Abstract
Loss-of-function mutations in the PTEN-induced kinase 1 (PINK1) or parkin genes, which encode a mitochondrially localized serine/threonine kinase and a ubiquitin-protein ligase, respectively, result in recessive familial forms of Parkinsonism. Genetic studies in Drosophila indicate that PINK1 acts upstream of Parkin in a common pathway that influences mitochondrial integrity in a subset of tissues, including flight muscle and dopaminergic neurons. The mechanism by which PINK1 and Parkin influence mitochondrial integrity is currently unknown, although mutations in the PINK1 and parkin genes result in enlarged or swollen mitochondria, suggesting a possible regulatory role for the PINK1/Parkin pathway in mitochondrial morphology. To address this hypothesis, we examined the influence of genetic alterations affecting the machinery that governs mitochondrial morphology on the PINK1 and parkin mutant phenotypes. We report that heterozygous loss-of-function mutations of drp1, which encodes a key mitochondrial fission-promoting component, are largely lethal in a PINK1 or parkin mutant background. Conversely, the flight muscle degeneration and mitochondrial morphological alterations that result from mutations in PINK1 and parkin are strongly suppressed by increased drp1 gene dosage and by heterozygous loss-of-function mutations affecting the mitochondrial fusion-promoting factors OPA1 and Mfn2. Finally, we find that an eye phenotype associated with increased PINK1/Parkin pathway activity is suppressed by perturbations that reduce mitochondrial fission and enhanced by perturbations that reduce mitochondrial fusion. Our studies suggest that the PINK1/Parkin pathway promotes mitochondrial fission and that the loss of mitochondrial and tissue integrity in PINK1 and parkin mutants derives from reduced mitochondrial fission.
Keywords: drp1, opa1, parkinsonism
Parkinson's disease (PD) is a common movement disorder caused by the degeneration of dopaminergic neurons in the midbrain. The molecular mechanisms underlying neurodegeneration in PD remain unclear, although substantial evidence suggests that mitochondrial dysfunction is a major contributor: Several mitochondrial toxins induce PD-like symptoms in humans and animal models (1, 2); systemic mitochondrial dysfunction appears to be a feature of a large proportion of PD sufferers (3); and several genes involved in rare heritable forms of Parkinsonism have been implicated in mitochondrial biology, including the PTEN-induced kinase 1 (PINK1) and parkin genes (4, 5).
The PINK1 and parkin genes encode a mitochondrially localized serine/threonine kinase and an E3 ubiquitin-protein ligase, respectively (6–13). Although a number of substrates of PINK1 and Parkin have been described, these advances have led to dramatically varying models of pathogenesis (5, 14–19), making it unclear precisely how PINK1 and Parkin influence neuronal integrity. Genetic studies of highly conserved Drosophila orthologs of parkin and PINK1 indicate that PINK1 acts upstream of Parkin in a common pathway that influences the integrity of flight muscle, sperm, and a subset of dopaminergic neurons in the brain (20–24). Mitochondrial dysfunction is a prominent and early feature of the fly tissues that degenerate in parkin and PINK1 mutants (20–24), suggesting that the PINK1/Parkin pathway influences mitochondrial integrity. However, the mechanism by which the PINK1/Parkin pathway impacts mitochondrial integrity is currently unknown.
The Drosophila tissues that are most profoundly affected by mutations in PINK1 and parkin, flight muscle and sperm cells, exhibit distinctive mitochondrial morphologies, raising the possibility that PINK1 and Parkin function to regulate mitochondrial morphology. The dynamic regulation of mitochondrial morphology is critical to mitochondrial function, where a shift to either a fused reticulum or a fragmented state leads to disease (25). Although the regulation of mitochondrial morphology has been little studied in metazoans, recent work has led to the identification of evolutionarily conserved GTPase family members that play critical roles in the mechanics of mitochondrial fission and fusion (25, 26). Three of these GTPases, Mitofusin1 (Mfn1), Mitofusin2 (Mfn2), and optic atrophy 1 (OPA1), promote mitochondrial fusion (27, 28). Mfn1 and Mfn2 reside in the outer mitochondrial membrane and promote outer membrane fusion, whereas OPA1 resides in the intermembrane space, where it promotes inner membrane fusion. Mitochondrial fission depends on another GTPase, the dynamin-related protein 1 (Drp1) (29, 30). Drp1 is a cytoplasmic protein that assembles with mitochondria and promotes the fission event. However, it remains largely unknown how Drp1 is recruited to mitochondria, and how this and other known components of the mitochondrial fission and fusion machinery are regulated.
To test the hypothesis that the PINK1/Parkin pathway regulates mitochondrial morphology, we explored the effects of altering the gene dosage of known mitochondrial fission and fusion-promoting components on the Drosophila PINK1 and parkin mutant phenotypes. Our studies show that perturbations that reduce mitochondrial fission enhance the PINK1 and parkin mutant phenotypes. By contrast, perturbations that reduce mitochondrial fusion or increase mitochondrial fission suppress the PINK1 and parkin mutant phenotypes. Our findings indicate that the PINK1/Parkin pathway promotes mitochondrial fission and suggest that the tissue loss accompanying reduced PINK1 and Parkin activity derives from reduced mitochondrial fission.
Results
Genetic Interaction of PINK1 and Parkin.
As a prelude to the current study, we sought to independently verify and extend recent work indicating that Parkin acts downstream of PINK1 (22–24). By using a strong muscle-specific GAL4 driver that can rescue the muscle defects of parkin mutants when used in conjunction with a GAL4-responsive Drosophila or Human parkin transgene (20) [supporting information (SI) Fig. 5A], we confirmed that Parkin overexpression was capable of rescuing the muscle defects of PINK1 mutants (SI Fig. 5 B and C). We also verified that PINK1 overexpression was unable to rescue the flight muscle degeneration of parkin mutants and that PINK1;parkin double mutants are phenotypically similar to the respective single mutants (data not shown). However, parkin mutants are shorter-lived, eclose from the pupal case at a lower frequency and are developmentally delayed relative to PINK1 mutants (refs. 20–22, 24, and 31 and data not shown), raising the possibility that Parkin may play additional biological roles not shared by PINK1 and vice versa.
Our experiments to address the effects of PINK1 overexpression on the parkin phenotypes revealed that abundant expression of PINK1 yielded only a small number of viable progeny, suggesting that PINK1 overexpression is toxic. In further support of this conclusion, we found that overexpression of human PINK1 or Drosophila PINK1 in the visual system resulted in a rough eye phenotype (SI Fig. 6B and data not shown). To test whether the toxicity associated with PINK1 overexpression derives from excessive signaling through the PINK1/Parkin pathway, we performed several experiments. First, we tested whether mutations in parkin could suppress the PINK1 overexpression phenotypes. We found that, although use of the strong Dmef2-GAL4 driver to overexpress PINK1 in WT flies was completely lethal, use of this same driver to overexpress PINK1 in a parkin null background yielded a small number of viable flies. Moreover, the eye phenotype associated with PINK1 overexpression is substantially attenuated in a parkin null background (SI Fig. 6 C and E). Finally, coexpression of PINK1 and Parkin in the eye caused a severe synergistic toxicity, resulting in dramatic loss of eye tissue and partial lethality (SI Fig. 6G). These findings provide independent confirmation that Parkin acts downstream of PINK1 in a linear pathway and indicate that the toxicity associated with PINK1 overexpression results from enhanced signaling through the PINK1/Parkin pathway.
Loss-of-Function Mutations of drp1 Enhance the parkin and PINK1 Mutant Phenotypes.
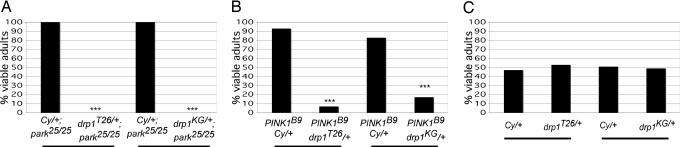
To explore the hypothesis that the PINK1/Parkin pathway promotes mitochondrial fission, we first tested whether PINK1 and parkin interact genetically with drp1, which encodes a key mitochondrial fission-promoting component. We found that large deletions (Df(2L)D20 and Df(2L)Excel6008) and specific loss-of-function alleles of drp1 (drp1T26 and drp1KG03815) were fully lethal in a parkin null background (Fig. 1A and data not shown). Lethal phase analyses indicate that parkin mutants bearing a heterozygous mutation of drp1 were developmentally delayed and died primarily during the larval stages of development. We also found that introducing heterozygous loss-of-function alleles of drp1 into a PINK1 mutant background resulted in nearly complete lethality (Fig. 1B). The few surviving adult PINK1 mutants bearing a heterozygous drp1 mutation emerged from the pupal case 2–5 days later than PINK1 mutants bearing WT alleles of drp1, and were substantially smaller and shorter-lived than PINK1 mutants in a WT drp1 background. By contrast, all of the drp1 alleles tested were fully viable as heterozygotes in a WT, parkin heterozygous, or PINK1 heterozygous background, indicating that reduced drp1 gene dosage does not generally confer loss of viability (Fig. 1C and data not shown).
Fig. 1.
drp1 is a parkin and PINK1 enhancer. (A) A stock bearing the parkin null allele park25 was mated to another stock that also carried the park25 allele and a given drp1 allele in trans to a recombination-suppressing chromosome bearing the dominant marker Cy (designated CyO). For each drp1 allele tested, the percentage of Cy (drp1+/+) and non-Cy (drp1+/−) park25/park25 offspring that resulted from the cross is shown. (B) Female flies heterozygous for the PINK1B9 deletion allele were mated to flies bearing a given drp1 allele in trans to the CyO chromosome and the percentage of Cy (drp1+/+) and non-Cy (drp1+/−) PINK1B9 hemizygous offspring (the PINK1 gene is on the X chromosome) is shown. (C) A WT fly stock was mated to flies bearing a given drp1 allele in trans to the CyO chromosome, and the percentage of Cy (drp1+/+) and non-Cy (drp1+/−) offspring that resulted is shown. Note that no significant difference from Mendelian expectations is detected in offspring frequency in crosses carried out with the drp1 alleles in a WT parkin and PINK1 background (P = 0.16 for drp1T26 in a WT background; P = 0.67 for drp1KG in a WT background). Genotypes: drp1KG = drp1KG03815. The number of offspring scored from each cross was >100. ***, P < 1 × 10−5. All statistical analyses were performed by using χ2 analysis.
Increased Drp1 Activity or Decreased OPA1 or Mfn2 Activity Suppress the parkin and PINK1 Mutant Phenotypes.
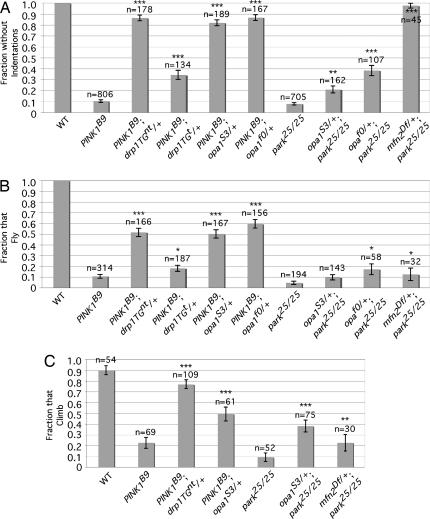
The genetic interaction of drp1 with parkin and PINK1 may indicate that Parkin and PINK1, like Drp1, act to promote mitochondrial fission. Alternatively, our findings may simply reflect a non-specific additive effect of two different insults to mitochondrial integrity. Several experiments were performed to distinguish these possibilities. First, we tested whether increasing the drp1 gene dosage would suppress the muscle degeneration phenotype of PINK1 mutants. To perform this analysis, a single copy of a transgene consisting of the drp1 gene under control of its natural promoter was introduced into a PINK1 mutant background (the drp1 transgenes used in this work appear to be tightly linked to the parkin gene and thus were not used in experiments with parkin mutants). Both of the drp1 transgenes conferred substantial suppression of the thoracic indentations of PINK1 mutants (Fig. 2A). The drp1 transgenes also rescued the flight and climbing defects that accompany muscle degeneration in PINK1 mutants (Fig. 2 B and C). These findings support the model that the PINK1/Parkin pathway acts to promote mitochondrial fission.
Fig. 2.
Increased drp1 gene dosage and loss-of-function mutations in opa1 and mfn2 suppress the parkin and PINK1 mutant phenotypes. (A) The frequency of adult flies of the indicated genotypes that lack thoracic indentations. (B) The frequency of adult flies of the indicated genotypes capable of flight. (C) The climbing ability of adult flies of the given genotypes. Genotypes: drp1TGnt = drp1 transgene; drp1TGt = epitope-tagged drp1 transgene; opa1S3 = opa1l(2)S3475; opa1f0 = opa1f02779; mfn2Df = Df (1)Excel6239. n = number of animals of the given genotypes analyzed. Statistical analysis was done by using Student's t test. *, P < 0.05; **, P < 0.001; ***, P < 1 × 10−5. Error bars indicate the SEM.
Previous work indicates that mutations in mitochondrial fusion-promoting components can partially suppress the excessive mitochondrial fusion resulting from mutations in mitochondrial fission components (25, 26, 32). Therefore, we further explored the model that the PINK1/Parkin pathway promotes mitochondrial fission by testing whether mutations in opa1 (CG8479), which encodes an essential component of the mitochondrial inner membrane fusion apparatus, could suppress the parkin and PINK1 mutant phenotypes. We also tested whether a deletion that removes the Drosophila mfn2 gene (CG3869), which encodes an essential component of the mitochondrial outer membrane fusion apparatus could suppress the parkin phenotypes (the mfn2 and PINK1 genes both reside on the X chromosome, and the phenotypes conferred by alleles of these genes prevented the generation of appropriate stocks to study genetic interactions between these factors). We found that heterozygous loss-of-function mutations of opa1 strongly suppressed the frequency and severity of thoracic indentations, and the flight and climbing defects of PINK1 mutants (Fig. 2 and SI Fig. 7). Heterozygous loss-of-function mutations in opa1 and a heterozygous mfn2 deletion also significantly suppressed the frequency of thoracic indentations and climbing defects of parkin mutants (Fig. 2 A and C), although the suppressive effects of the opa1 alleles on the parkin mutant phenotypes were of lesser magnitude than their effects on the PINK1 mutant phenotypes, and only one of the opa1 alleles was able to significantly suppress the flight defect of parkin null mutants (Fig. 2B). However, both opa1 alleles decreased the severity of thoracic indentations in a parkin null background and strongly suppressed the thoracic indentation frequency and flight defect in a parkin hypomorphic background (SI Fig. 7). Together, these findings offer further support for the model that the PINK1/Parkin pathway acts to promote mitochondrial fission.
The PINK1 Overexpression Phenotype Is Modified by Genetic Manipulation of Mitochondrial Fission/Fusion Components.
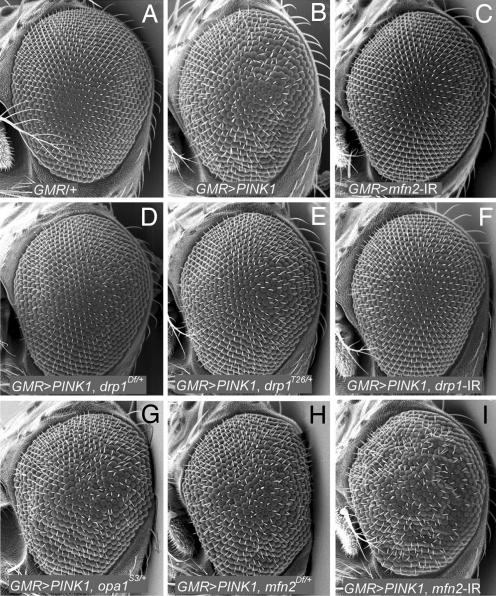
To further test the hypothesis that the PINK1/Parkin pathway promotes mitochondrial fission, we made use of the PINK1 eye overexpression phenotype. If the increased PINK1/Parkin pathway signaling that results from PINK1 overexpression involves excessive activation of mitochondrial fission, then genetic alterations that reduce mitochondrial fission should suppress this phenotype. Conversely, genetic alterations that decrease mitochondrial fusion should enhance the PINK1 overexpression phenotype. To test these predictions, we introduced heterozygous loss-of-function alleles of the drp1, opa1, and mfn2 genes into a PINK1 eye overexpression background. Because the compound eye is dispensable for viability, we also used an RNA interference (RNAi) approach to explore the effects of attenuating Drp1 and Mfn2 activity on the PINK1 eye overexpression phenotype. Results of our studies demonstrated that reduced Drp1 activity strongly suppressed the PINK1 eye overexpression phenotype (Fig. 3 D–F). Conversely, reduced Opa1 and Mfn2 activity enhanced the PINK1 eye overexpression phenotype (Fig. 3 G–I). These findings provide additional evidence that the PINK1/Parkin pathway acts to promote mitochondrial fission.
Fig. 3.
Genetic perturbations of mitochondrial fission and fusion components modified the PINK1 eye overexpression phenotype. (A) Compound eye from a WT fly showing regular arrangement of ommatidia. (B) Compound eye from a fly overexpressing PINK1 showing disruption of ommatidial structure. (C) Compound eye from a fly expressing a mfn2-inverted repeat (mfn2-IR) demonstrates that perturbation of Mfn2 had no effect on eye morphology. (D–F) A heterozygous deletion that removes the drp1 gene (D), a point mutation of drp1 (E), and an inverted repeat targeting the drp1 transcript (F) all suppress the PINK1 eye overexpression phenotype. (G–I) A heterozygous mutation of opa1 (G), a deletion of the mfn2 gene (H), and an inverted repeat targeting the mfn2 transcript (I) all enhance the PINK1 eye overexpression phenotype. Genotypes: drp1Df = Df(2L)C144; opa1S3 = opa1l(2)S3475; mfn2Df = Df (1)Excel6239.
Increased Drp1 and Decreased OPA1 Activity Suppress the Mitochondrial Morphological Defects of PINK1 and parkin Mutants.
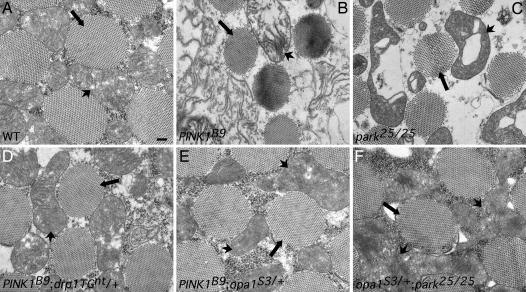
To investigate the effects of altered drp1 and opa1 gene dosage on mitochondrial morphology in PINK1 and parkin mutants, we subjected flight muscle from the appropriate animals to transmission electron microscopy. Given the severe mitochondrial morphological alterations of parkin and PINK1 mutants alone, we focused exclusively on genetic alterations that appear to suppress the flight muscle degeneration phenotypes in our studies. Specifically, we examined the effects of increased drp1 gene dosage in PINK1 mutants and decreased opa1 gene dosage in PINK1- and parkin-deficient genetic backgrounds. As reported in refs. 20–24, the flight muscle of 1-day old PINK1 and parkin mutants exhibited swollen mitochondria with disorganized and fragmented cristae relative to flight muscle from WT flies (Fig. 4 A–C and SI Fig. 8). Genetic perturbations that increase drp1 gene dosage in a PINK1 mutant background or that reduce opa1 gene dosage in a PINK1 or parkin mutant background conferred substantial rescue of the mitochondrial morphological defects in flight muscle (Fig. 4 D–F and SI Fig. 8). Specifically, mitochondria from flies bearing alterations that increase drp1 gene dosage or that decrease opa1 gene dosage in a parkin or PINK1 null background were less swollen and had substantially more intact cristae structures relative to PINK1 or parkin mutants alone (Fig. 4 and SI Fig. 8).
Fig. 4.
Increased drp1 gene dosage and decreased opa1 gene dosage suppress the mitochondrial morphological defects of PINK1 and parkin mutants. For each of the designated genotypes, transverse sections of one-day old adult flight muscle were examined using transmission electron microscopy. (A) Flight muscle from WT flies show regular arrangement of myofibrils (arrows) and densely packed mitochondria (arrowheads) with intact cristae. (Scale bar, 200 nm. All other images are at the same scale.) (B–F) Both increased drp1 gene dosage (D) and decreased opa1 gene dosage (E and F) substantially suppress the morphological defects seen in PINK1 (B) and parkin (C) mutants alone.
To further evaluate the effects of parkin and PINK1 deficiency on mitochondrial morphology, we used RNAi to inactivate parkin and PINK1 in S2 cells. Coinactivation of parkin and PINK1 resulted in a dramatic increase in mitochondrial interconnectivity and tubule structure relative to untreated S2 cells (SI Fig. 9). Similar but less dramatic changes in mitochondrial structure were also seen in S2 cells, in which parkin and PINK1 were individually silenced with RNAi (data not shown). Consistent with the results of our studies in flight muscle, the mitochondrial morphological alterations induced by coinactivation of parkin and PINK1 were suppressed by RNAi inactivation of either opa1 or mfn2 (SI Fig. 9). These findings, together with the other observations described in our article, offer strong evidence that the PINK1/Parkin pathway promotes mitochondrial fission.
Discussion
In ref. 20, we showed that loss-of-function mutations in the Drosophila parkin ortholog result in profound mitochondrial pathology, leading us to argue that Parkin acts to promote mitochondrial integrity. Mitochondrial defects have subsequently been detected in Parkin-deficient worms (33), mice (34), and humans (35, 36), and recent studies in Drosophila have shown that PINK1 and Parkin act in a common pathway that influences mitochondrial integrity (22–24). Our current work advances these findings by providing evidence that the PINK1/Parkin pathway acts to promote mitochondrial fission. Five major findings support this conclusion: (i) Loss-of-function mutations of PINK1 and Parkin result in enlarged or swollen mitochondria in Drosophila tissues and cell lines; (ii) loss-of-function mutations of drp1 enhance the PINK1 and parkin mutant phenotypes; (iii) loss-of-function mutations of opa1 and mfn2 or increased Drp1 activity suppress the parkin and PINK1 mutant phenotypes; (iv) perturbations that reduce Drp1 activity suppress the eye phenotype associated with PINK1/Parkin pathway activation; and (v) perturbations that reduce opa1 and mfn2 activity enhance the eye phenotype associated with PINK1/Parkin pathway activation.
Although our work advances our understanding of the PINK1/Parkin pathway, it also raises a number of new questions. One important question is whether insights gained from studies of the PINK1/Parkin pathway in Drosophila are relevant to the mechanisms underlying neuron loss in PD. Although mitochondrial defects have been documented in patients with mutations in PINK1 and parkin, and recent work has shown that the mitochondrial morphological alterations accompanying mutations in the human PINK1 gene can be rescued by Parkin overexpression, the specific mitochondrial morphological alterations reported in PINK1-deficient human cells differ from those documented in our current study (37). Also, there is currently no direct evidence for the involvement of dysfunctional mitochondrial dynamics in PD. Although further work will be required to resolve differences in the effects of mutations in PINK1 on mitochondrial morphology in Drosophila and human cell lines, recent work has shown that several different nervous system disorders result from impairments in mitochondrial dynamics (25, 28, 38, 39), suggesting that the nervous system is selectively vulnerable to perturbations in mitochondrial dynamics. Studies with cultured cells also indicate that defects in mitochondrial fission confer increased reactive oxygen species production and DNA damage (40, 41). Because oxidative stress is a prime suspect in the mechanism of neuronal loss in sporadic and heritable forms of PD (1, 3–5), our current findings raise the possibility that impaired mitochondrial fission in parkin and PINK1 mutants increases oxygen radical stress, resulting ultimately in dopamine neuron cell death. Alternatively, Parkin and PINK1 may promote fission as part of a mechanism to segregate small damaged mitochondrial units for elimination through autophagy. Future work will be required to test these models.
Our current findings also raise questions about the apoptotic mechanism of cell death in Parkin and PINK1 deficient tissues. Recent studies have shown that mitochondrial fission promotes the cristae remodeling that facilitates cytochrome c release in some forms of apoptosis (42, 43). Thus, our findings suggest that the cell death mechanism in Parkin and PINK1-deficient tissues is a fission-independent form of apoptosis. Indeed, if there is already sufficient cytochrome c available to activate the downstream caspase cascades, then the requirement for Drp1 and cristae remodeling may not be absolute (44). Because mitochondria in PINK1 and parkin mutants appear to be swollen and ruptured, these morphological alterations may negate the requirement for further Drp1-mediated remodeling during apoptosis. Further work will be required to distinguish these models and define the apoptotic mechanism of cell death in PINK1 and parkin mutants.
Finally, an extremely important question for future study concerns the mechanism by which the PINK1/Parkin pathway influences mitochondrial morphology. Given that PINK1 appears to act upstream of Parkin and that most previous work places PINK1 within the mitochondrial inner membrane space and Parkin in the cytoplasm, we propose that PINK1 acts through a signal transduction cascade to promote Parkin to ubiquitinate particular cytoplasmic targets of the mitochondrial morphogenesis machinery. One possible target of the PINK1/Parkin pathway is Drp1. It is currently unclear how Drp1 is recruited to mitochondria from the cytoplasm to promote the fission event (25), although Drp1 is subjected to multiple posttranslational modifications, including phosphorylation (45, 46), SUMOylation (47–49), and ubiquitination (50, 51), raising the possibility that one or more of these posttranslational modifications regulates the recruitment of Drp1 to mitochondria. Because Parkin can apparently promote both degradative and nondegradative forms of ubiquitination (52, 53), the ubiquitination of Drp1 by Parkin could act in a nondegradative fashion to promote Drp1 to assemble with mitochondria to activate fission. Alternatively, the PINK1/Parkin pathway may indirectly promote mitochondrial fission by inhibiting a key component of the mitochondrial fusion machinery. In this potential model, the mitofusins, which reside in the mitochondrial outer membrane, represent attractive candidate targets of a degradative form of Parkin-mediated ubiquitination. These and other potential models by which the PINK1/Parkin pathway promotes mitochondrial fission can be readily addressed in future work.
Materials and Methods
Drosophila Strains and Culture.
Drosophila stocks were maintained on standard cornmeal molasses food at 25°C. The drp1, UAS-PINK1, and UAS-hparkBWL transgenic lines are described in refs. 54, 24, and 55 and were obtained from H. J. Bellen (Baylor College of Medicine, Houston), J. K. Chung (Korea Advanced Institute of Science and Technology, Taejon, Korea), and B. Lu (Stanford University School of Medicine, Stanford, CA), respectively. The park25 (20), parkZ472 (20, 56), Df(2L)Excel6008 (54), drp1T26 (54), drp1KG03815 (54, 57), and opa1l(2)S3475 (58) alleles have been described and were obtained from the Bloomington Stock Center or were generated in house. The PINK1B9 allele is described in ref. 24 and was obtained from J. K. Chung. Inducible RNAi lines that express short inverted repeats (IR) UAS-drp1-IR and UAS-mfn2-IR were obtained from the Vienna Drosophila Resource Centre. The opa1f02779 mutant stock was obtained from the Exelixis collection at Harvard, and the Df(2L)D20, Df(1)Excel6239, and Df(2L)C144 deletion stocks were obtained from the Bloomington Stock Center. The ey-GAL4 (59) and gmr-GAL4 (60) driver lines have been characterized and were obtained from the Bloomington Stock Center. The dmef2-GAL4 driver line is described in ref. 61 and was obtained from R. Ordway (Pennsylvania State University, University Park, PA).
Behavioral and Morphological Assays.
One- to 2-day-old adult flies of the appropriate genotypes were individually analyzed under a microscope for the presence of thoracic indentations. If any indentation was present, the fly was assigned a score of zero. If no indentations were visible, the fly was assigned a score of one. Severe indentations were defined as those that could be readily detected using a low power microscope objective, whereas mild indentations required a higher power objective for confirmation. Student's t test was used to assess whether the mean scores of the experimental and control genotypes were statistically different from each other.
Flight assays were performed by using a modified version of a procedure described in ref. 54. Flight assays were carried out by gently tapping 1- to 2-day-old flies through a funnel placed 38 cm above a 14-cm-diameter Petri dish containing a small amount of mineral oil. Flies that landed in the mineral oil were assigned a score of zero, whereas flies that managed to avoid the mineral oil were assigned a score of one. Student's t test was used to assess whether the mean scores of the experimental and control genotypes were statistically different from each other. Climbing assays were carried out as described in ref. 20.
RNAi Treatment of Drosophila S2 Cells.
S2 cells in a 24-well plate were transfected with a pMT plasmid (Invitrogen) encoding a mitochondrially targeted PAGFP (47) and then treated with 2 μg of the appropriate double stranded RNA for 3–4 days as described in ref. 31. Further details provided in SI Materials and Methods.
Microscopy.
For transmission electron microscopy, thoraces were dissected from 1-day-old flies of the appropriate genotypes and fixed in 2% paraformaldehyde and 2% glutaraldehyde overnight. After rinsing in 0.1 M sodium cacodylate with 0.001% calcium chloride, samples were postfixed in 1% OsO4 in cacodylate buffer for 1 h. Samples were rinsed, dehydrated in an ethanol series, and embedded in Epon. Transverse sections were examined with a JEOL JEM 1200EXII transmission electron microscope. Quantification of aberrant mitochondria involved two animals of each genotype analyzed. For scanning electron microscopy, heads were fixed in 4% paraformaldehyde overnight, dehydrated in an ethanol series, gold-coated, and analyzed on a XL-20 scanning electron microscope. Mitochondrial morphology of S2 cells was documented with an Olympus FV-1000 confocal microscope.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Josh Akey for help with statistics; Nishi Gill and Stephanie Lara for technical assistance with electron microscopy; Dr. Hugo J. Bellen for drp1 transgenic lines; Dr. J. K. Chung for PINK1 mutant and transgenic stocks; Dr. Bingwei Lu for a UAS-responsive human Parkin transgene; Dr. Richard Ordway for the Dmef2-GAL4 line; and the Bloomington, Exelixis, and Vienna Drosophila stock centers for other fly stocks used in our studies. We received core facilities support to perform transmission electron and confocal microscopy from the University of Washington Center for Human Development and Disabilities, which is funded by National Institute of Neurological Disorders and Stroke Grant P30-HD02774. This work was supported in part by Genetic Approaches to Aging Training Grant 5T32AG00057–28a (to A.C.P.), National Institutes of Health Grants 1RO1NS41780-01 and 1R21NS053762-01 (to L.J.P.), and the Parkinson's Disease Society and Wellcome Trust (A.J.W.). The MRC Centre for Developmental and Biomedical Genetics is supported by Medical Research Council Grant G070091.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709336105/DC1.
References
- 1.Sherer TB, Betarbet R, Greenamyre JT. Neuroscientist. 2002;8:192–197. doi: 10.1177/1073858402008003004. [DOI] [PubMed] [Google Scholar]
- 2.Bove J, Prou D, Perier C, Przedborski S. NeuroRx. 2005;2:484–494. doi: 10.1602/neurorx.2.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schapira AH. Cell Death Differ. 2007;14:1261–1266. doi: 10.1038/sj.cdd.4402160. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Sleiman PM, Muqit MM, Wood NW. Nat Rev Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- 5.Abeliovich A, Flint Beal M. J Neurochem. 2006;99:1062–1072. doi: 10.1111/j.1471-4159.2006.04102.x. [DOI] [PubMed] [Google Scholar]
- 6.Imai Y, Soda M, Takahashi R. J Biol Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 7.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima A, Kataoka K, Hong M, Sakaguchi M, Huh NH. Cancer Lett. 2003;201:195–201. doi: 10.1016/s0304-3835(03)00443-9. [DOI] [PubMed] [Google Scholar]
- 10.Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Proc Natl Acad Sci USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G. Hum Mol Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 12.Sim CH, Lio DS, Mok SS, Masters CL, Hill AF, Culvenor JG, Cheng HC. Hum Mol Genet. 2006;15:3251–3262. doi: 10.1093/hmg/ddl398. [DOI] [PubMed] [Google Scholar]
- 13.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, et al. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 14.Wang HQ, Takahashi R. Antioxid Redox Signal. 2007;9:553–561. doi: 10.1089/ars.2006.1524. [DOI] [PubMed] [Google Scholar]
- 15.Feng J. Neuroscientist. 2006;12:469–476. doi: 10.1177/1073858406293853. [DOI] [PubMed] [Google Scholar]
- 16.Dawson TM. J Neural Transm Suppl. 2006:209–13. doi: 10.1007/978-3-211-45295-0_32. [DOI] [PubMed] [Google Scholar]
- 17.Cookson MR. Neuromol Med. 2003;3:1–13. doi: 10.1385/NMM:3:1:1. [DOI] [PubMed] [Google Scholar]
- 18.Um JW, Min DS, Rhim H, Kim J, Paik SR, Chung KC. J Biol Chem. 2006;281:3595–3603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- 19.Abeliovich A. Nature. 2007;448:759–760. doi: 10.1038/448759a. [DOI] [PubMed] [Google Scholar]
- 20.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, Zhou Y, Harding M, Bellen H, Mardon G. Development. 2004;131:2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 22.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 25.Chan DC. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 26.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legros F, Lombes A, Frachon P, Rojo M. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Chan DC. Hum Mol Genet 14 Spec No. 2005;2:R283–9. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- 29.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 30.Hoppins S, Lackner L, Nunnari J. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 31.Greene JC, Whitworth AJ, Andrews LA, Parker TJ, Pallanck LJ. Hum Mol Genet. 2005;14:799–811. doi: 10.1093/hmg/ddi074. [DOI] [PubMed] [Google Scholar]
- 32.McBride HM, Neuspiel M, Wasiak S. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 33.Ved R, Saha S, Westlund B, Perier C, Burnam L, Sluder A, Hoener M, Rodrigues CM, Alfonso A, Steer C, et al. J Biol Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 35.Muftuoglu M, Elibol B, Dalmizrak O, Ercan A, Kulaksiz G, Ogus H, Dalkara T, Ozer N. Mov Disord. 2004;19:544–548. doi: 10.1002/mds.10695. [DOI] [PubMed] [Google Scholar]
- 36.Hoepken HH, Gispert S, Morales B, Wingerter O, Del Turco D, Mulsch A, Nussbaum RL, Muller K, Drose S, Brandt U, et al. Neurobiol Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, et al. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 39.Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. J Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Jeong SY, Lim WC, Kim S, Park YY, Sun X, Youle RJ, Cho H. J Biol Chem. 2007;282:22977–22983. doi: 10.1074/jbc.M700679200. [DOI] [PubMed] [Google Scholar]
- 41.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, Rossignol R. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 42.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 43.Germain M, Mathai JP, McBride HM, Shore GC. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parone PA, James DI, Da Cruz S, Mattenberger Y, Donze O, Barja F, Martinou JC. Mol Cell Biol. 2006;26:7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 46.Chang CR, Blackstone C. J Biol Chem. 2007;282(30):21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 47.Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM. J Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- 48.Wasiak S, Zunino R, McBride HM. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harder Z, Zunino R, McBride H. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, et al. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim KL, Dawson VL, Dawson TM. Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 53.Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, Corera AT, Brice A, Seguela P, Fon EA. Mol Biol Cell. 2007;18:3105–3118. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Nishimura I, Imai Y, Takahashi R, Lu B. Neuron. 2003;37:911–924. doi: 10.1016/s0896-6273(03)00143-0. [DOI] [PubMed] [Google Scholar]
- 56.Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. Proc Natl Acad Sci USA. 2005;102:8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rikhy R, Kamat S, Ramagiri S, Sriram V, Krishnan KS. Genes Brain Behav. 2007;6:42–53. doi: 10.1111/j.1601-183X.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- 58.McQuibban GA, Lee JR, Zheng L, Juusola M, Freeman M. Curr Biol. 2006;16:982–989. doi: 10.1016/j.cub.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 59.Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- 60.Freeman M. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 61.Ranganayakulu G, Schulz RA, Olson EN. Dev Biol. 1996;176:143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.