Abstract
We report the identification and functional analysis of nine genes from Gram-positive and Gram-negative bacteria and their phages that are similar to lambda (λ) bet or Escherichia coli recT. Beta and RecT are single-strand DNA annealing proteins, referred to here as recombinases. Each of the nine other genes when expressed in E. coli carries out oligonucleotide-mediated recombination. To our knowledge, this is the first study showing single-strand recombinase activity from diverse bacteria. Similar to bet and recT, most of these other recombinases were found to be associated with putative exonuclease genes. Beta and RecT in conjunction with their cognate exonucleases carry out recombination of linear double-strand DNA. Among four of these foreign recombinase/exonuclease pairs tested for recombination with double-strand DNA, three had activity, albeit barely detectable. Thus, although these recombinases can function in E. coli to catalyze oligonucleotide recombination, the double-strand DNA recombination activities with their exonuclease partners were inefficient. This study also demonstrated that Gam, by inhibiting host RecBCD nuclease activity, helps to improve the efficiency of λ Red-mediated recombination with linear double-strand DNA, but Gam is not absolutely essential. Thus, in other bacterial species where Gam analogs have not been identified, double-strand DNA recombination may still work in the absence of a Gam-like function. We anticipate that at least some of the recombineering systems studied here will potentiate oligonucleotide and double-strand DNA-mediated recombineering in their native or related bacteria.
Keywords: bacteriophage lambda, Beta, oligonucleotide recombination, RecT, ssDNA annealing proteins
Recombineering is a simple and efficient way to engineer DNA molecules in vivo without the in vitro use of restriction enzymes and DNA ligase (1, 2). In Escherichia coli, it allows genetic modifications, from point mutations to substitutions and deletions. Recombineering is mediated by phage-derived proteins, either the Red proteins of phage λ (1–3) or RecET from the Rac prophage of E. coli (4), which are particularly efficient in catalyzing homologous recombination between short (≈50 bp) homology segments. The λ Red recombination functions are encoded by three adjacent genes, gam, bet and exo, in the λ pL operon (1). RecET functions are encoded by two adjacent genes, recE and recT, present on the cryptic lambdoid Rac prophage found in the genome of many E. coli K-12 strains (5). A variety of studies have concluded that the Exo/Beta and RecE/RecT protein pairs are functionally equivalent although not related at the sequence level (6, 7). Exo and RecE are 5′–3′ exonucleases that degrade the 5′ ends of linear duplex DNA, creating 3′ single-strand (ss) DNA overhangs (8, 9). Beta and RecT are single-strand annealing proteins that bind to these ssDNA overhangs and pair them with complementary ssDNA targets (10–12). λ Gam inhibits two potent host nucleases, RecBCD and SbcCD (13, 14), thereby preventing the degradation of linear double-strand (ds) DNA introduced into the cell. The Rac prophage does not encode a known functional analogue of Gam, but linear DNA recombination studies with RecET have used λ Gam to inhibit nucleases (4).
For recombineering, either a dsDNA PCR product (1, 4, 15–17) or an oligonucleotide (oligo) (18–21) carrying short (≈50-bp) segments homologous to the target sequences can be used. These linear DNA substrates are precisely recombined in vivo by the phage proteins to target DNA on any replicon. When linear dsDNA is used for recombination, both the exonuclease and single-strand annealing protein are required. For optimal recombination, RecBCD should be inactivated, either by mutation or by λ Gam (1, 15, 22). In contrast, recombineering with an oligo requires only the expression of single-strand annealing protein, Beta, or RecT (18, 21, 22). The oligo recombination is mechanistically simpler and more efficient than dsDNA recombination and is the assay we have used to define the new recombineering systems described here.
We have identified and characterized genes similar to bet or recT from Gram-positive and other Gram-negative bacteria and their phages. For simplicity, we refer to these putative single-strand annealing proteins as recombinases. To initially characterize the proteins, we have expressed these foreign genes in E. coli under λ pL control and assayed them, using our optimized oligo recombination system (20). We report here that every recombinase we assayed catalyzes oligo recombination in E. coli, some with high efficiency. Most of the newly identified recombinases lie next to putative exonucleases genes, as do bet and recT. Four of these other recombinase–exonuclease pairs were also tested in E. coli for dsDNA recombination activity. This study provides a solid foundation for our goal of developing recombineering in other bacterial species.
Results
Recombinases Selected as Potential Recombineering Functions.
We searched for putative DNA recombination proteins from different bacteria and their phages with the BLAST search engine of the National Center for Biotechnology Information database, using the amino acid sequences of either λ Beta or RecT as queries. Nine different putative recombinases were selected for functional comparison with Beta and RecT [supporting information (SI) Table 3]. These include proteins from phages or prophages of Gram-positive bacteria Bacillus subtilis, Mycobacterium smegmatis, Listeria monocytogenes, Lactococcus lactis, Staphylococcus aureus, and Enterococcus faecalis and also from the Gram-negative bacteria Vibrio cholerae, Legionella pneumophila, and Photorhabdus luminescens. Sequence annotation revealed that at least seven of the recombinase genes are adjacent to a known or putative exonuclease gene, just as λ bet is located next to exo, and recT is next to recE. Such recombinase–exonuclease pairs are seen in phages or prophages of B. subtilis (gp 34.1/gp35), M. smegmatis (gp60/gp61), L. monocytogenes (orf47/orf48), V. cholerae (s066/s065), P. luminescens (plu2936/plu2935), L. pneumophila (orfB/orfC), and E. faecalis (EF2131/EF2132) (SI Table 4).
Placing the Different Recombinase Genes and Exonuclease–Recombinase Gene Pairs into the Defective λ Prophage.
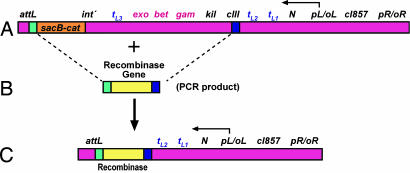
The 11 different recombinase genes, including bet and recT, were introduced into the chromosome of SIMD30 on the defective λ prophage in place of the red genes, using recombineering to create SIMD40–50. Each of these genes was fused to the λ cIII translation initiation signal, and expressed from the λ pL promoter under the control of the temperature inducible CI857 repressor (Fig. 1). Likewise, four different recombinase–exonuclease pairs were also inserted in the same location and in their native orientation.
Fig. 1.
Replacement of λ red genes by individual recombinase genes. (A) Strain SIMD30 carries a defective λ prophage and a cat-sacB cassette. int′ indicates deletion of the 5′ end of the int gene by cat-sacB. The red genes exo, bet, and gam are expressed from the pL promoter under the control of the CI857 repressor. The arrow at pL indicates the direction of transcription. Transcription terminators tL1, tL2, and tL3 and the anti-termination N gene are indicated. (B) The putative recombinase genes were PCR-amplified by using 70 base primers whose 5′ ends contain 50 bases identical to the sequence at the target sites, where the PCR product will be recombined. The target homologies, indicated by green and blue rectangles, are in the att region and upstream of the cIII start codon, respectively. (C) Recombination between the homologous sequences on the PCR product and the target replaces the target segment comprising cat-sacB, exo, bet, gam, kil, and cIII by the recombinase gene (yellow).
Recombineering with an Oligo.
Methyl-directed mismatch repair (MMR) reduces the apparent oligo recombination frequency nearly 100-fold (20); therefore, MMR was eliminated in SIMD40–50 by deleting mutS and creating strains SIMD80–90, which were used in the oligo recombination assay. Two complementary 70 base oligos (oligos 100 and 101) were used individually to correct the galKTYR145UAG amber mutation to the native tyrosine codon TAT (18, 20), and the Gal+ recombinant frequency was scored. Table 1 contains the strains used and the results of these assays.
Table 1.
Oligo recombineering using different recombinases
| Strain† | Recombinase | Bacterial source | Gal+ per 108 viable cells‡ |
|
|---|---|---|---|---|
| Oligo 100 | Oligo 101 | |||
| SIMD90 | Beta | E. coli | 1.8 × 107 | 1.3 × 106 |
| SIMD84 | EF2132 | E. faecalis | 2.1 × 107 | 1.8 × 106 |
| SIMD87 | OrfC | L. pneumophila | 1.0 × 107 | 6.0 × 105 |
| SIMD85 | s065 | V. cholerae | 4.6 × 106 | 7.0 × 104 |
| SIMD86 | plu2935 | P. luminescens | 3.3 × 106 | 2.4 × 105 |
| SIMD89 | RecT | E. coli | 5.0 × 105 | 4.0 × 104 |
| SIMD82 | Orf48 | L. monocytogenes | 2.0 × 105 | 1.5 × 104 |
| SIMD83 | Orf245 | L. lactis | 2.2 × 105 | 2.2 × 104 |
| SIMD80 | GP35 | B. subtilis | 6.4 × 103 | 1.0 × 103 |
| SIMD81 | GP61 | M. smegmatis | 1.0 × 104 | 2.0 × 103 |
| SIMD88 | GP20 | S. aureus | 5.4 × 104 | 2.5 × 103 |
†These strains are mutS< >cat derivatives of SIMD40–50 in SI Table 3.
‡Values indicated are the average of three or more experiments. The background Gal+ recombination in strain HME75 lacking the defective λ and Rac prophage is 3 × 102 recombinants per 108 viable cells.
Our standard with which to compare the other recombinases was λ Beta (in SIMD90), which generated 1.8 × 107 Gal+ recombinants per 108 viable cells with oligo 100. E. coli RecT, which has also been widely used in linear dsDNA recombineering studies (4), was ≈40-fold less efficient than Beta, yielding 5.0 × 105 recombinants per 108 viable cells. EF2132* of E. faecalis, OrfC of L. pneumophila, s065 of V. cholerae, and plu2935 of P. luminescens generated recombinants at nearly the same frequency as Beta, whereas Orf245 from L. lactis phage and Orf48 of the A118 phage of L. monocytogenes had recombinogenic potential similar to that of RecT. Under identical experimental conditions, GP35 of B. subtilis, GP61 of M. smegmatis, and GP20 of S. aureus were relatively inefficient as they could only generate ≈104 recombinants per 108 viable cells. Nonetheless, even this lower frequency is well above the background recombination of ≈102 Gal+ recombinants per 108 viable cells obtained in strain HME75 that lacks both the Rac and Red recombination systems.
When the complementary oligo 101 was used, the level of recombination was reduced by ≈10-fold in each case (Table 1). As reported (18, 20), the direction of replication through the galK gene determines the efficiency of oligo recombination, such that the oligo corresponding to lagging-strand replication (oligo 100) is more efficient than the oligo corresponding to the leading-strand sequence (oligo 101).
Another oligo was used to repair the galK mutation; oligo144 corrects the tyrosine to TAC rather than TAT. When annealed, the oligo 144 TAC sequence forms a C:C mismatch with the parental strand. Such mismatches are not repairable by the MMR system (23), and, thus, high recombination frequencies are obtained even in MMR+ conditions (20). Recombineering with oligo 144 in strains SIMD40–50 gave essentially the same recombination frequency as oligo 100 in their mutS derivatives (data not shown).
We also assayed the ability of two of the foreign recombinases to delete a large DNA segment in a cross in which oligo 144 was used to remove the 3.3 kb cat-sacB insertion in galK to generate Gal+ recombinants. The two high-efficiency recombinases EF2132* in SIMD67 and OrfC in SIMD69, produced ≈2 × 105 Gal+ recombinants per 108 viable cells as did λ Beta in SIMD71.
λ Gam Function Not Essential for Red-Mediated Recombination with Linear dsDNA.
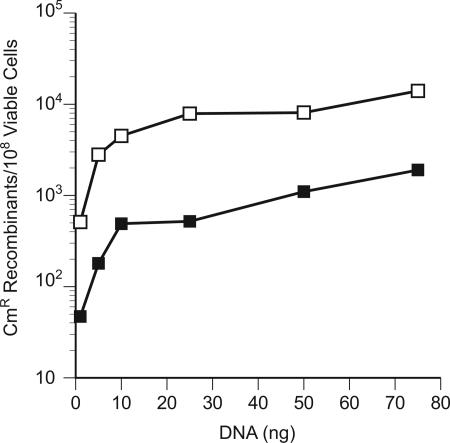
It has been stated that, in wild-type E. coli, recombination with long (≈1-kb), linear dsDNA carrying flanking target homologies requires all three λ Red functions, Gam, Exo, and Beta (3, 4, 15). We had observed that, with shorter dsDNA, Gam had only a small positive effect on dsDNA recombination (22). This study further corroborates the finding that Gam is not absolutely required for recombineering with dsDNA. Strain SIMD61 expressing λ bet and exo, but not gam, was used in dsDNA recombineering to replace the galK gene with a ≈1-kb cat cassette. Even in the absence of Gam, many CmR recombinants were generated with the a galK< >cat PCR product. Providing Gam function from plasmid pSIM22 increased the recombinant yield by ≈10-fold at each DNA concentration tested (Fig. 2). The < > symbol designates that the genetic change was created by recombineering methods (15).
Fig. 2.
Effect of Gam function on linear dsDNA recombination. The strain SIMD61 expresses only λ exo and bet from pL in the defective prophage. The plasmid pSIM22 provides λ Gam function in trans. SIMD61 along with SIMD61[pSIM22] were made competent for recombineering and different amounts of the galK< >cat PCR product were targeted to the galK gene. The number of CmR recombinants per 108 viable cells was plotted vs. the amount of DNA used. black rectangles, SIMD61; white rectangles, SIMD61[pSIM22].
dsDNA Recombineering by Cognate Exonuclease–Recombinase Pairs.
Four different recombinase–exonuclease pairs, GP35+GP34.1 (B. subtilis), Orf48+Orf47 (L. monocytogenes), OrfC+OrfB (L. pneumophila), and plu2935+plu2936 (P. luminescens), were tested for dsDNA recombination activity with the galK< >cat PCR product in the presence of Gam. All but the L. monocytogenes gene pair could catalyze dsDNA recombination, but only at very low efficiency (Table 2). With B. subtilis and L. monocytogenes recombinase–exonuclease pairs, the dsDNA recombination levels were expected to be low, because, in these cases, oligo recombination was almost 103-fold lower than that observed with λ Beta. Unexpectedly, for the L. pneumophila and P. luminescens protein pairs, where the recombinase-mediated oligo recombination frequency was almost as high as that of λ Beta, dsDNA recombination levels were found to be nearly1,000-fold lower than those of the λ Exo/Beta-mediated reaction (Table 2). Each of the recombinases in the four combinations exhibited oligo recombineering activity similar to that seen when expressed alone in Table 1.
Table 2.
Recombineering with different recombinase–exonuclease pairs
| Strains† | Recombinase–exonuclease | galK< >cat‡ | Oligo 144§ |
|---|---|---|---|
| SIMD61 | Beta + Exo | 1.7 × 104 | 1.1 × 107 |
| SIMD51 | GP35 + GP34.1 | 2.3 × 101 | 3.0 × 104 |
| SIMD53 | Orf48 + Orf47 | <1 | 3.4 × 105 |
| SIMD54 | OrfC + OrfB | 1.7 × 101 | 2.4 × 107 |
| SIMD55 | plu2935 + plu2936 | 7.2 × 101 | 1.7 × 106 |
†The recombinase–exonuclease gene pairs in these strains replace int through cIII in HME6 background. All strains carry pSIM22 that expresses λ Gam.
‡CmR recombinants per 108 viable cells recombinants generated from galK< >cat PCR product. The control strain HME6 expressing exo, bet, and gam from the λ prophage generated 2.0 × 104 CmR recombinants per 108 viable cells.
§Gal+ recombinants per 108 viable cells generated with oligo 144.
Testing the Abilities of Different Recombinases to Work with λ Exo in dsDNA Recombination.
To test whether the heterologous recombinases could work in conjunction with λ Exo in mediating dsDNA recombination, we assayed strains in which the λ exo gene was next to four of the highly efficient recombinase genes, so65, orfC, EF2132*, and recT. Gam was provided from plasmid pSIM22. None of the heterologous combinations with λ Exo generated recombinants, using the linear dsDNA galK< >cat DNA substrate. In each of these combinations, however, the recombinase remained active as judged by its ability to carry out oligo recombination at the level observed in Table 1.
Discussion
We have identified several genes encoding putative single-strand DNA annealing proteins from both Gram-negative and Gram-positive bacteria and their phages. Analysis of their recombination activities in E. coli demonstrates that they all catalyze oligo-mediated recombination despite their varied origins. It seems likely that, because these recombinases function in E. coli, they will also perform at least as well, and probably better, in their native hosts, where specific molecular interactions with host factors may be required. This bodes well for using single-strand oligo recombineering catalyzed by these and other recombinases as a general tool for initiating genetic studies in distantly related bacteria.
In the native hosts, these recombinases are encoded by bacterial phages, prophages, or their remnants, as are Beta and RecT in E. coli. The amino acid sequences of Beta and RecT and the other nine recombinases were aligned and a relatedness tree was generated (SI Fig. 3 and 4). Although functionally similar, Beta and RecT share almost no sequence identity. However, aligning all 11 recombinases reveals a continuum of similarities and identities spanning from Beta to RecT. Most of the other recombinases are also accompanied by known or putative exonucleases and, thus, resemble the viral recombination modules seen in λ and the Rac prophage.
To create the test strains for functional analysis, the red genes (exo, bet, and gam) present in the defective λ prophage were replaced by the putative recombinase genes. The pL promoter provides strong transcription and expression of these recombinases and at the same time, expression is tightly controlled by the λ repressor (24).
Oligo recombination was used as the defining assay for the various recombinases because it is simple and efficient, and the reaction can be accomplished by the recombinase function alone (18). Four of the recombinases exhibited high activities similar to λ Beta. Not surprisingly, this included s065 of V. cholerae, which is most closely related to Beta by amino acid sequence. M. Waldor has also demonstrated s065 has oligo activity in E. coli (M. Waldor, personal communication). The other three highly active recombinases, EF2132*, OrfC, and plu2935, diverge more from Beta, and one (EF2132*) derives from the Gram-positive bacteria E. faecalis.
Both Beta and RecT are widely used for recombineering (4, 15, 18). We were surprised to find that oligo recombination catalyzed by RecT was ≈40-fold lower than Beta. Two other RecT-like proteins, GP35 and GP61, were even less efficient than RecT. However, not all RecT-like proteins were inefficient; OrfC, which is closely related to RecT, was as competent as λ Beta. The three poorest of the recombinases, GP35, GP61, and GP20, are found in the phages of Gram-positive bacteria B. subtilis, M. smegmatis, and S. aureus, respectively. We do not know why some recombinases are better than the others under the same experimental conditions. Cellular levels and decay rates of GP61 were similiar to those of λ Beta by Western Blot analysis. No improvement in GP35- or GP61-mediated oligo-recombineering was observed in the presence of pRARE, a plasimd that supplies tRNAs, which should alleviate codon-bias problems (25) (data not shown).
To gain insight into the molecular mechanisms of these divergent recombinases, we studied oligo recombination with both leading- and lagging-strand oligos in the absence of mismatch repair. For all of the recombinases, the more efficient oligo corresponded to the lagging-strand of DNA replication, which generated ≈10-fold or more recombinants than did the leading-strand oligo (Table 1). This leading-strand vs. lagging-strand bias suggests that mechanistically these recombinases function like λ Beta, for which a similar phenomenon was first observed (18, 20). The two most efficient recombinases, EF2132* and OrfC, corrected both the point mutation and a 3.3-kb insertion mutation in galK as efficiently as λ Beta by oligo recombination.
The oligo-directed recombination is likely to occur at the DNA replication fork as the fork passes through the targeted region (1, 18). An intriguing question is what molecular interactions are mediated by these foreign recombinases with the E. coli replication machinery to successfully anneal the oligo to the appropriate target. This work demonstrates that several of the recombinases function efficiently across a species barrier, suggesting they may have evolved in diverse systems. By this logic, λ Beta should also function in other bacterial species to catalyze oligo recombination.
The success of oligo recombination with these recombinases prompted us to examine their ability to mediate linear dsDNA recombination in conjunction with λ exonuclease. None of the recombinases tested except λ Beta could achieve linear dsDNA recombination when coupled with λ Exo, although all of the recombinases were competent for oligo recombination in this configuration. When present with their canonical exonucleases, three of the four foreign recombinase–exonuclease pairs tested were able to carry out low level recombination with linear dsDNA. These results support earlier studies showing that neither heterologous combination of λ Exo with RecT nor RecE with λ Beta can catalyze dsDNA recombination (6), probably because a physical and specific coupling is required between the cognate exonuclease and single-strand annealing proteins.
Our model for Red-mediated dsDNA recombination, proposes that λ Exo processes the 5′ end of the dsDNA and that the associated Beta protein loads immediately on the 3′ ssDNA overhangs to form a dsDNA intermediate. This dsDNA intermediate may be annealed by Beta to the appropriate target at the replication fork. The functions of a host DNA polymerase like PolA, and DNA ligase are likely required to complete the recombination process (1, 22). It is not known which host proteins, if any, interact with Exo or Beta during the process, but RecE can interact with DNA ligase (26). Because the different recombinases function efficiently, it seems probable that for linear dsDNA recombination, it is the exonucleases that are less able to work in conjunction with the E. coli proteins. Thus, the exonuclease appears more species-specific, whereas the recombinase can function in diverse hosts. Based on this logic, we expect that some of the recombinase–exonuclease pairs will mediate recombination more efficiently in their native hosts. Linear dsDNA recombination has been attempted in Mycobacteria (27), using the same phage Che9c recombination genes we have examined here. In Mycobacteria, dsDNA recombination was catalyzed by GP60 and GP61, using 500 bp of target homology (27). We found that the GP61 recombinase performs relatively poorly in E. coli, using an oligo. This recombinase may have an inherently poor activity or a species-specific defect in E. coli.
This study demonstrates the ability of λ Exo and Beta to generate recombinants with dsDNA even in the absence of Gam function. Although Gam improves the recombination frequency by protecting the dsDNA from degradation, it is not essential, at least when λ Exo and Beta are optimally expressed. This finding suggests that recombineering with PCR products may also be successful in other bacterial species where Gam homologs are not easy to identify. Gam protection of linear dsDNA for the process of Red-mediated recombination conferred the same (10-fold) stimulation at both low and high DNA concentrations. It appears that dsDNA itself does not titrate RecBCD nuclease in the absence of Gam, because, if that was the case, Gam would have a greater effect at low DNA concentrations. Our results suggest that the recombinase–exonuclease complex itself may rapidly bind to the dsDNA and protect it from RecBCD.
This work demonstrates functionality in E. coli of a number of recombinases from diverse sources. For recombineering to succeed in other bacteria, several other factors must be considered. These include choice of expression system, method of introducting substrate DNA, and identification of recombinants. Once adapted, such systems would facilitate in vivo genetic engineering in both Gram-negative and Gram-positive bacteria and contribute to the development of functional genomics in those organisms.
Materials and Methods
Bacterial Strains and Their Construction.
Bacterial strains used in this work are listed in SI Table 4. Standard protocols for recombineering are described in detail elsewhere (28, 29). HME6 (18) carries a galKtyr145UAG amber mutation and a defective λ prophage. SIMD30 was made from HME6 by inserting the selectable-counterselectable cat-sacB cassette between nucleotide (nt) 27810–28805 (30) in the λ int gene (Fig. 1A). Thus, SIMD30 carries a defective λ prophage and is resistant to chloramphenicol (Cm) but sensitive to sucrose. This strain was used to express the Red functions for recombineering, and its prophage region was also the target for substituting the different recombinase genes so as to replace in entirety cat-sacB, int′, the native λ red genes, kil, and cIII to its ATG start codon (Fig. 1 B and C). Recombinants were selected on l-sucrose plates, purified, and shown to be Cm-sensitive. These isolates were analyzed by PCR using recombinase gene-specific and flanking primers; final constructs were confirmed by sequencing. Ten SIMD30 derivatives (SIMD40–49), each carrying a different recombinase gene in place of the int to cIII λ segment, are listed in SI Table 4. Four other derivatives of SIMD30 (SIMD51, 53, 54, 55) were also made, each of which carries different recombinase–exonuclease gene pairs at the same location.
We also placed the λ bet gene in the pL operon in the exact configuration as the foreign recombinase genes to make strain SIMD50. First, a deletion of the gam through cIII region in the pL operon (Fig. 1) of SIMD30 was created by recombination with the oligonucleotide TAACGCTTCACTCGAGGCGTTTTTCGTTATGTATAAATAAGGAGCACACC/ATGAGTACTGCACTCGCAACGCTGGCTGGGAAGCTGGCTG, where the slash represents the deletion junction. This moved the λ bet gene AUG initiation codon to the position of the AUG of gene cIII. Recombinants were be selected at 42°C as temperature-resistant survivors because the deletion removes the toxic kil gene between bet and cIII. This deletion mutant was then used in a second round of recombineering with a second oligonucleotide (GCTTTTTTATACTAAGTTGGCATTATAAAAAAGCATTGCTTATC-AATTTG/TCATGCTGCCACCTTCTGCTCTGCGGCTTTCTGTTTCAGG) to create another deletion from int through exo, removing the counterselectable cat-sacB cassette located in int, generating SIMD50.
P1 phage was grown on HME68 (29) and transduction was used to move the mutS< >cat gene replacement into strains SIMD40–50 to create derivatives (SIMD80–90) that are defective in methyl-directed mismatch repair (MMR).
The λ exo gene was inserted just beyond five different recombinase genes in the position and orientation as in phage λ. For this, SIMD31 was first constructed from HME6 by introducing an ampicillin resistance (ApR) cassette (amp) (15) downstream of exo just beyond the tL3 terminator between nucleotides 29847 and 31231 of λ (30). The λ exo and the now adjacent amp cassette were PCR-amplified from SIMD31 with specific primers, one of which contained 50 bases of homology to the att region at its 5′ end (see below), while the other primer had homology to the 3′ ends of the five recombinase genes. The strains SIMD44, 45, 47, 49, and 50 were transformed with the Red-expression plasmid pSIM5 (31), induced for Red function, and the amp-exo PCR products electroporated into the cells. The electroporation mix was diluted with 20 ml of L broth and grown nonselectively overnight at 37°C, causing the loss of the temperature-sensitive pSIM5 plasmid from the cells (31). The cultures were plated on l-ampicillin to select recombinants, which were colony purified and verified by PCR and sequence analyses. The new strains, SIMD57 to 61, carry the λ exo downstream of the recombinase genes and are listed in SI Table 4. The galK< >cat-sacB insertion was moved from strain HME31 (18) to SIMD57, 59, and 61 to create SIMD67, 69, and 71, respectively, by P1 transduction.
Amplification of Putative Phage Recombinase and Exonuclease Genes.
Standard PCR conditions were used to amplify either the putative recombinase genes or the recombinase–exonuclease pairs, using a high fidelity TaqDNA Polymerase (Invitrogen). Chromosomal DNA or plasmids carrying these different genes were used as templates. When plasmid DNA was used, the PCR product was digested with the restriction enzyme DpnI to eliminate the parental methylated plasmid DNA before use in recombineering. Each primer used for PCR was a hybrid in which the 5′ 50 bases contained the λ target homology and the 3′ end primed the recombinase or exonuclease genes as appropriate. In all cases one primer had 50 base homology to the λ att region (5′-GCTTTTTTATACTAAGTTGGCATTATAAAAAAGCATTGCTTATCAATTTG) corresponding to nucleotides 27725–27774 of λ (30), whereas the other primer had 50 base target homology immediately upstream of the cIII gene start codon, including nucleotides 33465–33514 of λ (5′-TAACGCTTCACTCGAGGCGTTTTTCGTTATGTATAAATAAGGAGCACACC). After PCR amplification, each foreign recombinase gene or recombinase–exonuclease pair is flanked by homology to these λ sequences to target the pL operon in the λ prophage (Fig. 1). The recombination event replaces the λ red genes and the cat-sacB segment with the foreign genes. The PCR products were purified with a PCR clean-up kit (Qiagen).
The putative recombinase EF2132 gene was amplified from the unsequenced E. faecalis strain CRMEN 19, and sequencing results showed amino acid coding differences at positions 170 (Y to D) and 229 (P to Q) with respect to EF2132 of the sequenced E. faecalis strain V583 (GenBank accession no. AE016830). We will refer to this recombinase as EF2132*.
Plasmids.
The plasmid pSIM22, carrying the λ gam gene, was constructed from the hygromycin resistant pSIM18 plasmid (31) by recombineering with the oligo (GTGATTGCGCCTACCCGGATATTATCGTGAGGATGCG/TCGTTTTATACCTCTGAATCAATATCAACCTGGTGGTGAGCAA), to precisely delete the exo and bet genes, fusing nucleotides 31348–32812 of λ (30) at the position indicated by the slash in the oligo sequence. A unique SalI site is present in the λ bet gene in pSIM18 but is absent in the recombinant pSIM22. After recombineering with the oligo, plasmid DNA was purified and digested with SalI to eliminate parental pSIM18. This digested DNA was retransformed, and a purified plasmid, pSIM22, was isolated and sequenced.
The pRARE plasmid was obtained from Novagen and used to transform strains SIMD40, 41, and 49.
Media and Chemicals.
L broth contains 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter. For selecting against sacB, modified L plates, lacking any added NaCl, were supplemented with 6% (wt/vol) sucrose (32). Gal+ recombinants were selected on M63 minimal agar plates with galactose (0.2%) and biotin (0.001%) (18). For drug resistant selection, each milliliter of L medium contained 10 μg of chloramphenicol, 30 μg of ampicillin, or 50 μg of hygromycin B (Invitrogen). All of the oligonucleotides were purchased from Integrated DNA Technologies as salt-free without further purification.
Recombination Assays with Oligo and dsDNA.
The 70 base oligos 144, 100, and 101 were used to correct either the galKtyr145UAG amber mutation or the galK< >cat-sacB insertion to gal+ by recombineering as described in ref. 20. All of the oligos correct the TAG stop codon of the galK to a tyrosine codon (either TAC or TAT) allowing selection of Gal+ recombinant colonies on minimal galactose plates. For recombineering with dsDNA, a cat cassette was used to replace the galK gene of E. coli as described in ref. 15. The strains SIMD57–61 were transformed with the Gam-plasmid pSIM22, induced at 42°C to express the recombinase–exonuclease gene pairs, electroporated with the galK< >cat PCR product, and Cm-resistant recombinants were selected at 32°C. For both assays, total viable cells were counted on L agar (20).
Predicting Putative Recombinases and Associated Exonucleases from Gram-Negative and Gram-Positive Bacteria and Their Phages.
BLASTP searches of the National Center for Biotechnology Information nonredundant database were performed using the amino acid sequences of λ Beta, encoded by nucleotides 32028–32810 of λ (30) or E. coli RecT (nucleotides 1412008–1412817) (GenBank accession no. NC_000913). A list of the candidate recombinases identified by the BLAST searches and included in this study is given in SI Table 3 along with the source of the genetic material used for amplification. Sequence annotation revealed that most of these recombinases lie next to bacteriophage-like exonucleases. The relative positions of the two genes with respect to the promoter are indicated in SI Table 3.
All amino acid sequences of the recombinases were aligned with CLUSTAL W (33) (SI Fig. 3 A–C), and a relatedness tree (SI Fig. 4) was also generated with PHYLIP software, Version 3.6 (34).
ACKNOWLEDGMENTS.
We thank L. Thomason, J. Sawitzke, and M. Bubunenko for many discussions and help with the manuscrip; those who sent us material for this work and who are acknowledged further in SI Table 3; K. Murphy (University of Massachusetts Medical School, Worcester, MA) for Beta antibody; G. Hatfull (University of Pittsburgh, Pittsburgh) for GP61 antibody; Gary W. Smythers (Advanced Biomedical Computing Center, Science Applications International Corporation-Frederick, Frederick Cancer Research Facility) for assistance with bioinformatic analysis and discussions; and Marti Welch (Scientific Publications, Graphics, and Media, Science Applications International Corporation-Frederick) for help in illustrations. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and in part by a Trans National Institutes of Health/Food and Drug Administration Intramural Biodefense Program Grant of National Insitutes of Allergy and Infectious Diseases (to D.L.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709089105/DC1.
References
- 1.Court DL, Sawitzke JA, Thomason LC. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 2.Copeland NG, Jenkins NA, Court DL. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 3.Murphy KC. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 5.Clark AJ, Sharma V, Brenowitz S, Chu CC, Sandler S, Satin L, Templin A, Berger I, Cohen A. J Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer LM, Koonin EV, Aravind L. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Little JW. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 9.Joseph JW, Kolodner R. J Biol Chem. 1983;258:10418–10424. [PubMed] [Google Scholar]
- 10.Karakousis G, Ye N, Li Z, Chiu SK, Reddy G, Radding CM. J Mol Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 12.Hall SD, Kolodner RD. Proc Natl Acad Sci USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unger RC, Clark AJ. J Mol Biol. 1972;70:539–548. doi: 10.1016/0022-2836(72)90558-x. [DOI] [PubMed] [Google Scholar]
- 14.Kulkarni SK, Stahl FW. Genetics. 1989;123:249–253. doi: 10.1093/genetics/123.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy KC, Campellone KG, Poteete AR. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 18.Ellis HM, Yu D, DiTizio T, Court DL. Proc Natl Acad Sci USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan S, Ellis HM, Waters LS, Yu D, Lee EC, Court DL, Sharan SK. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 20.Costantino N, Court DL. Proc Natl Acad Sci USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. BMC Mol Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, Sawitzke JA, Ellis H, Court DL. Proc Natl Acad Sci USA. 2003;100:7207–7212. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modrich P. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 24.Court DL, Oppenheim AB, Adhya SL. J Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Martinez I, Navarro-Fernandez J, Lozada-Ramirez JD, Garcia-Carmona F, Sanchez-Ferrer A. Biotechnol Prog. 2006;22:647–652. doi: 10.1021/bp050397g. [DOI] [PubMed] [Google Scholar]
- 26.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 27.van Kessel JC, Hatfull GF. Nat Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 28.Thomason LC, Bubunenko M, Costantino N, Wilson H, Oppenheim A, Datta S, Court DL. Current Protocols in Molecular Biology. Hoboken, NJ: Wiley; 2005. pp. 1–21. [DOI] [PubMed] [Google Scholar]
- 29.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Methods Enzymol. 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 30.Daniels DL, Schroeder JL, Szybalski W, Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB, Blattner FR. In: Lambda II. Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1983. pp. 519–676. [Google Scholar]
- 31.Datta S, Costantino N, Court DL. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Blomfield IC, Vaughn V, Rest RF, Eisenstein BI. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felsenstein J. PHYLIP, Phylogeny Interface Package. Seattle: Departments of Genome Sciences and Biology, University of Washington; 2005. Version 3.6. [Google Scholar]