have defective T cell development.
We show by bone marrow transfer and fetal liver coculture experiments that the Notch112f/12f T cell defect is cell-autonomous. A similar phenotype is obtained when Notch1 is deleted by lck-Cre (12). In both cases, the generation of TCRγδ T cells is not altered, but the production of TCRαβ DP T cells is markedly reduced (≈80% with lck-Cre and ≈50% in Notch112f/12f mice). Although the absolute number of DN cells is unaltered in both mutants, the DN2 and DN3 subsets are significantly increased when Notch1 is deleted by lck-Cre, whereas only the DN3 subset is slightly increased in Notch112f/12f mice. TCRαβ T cells are aberrant when Notch1 is deleted by lck-Cre as they mature to DN4 without pre-TCR signaling and die before becoming DP T cells (12). In Notch112f/12f mice, the number of DN4 T cells is reduced, but the remaining DN4 cells proliferate to become DP T cells. When RBP-Jκ is deleted by lck-Cre, there is a more severe defect in T cell development with an ≈80–90% decrease in DP and SP T cells (30). Survivors are probably due to inefficient deletion by Cre recombinase because dominant-negative mastermind-like 1 causes a complete block in T cell development (13), precluding further study. By contrast, Notch112f/12f mutant T cells can be investigated at all stages of T cell development. At later stages, Notch112f/12f thymus has an increased proportion of apoptotic DP cells, an increase in CD4+/DP and CD8+/DP ratios, and an increased proportion of more mature SP T cells. The data suggest that the O-fucose glycan of Notch1 EGF12 normally functions to inhibit apoptosis of DP cells and may function to delay the DP-to-SP transition.
The T/B lineage decision was unimpaired in Notch112f/12f mice. Although deletion of Notch1 in bone marrow with Mx-Cre causes an ≈200-fold increase in thymic B cells and a block in T cell development at the DN1-to-DN2 transition (11), and a similar production of thymic B cells is found by overexpressing Lfng with an lck proximal promoter (17, 31), deletion of Notch1 by lck-Cre or CD4-Cre causes no increase in thymic B cells (12, 32). Notch2 and Notch3 are also expressed in T cells (33), but lck-Cre-induced deletion of RBP-J (30), which inhibits global Notch signaling, gives only a marginally more severe phenotype than deletion of Notch1 by lck-Cre (12). However, this may be due to differential effects on the timing and efficiency of deletion of different genes by Cre recombinase (34). Thus, deletion of presenilins 1 and 2 by CD4-Cre occurs at the DN2–DN3 stage, earlier than deletion of RBP-J by the same transgene (34). Inhibition of global Notch signaling by deleting the presenilins reduces the production of SP T cells, but the effect is rather weak for CD8+ T cells unless TCR signaling is artificially strengthened (34).
Competition experiments using Notch1+/− heterozygous cells show that the level of Notch1 signaling must be regulated for optimal T cell development to occur (35, 36). This is presumably why enhancing Notch1 signaling by overexpression of Notch1 intracellular domain (NICD) has led to several different proposals on the role of Notch1 signaling in the DP-to-SP transition (37). Nevertheless, overexpression of NICD gives a complete block in the production of SP T cells (38), a result consistent with our observation that the DP-to-SP transition is promoted in Notch112f/12f thymus (Fig. 6). The fact that CD4-Cre-induced deletion of Notch1 at the DN3–DN4 stage did not lead to an alteration in the numbers or proportions of SP thymocytes (32) may reflect inefficient deletion by CD4-Cre. In terms of peripheral T cell function, deletion of Notch1 by CD4-Cre in thymus does not affect the emigration or activation of T cells to the spleen (39), as observed here with Notch112f/12f mice.
In summary, the O-fucose glycan in the Notch1 ligand-binding domain regulates T cell development at multiple stages in the thymus by enhancing Notch ligand binding and Notch1 signaling. Although it does not affect the T/B or other lineage decisions, nor thymocyte emigration from thymus to spleen, nor the activation of splenic T cells, the presence of this O-fucose glycan promotes T cell development at early DN stages, keeps DP T cells from apoptosing, and may regulate the maturation of SP T cells. Because Notch112f/12f mice are viable and fertile, this hypomorphic Notch1 germ-line mutation can provide insights into roles for Notch1 and the effects of altering the strength of Notch1 signaling on the downstream pathways engaged during development and differentiation of many cell lineages.
Materials and Methods
Mice.
The Notch1 O-fucosylation site Thr-466 in EGF12 (ACT in exon 8) was replaced by Ala (GCA) by using site-directed mutagenesis (Stratagene), that also introduced an SphI site. Notch1 exons 6–8 with the Notch112f mutation (1.6 kb) were cloned into the pFlox vector (provided by Jamey Marth, University of California at San Diego) and flanked by genomic DNA. Homologous recombination was performed in WW6 embryonic stem cells (40), and G418-resistant isolates were injected into C57BL/6 blastocysts to obtain chimeras. Genomic DNA deletions to generate Notch112f or Notch1lbd were achieved by crossing to mice expressing MeuCre40 recombinase (19). Mice were back-crossed 5–7 generations onto C57BL/6 and intercrossed and housed under barrier protection, and protocols were approved by the Albert Einstein College of Medicine Animal Institute Committee. Southern blot analysis of BamHI-digested genomic DNA used Notch1 probe 1415 (Fig. 1). PCR genotyping used primers 5F: GTATGTATATGGGACTTGTAGGCAG and 6R: CTATGAGGGGTCACAGGACCAT for the Notch112f and Notch1+ alleles. Notch1lbd genotyping will be described elsewhere. RT-PCR was performed by using primers 7F: GCGTGCATCAGCAACCCCTGCAACGAGG and 11R: CATAAGCAGAGGTAGGAGTTGTCAC on total RNA from thymus.
Western Blot Analysis.
Thymocytes were lysed in 200 μl of RIPA buffer supplemented with complete protease inhibitor mixture (Roche) for 30 min on ice. After centrifugation, lysates were resolved by SDS/PAGE, transferred to polyvinyldifluoride membrane, and probed with Notch1 antibody 8G10 (UpstateBiotechnology) for detection of full-length Notch1 or with Notch1 antibody Val-1744 (Cell Signaling Technology) for detection of cleaved, activated Notch1 or with β-tubulin III-specific antibody (Sigma), followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch). Bands were visualized with Enhanced Chemiluminescence reagent (Amersham Pharmacia Biotech).
RT-PCR.
Total RNA was isolated from thymocytes with TRIzol (Invitrogen), followed by DNase I digestion. cDNA was prepared by using RNA PCR Kit ver. 3.0 (Takara Mirus Bio) and normalized for equivalent template amounts by 5-fold serial dilution and comparison with Gapdh transcripts. Primers are given in SI Text. PCR was performed at 55°C except for Bcl-2 at 60°C. PCR products visualized by ethidium bromide were 306 bp (Hes1), 352 bp (Deltex1), 293 bp (Bcl-2), 344 bp (Bcl-xL), 194 bp (Bax), and 452 bp (Gapdh). Bands were imaged by using the Gel Logic 2000 image system (Kodak) and quantitated by using NIH ImageJ software (http://rsb.info.nih.gov/ij/). Histograms compare relative intensities from the 1:5 dilution.
Flow Cytometry.
Fluorescein (FITC)-, Phycoerythrin (PE)-, Peridinin-chlorophyll-protein (PerCP)-, Phycoerythrin-Cy7 (PE-Cy7)-, Phycoerythrin-Cy5.5 (PE-Cy5.5)-, and Allophycocyanin (APC)-conjugated monoclonal antibodies specific for mouse CD4 (GK1.5), CD8α (53–6.7), CD25 (7D4), CD44 (1M7), CD24 (M1/69), CD5 (53–7.3), CD43 (S7), CD69 (H1.2F3), TCRβ (H57–597), CD3ε (145–2C11), anti-CD45.1 (A20), and anti-CD45.2 (104) were purchased from BD Pharmingen or eBiosience. Single-cell suspensions from thymus or spleen were stained with fluorochrome-conjugated antibodies in HBSS, 3% BSA, and 0.05% sodium azide (binding buffer) according to standard protocols. Dying cells were excluded by 7-AAD except in intracellular staining and BrdU incorporation experiments. Intracellular antigens were detected after binding of antibodies to surface markers by fixation, permeablization, and incubation with antibody to the intracellular antigen. Surface Notch1 was determined by using antibody 8G10 (Upstate Biotechnology) versus control secondary antibody incubated in binding buffer with azide at room temperature for 1 h or at 4°C. After washing, incubation with Alexa Fluor 488 anti-hamster IgG (Invitrogen) and anti-CD4-APC and anti-CD8α-PE antibodies was for 30–60 min. Immunofluorescence was analyzed on a FACS Calibur (BD Biosciences). Data files were analyzed by using Flowjo software (Tree Star).
Bone Marrow (BM) Transfer.
BM cells (3 × 106) from 8- to 10-week-old Notch1+/12f and Notch112f/12f mice (CD45.2+) were injected into 6-week-old C57BL/6.SJL (CD45.1+) mice via the tail vein. Recipients received 550 rad twice, 16 h apart, before injection. After 6 weeks, thymi were weighed, and total thymocytes were counted. Thymocytes were analyzed by flow cytometry using anti-CD45.2-FITC or anti-CD45.1-PE-Cy7, anti-CD4-APC, and anti-CD8α-PE antibodies.
Notch Ligand-Binding Assay.
Soluble Delta1-Fc and Jagged1-Fc plasmids were from Gerry Weinmaster (University of California, Los Angeles) (41). HEK-293T cells stably expressing either plasmid were cultured in DMEM 10% FBS, followed by 293 SFM II serum-free medium (Invitrogen). After 3 days, conditioned medium was subjected to Western blot analysis using anti-human IgG antibody-HRP (Jackson ImmunoResearch). For binding assays, 106 thymocytes were incubated with Delta1-Fc (2 μg/ml) or Jagged1 (0.5 μg/ml) in HBSS containing 2% BSA, 0.05% azide, and 1 mM Ca2+ for 1 h at 4°C, followed by incubation with 1:100 PE-conjugated anti-human Fc antibody (Jackson ImmunoResearch) for 30 min at 4°C before being analyzed by flow cytometry.
BrdU Incorporation.
Mice (6–8 weeks of age) received two i.p. injections of BrdU (1 mg) 2 h apart and were subsequently injected with 1 mg of BrdU every 12 h. Thymocytes were isolated at different times and examined by flow cytometry for cell surface expression of CD4 and CD8 and intracellular BrdU. Cell surface binding of anti-CD4-APC and anti-CD8-PE was performed as described above, cells were then fixed, permeabilized, treated with DNase I, and stained with anti-BrdU-FITC according to the manufacturer's instructions (BrdU Flow Kit, BD Pharmingen).
Apoptosis.
Thymocytes from 6- to 8-week-old mice were analyzed by flow cytometry after incubation of 106 cells with anti-CD4-FITC, and anti-CD8-APC, followed by annexin V-PE, and 7-AAD according to the manufacturer's instructions (BD Biosciences). For culturing, thymocytes were incubated at 4 × 106 per well in 24-well plates in DMEM containing 10% FBS for 24 h before analysis. Direct TUNEL labeling of thymocytes was performed by the fluorescein in situ cell death detection kit (Roche) and analysis by flow cytometry.
Statistics.
Statistical significance for all data were calculated by using the unpaired t test (two-tailed, unless noted otherwise) with Graphpad Prism (GraphPad Software).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Martin Holzenberger (Institut National de la Santé et de la Recherche Médicale U515, France) for MeuCre40 mice, Wen Dong and Weijun Liu for excellent technical assistance, and Thomas Graf, Hilda Ye, and Fernando Macian for helpful discussions. This work was supported by National Institutes of Health Grant R01 95022 (to P.S.) and Albert Einstein Cancer Center Grant P01 13330.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702846105/DC1.
References
- 1.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 2.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. J Biol Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 3.Bruckner K, Perez L, Clausen H, Cohen S. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 4.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 5.Xu A, Lei L, Irvine KD. J Biol Chem. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. [DOI] [PubMed] [Google Scholar]
- 6.Lei L, Xu A, Panin VM, Irvine KD. Development. 2003;130:6411–6421. doi: 10.1242/dev.00883. [DOI] [PubMed] [Google Scholar]
- 7.Rampal R, Arboleda-Velasquez J, Nita-Lazar A, Kosik KS, Haltiwanger RS. J Biol Chem. 2005;280:32133–32140. doi: 10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi S, Ge C, Luo Y, Hou X, Haltiwanger RS, Stanley P. J Biol Chem. 2007;282:20133–20141. doi: 10.1074/jbc.M702593200. [DOI] [PubMed] [Google Scholar]
- 9.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 10.Radtke F, Wilson A, MacDonald HR. BioEssays. 2005;27:1117–1128. doi: 10.1002/bies.20315. [DOI] [PubMed] [Google Scholar]
- 11.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 12.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 13.Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, Owen MJ. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt TM, Zuniga-Pflucker JC. Immunol Rev. 2006;209:95–102. doi: 10.1111/j.0105-2896.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- 16.Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. J Biol Chem. 2005;280:42454–42463. doi: 10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- 17.Visan I, Tan JB, Yuan JS, Harper JA, Koch U, Guidos CJ. Nat Immunol. 2006;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- 18.Shao L, Moloney DJ, Haltiwanger R. J Biol Chem. 2003;278:7775–7782. doi: 10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- 19.Leneuve P, Colnot S, Hamard G, Francis F, Niwa-Kawakita M, Giovannini M, Holzenberger M. Nucleic Acids Res. 2003;31:e21. doi: 10.1093/nar/gng021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl M, Ge C, Shi S, Pestell RG, Stanley P. Cancer Res. 2006;66:7562–7570. doi: 10.1158/0008-5472.CAN-06-0974. [DOI] [PubMed] [Google Scholar]
- 21.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 22.Conlon RA, Reaume AG, Rossant J. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 23.Anderson AC, Kitchens EA, Chan SW, St Hill C, Jan YN, Zhong W, Robey EA. J Immunol. 2005;174:890–897. doi: 10.4049/jimmunol.174.2.890. [DOI] [PubMed] [Google Scholar]
- 24.Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner LA, Kopan R. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 25.Ciofani M, Zuniga-Pflucker JC. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 26.Sade H, Krishna S, Sarin A. J Biol Chem. 2004;279:2937–2944. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- 27.Jehn BM, Bielke W, Pear WS, Osborne BA. J Immunol. 1999;162:635–638. [PubMed] [Google Scholar]
- 28.Deftos ML, He YW, Ojala EW, Bevan MJ. Immunity. 1998;9:777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hough MR, Takei F, Humphries RK, Kay R. J Exp Med. 1994;179:177–184. doi: 10.1084/jem.179.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 31.Koch U, Lacombe TA, Holland D, Bowman JL, Cohen BL, Egan SE, Guidos CJ. Immunity. 2001;15:225–236. doi: 10.1016/s1074-7613(01)00189-3. [DOI] [PubMed] [Google Scholar]
- 32.Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, Radtke F. Nat Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 33.Felli MP, Maroder M, Mitsiadis TA, Campese AF, Bellavia D, Vacca A, Mann RS, Frati L, Lendahl U, Gulino A, Screpanti I. Int Immunol. 1999;11:1017–1025. doi: 10.1093/intimm/11.7.1017. [DOI] [PubMed] [Google Scholar]
- 34.Laky K, Fowlkes BJ. J Exp Med. 2007;204:2115–2129. doi: 10.1084/jem.20070550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 36.Visan I, Yuan JS, Tan JB, Cretegny K, Guidos CJ. Immunol Rev. 2006;209:76–94. doi: 10.1111/j.0105-2896.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 37.Fowlkes BJ, Robey EA. J Immunol. 2002;169:1817–1821. doi: 10.4049/jimmunol.169.4.1817. [DOI] [PubMed] [Google Scholar]
- 38.Izon DJ, Punt JA, Xu L, Karnell FG, Allman D, Myung PS, Boerth NJ, Pui JC, Koretzky GA, Pear WS. Immunity. 2001;14:253–264. doi: 10.1016/s1074-7613(01)00107-8. [DOI] [PubMed] [Google Scholar]
- 39.Radtke F, Wilson A, Ernst B, MacDonald HR. Immunol Rev. 2002;187:65–74. doi: 10.1034/j.1600-065x.2002.18706.x. [DOI] [PubMed] [Google Scholar]
- 40.Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor SM, Stanley P. Proc Natl Acad Sci USA. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.