Abstract
Activating transcription factor 2 (ATF2) regulates transcription in response to stress and growth factor stimuli. Here, we use a mouse model in which ATF2 was selectively deleted in keratinocytes. Crossing the conditionally expressed ATF2 mutant with K14-Cre mice (K14.ATF2f/f) resulted in selective expression of mutant ATF2 within the basal layer of the epidermis. When subjected to a two-stage skin carcinogenesis protocol [7,12-dimethylbenz[a]anthracene/phorbol 12-tetradecanoate 13-acetate (DMBA/TPA)], K14.ATF2f/f mice showed significant increases in both the incidence and prevalence of papilloma development compared with the WT ATF2 mice. Consistent with these findings, keratinocytes of K14.ATF2f/f mice exhibit greater anchorage-independent growth compared with ATF2 WT keratinocytes. Papillomas of K14.ATF2f/f mice exhibit reduced expression of presenilin1, which is associated with enhanced β-catenin and cyclin D1, and reduced Notch1 expression. Significantly, a reduction of nuclear ATF2 and increased β-catenin expression were seen in samples of squamous and basal cell carcinoma, as opposed to normal skin. Our data reveal that loss of ATF2 transcriptional activity serves to promote skin tumor formation, thereby indicating a suppressor activity of ATF2 in skin tumor formation.
Keywords: β-catenin, keratinocyte, presenilin, cyclin D1, papilloma
Activating transcription factor 2 (ATF2) is a member of the bZIP family of transcription factors that is activated upon its phosphorylation by stress-activated kinases in response to stress and cytokine stimuli (1, 2). Transcriptional activity of ATF2 depends on its heterodimerization with members of the AP1 family, including c-Jun (3–5), or interaction with viral proteins, including v-Jun, E1A, and the Tax proteins (6–8). ATF2 target genes include AP1-responsive genes, such as cyclin A, IFN-β, and TNF-α (9–11). Intriguingly, ATF2 has also been implicated in the DNA damage response, through its phosphorylation by phosphoinositide-3-kinase-related protein kinase, including ATM (12). This phosphorylation is required for intra-S phase checkpoint control and for its colocalization with components of the Mre11–Rad50–Nbs1 (MRN) complex within DNA damage repair foci.
The role of ATF2 in stress and DNA damage response suggests that this protein could also play a role in tumorigenesis. Consistent with this possibility are earlier studies from our laboratory that have suggested an important role of ATF2 in melanoma development and progression. Nuclear localization of ATF2 coincides with poor prognosis in melanoma patients (13). In addition, peptides derived from the N-terminal region of ATF2 efficiently repressed ATF2 function and reduced growth and metastasis of melanoma tumor cells in mouse models (14–17).
The role of the AP1 transcriptional complex in skin carcinogenesis has been addressed. Specifically, c-Jun family members were shown to play an important role in skin cancer development because K14-driven expression of the TAM67 dominant-negative Jun family construct in the basal layer of the epidermis blocked phorbol 12-tetradecanoate 13-acetate (TPA) or UV-B induced tumors in a skin carcinogenesis model (18–20). In addition, mice in which c-Jun or JNK2 has been deleted exhibit marked reduction in skin cancer development (21–23).
Here, we directly assessed the role of ATF2 in a mouse skin carcinogenesis model, by using a conditional mutant of ATF2 in keratinocytes. Unlike the oncogenic role for c-Jun and JNK2, our work reveals that lack of ATF2 function contributes to accelerated development of papillomas, thereby suggesting a tumor suppressor role of ATF2 in keratinocytes.
Results
The mouse KO of ATF2 leads to early postnatal lethality (24). Thus, to study the function of ATF2 in the skin, we used the Cre-loxP system for disruption of the ATF2 gene in keratinocytes. Cre-dependent deletion of the ATF2 DNA-binding domain and a portion of its leucine zipper results in a transcriptionally inactive form of ATF2 (W.B. and N.J., unpublished results). Mice homozygous for the loxP-flanked (floxed) ATF2 gene (ATF2f/f) were born at the expected Mendelian ratios and presented no obvious abnormalities. In addition, in a number of tissues that were analyzed, the levels of ATF2 expression were comparable between wild-type (WT) and ATF2f/f (data not shown).
To elucidate the role of ATF2 in skin cancer, ATF2f/f mice were crossed with keratin14-cre transgenic mice (K14-Cre). The resulting ATF2f/f/K14-Cre (K14.ATF2f/f) mice expressed the transcriptional mutant ATF2 gene in keratinocytes. Immunoblot analysis confirmed that keratinocytes prepared from WT express a 70-kDa band corresponding to full-length ATF2, whereas keratinocytes of the K14.ATF2f/f mice express a 55-kDa band, corresponding to ATF2, which lacks DNA-binding and leucine zipper domains (Fig. 1b). Immunohistochemical analysis revealed that ATF2 is expressed throughout the nucleated layers of epidermis and the dermis (Fig. 1c). Importantly, the use of the K14-Cre transgene expression is limited to the basal layer of stratified squamous epithelia, thereby leaving ATF2 expression intact in neighboring tissues, including the dermis (Fig. 1c).
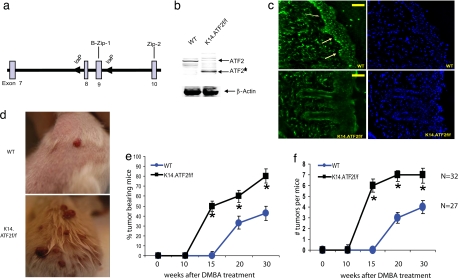
Fig. 1.
Targeted disruption of ATF2 in mouse skin increases susceptibility to papilloma formation in the two-stage chemical carcinogenesis. (a) Targeting strategy shows WT allele of ATF2 encompassing exons 8 and 9 (boxes) and flanking loxP sequences (arrowheads). (b) Expression of ATF2 by immunoblot in skin extracts from K14.ATF2wt/wt (WT) and K14.ATF2f/f mice. β-Actin was used as loading control. ATF2* indicates the fast migrating form of ATF2 released after deletion of exons 8 and 9. An ATF2 antibody that recognizes C-terminal epitopes was used for Western blotting. (c) Immunohistochemical analysis of the expression of ATF2 in the frozen skin sections of WT and K14.ATF2f/f mice. (Scale bars, 50 μm.) Arrows point to the expression of ATF2 in the epidermis of WT skin. (d) Representative pictures of mice bearing papillomas 15 weeks after DMBA treatment. (e) Tumor incidence in the WT and K14.ATF2f/f mice. Data represent the percentage of mice with skin papillomas. Bars indicate SE. *, P < 0.04, statistically different from the WT mice, as determined by Student's t test. (f) Average number of papillomas per mouse after DMBA/TPA treatment. Data represent an average number of skin papillomas per mouse. Bars indicate SE. *, P < 0.04, statistically different from WT mice, as determined by Student's t test.
Disruption of ATF2 Increases Susceptibility to Skin Carcinogenesis.
To address the role of ATF2 in de novo skin carcinogenesis, the two-stage skin carcinogenesis protocol was used (25). In this model, tumors are initiated in epidermal keratinocytes by single topical application of the chemical carcinogen 7,12-dimethylbenz[a]anthracene (DMBA) with subsequent addition of the tumor promoter TPA over a period of 8–12 weeks. This procedure results in the development of benign papillomas with a high incidence of H-Ras mutations (25, 26). Some of these tumors progress to squamous cell carcinomas (SCCs), which can undergo epithelial–mesenchymal transition to spindle cell carcinomas. Significantly, K14.ATF2f/f mice exhibited increased susceptibility to skin tumorigenesis with markedly accelerated kinetics of papilloma development (Fig. 1d). In K14.ATF2wt/wt mice (WT), papillomas started to appear ≈17 weeks after DMBA treatment (Fig. 1e), and by 30 weeks, ≈40% had developed tumors. This slow papilloma development in littermate controls is attributed to their genetic background, FVB/C57BL/6, because mice of C57BL/6 background are more resistant to chemical-induced skin cancer (27). In contrast, skin papillomas in K14.ATF2f/f mice were observed as early as 11 weeks after DMBA treatment, with 50% of mice having developed tumors between weeks 13 and 15. Thus, compared with WT mice, the median appearance of skin papillomas is 5–6 weeks earlier in the ATF2 mutant keratinocytes [average ± SE, 11.42 ± 0.36 weeks (K14.ATF2f/f; n = 32) vs. 17.63 ± 0.30 weeks (WT; n = 27); P < 0.002] (Fig. 1e). Importantly, the number of papillomas also increased in the K14.ATF2f/f mice. By 15 weeks, K14.ATF2f/f mice developed an average number of six tumors per mouse, whereas the WT mice had none (Fig. 1f). These findings strongly suggest that the absence of functional ATF2 in keratinocytes confers increased sensitivity to skin cancer development and imply that ATF2 may elicit tumor suppressor function in the skin. Of note, treatment of DMBA or TPA alone did not result in papilloma development in WT or ATF2 mutant mice. This finding implies that lack of transcriptionally active ATF2 is not sufficient to augment initiation or promotion phases per se but is important for the accelerated development of initiated lesions.
ATF2 Deficiency Increases Epidermal Hyperproliferation After Addition of TPA.
To determine the possible effect of ATF2 disruption on keratinocyte proliferation, possible changes in epidermal hyperplasia were assessed. The number of nucleated cell layers in the untreated epidermis of WT and K14.ATF2f/f mice was not significantly different (Fig. 2a Top and b). After TPA treatment, epidermal hyperplasia was induced in all genotypes, although the number of nucleated cell layers in the K14.ATF2f/f mice was higher compared with the WT mice. Three topical applications of TPA (at 10 μg each) induced the formation of a hyperplastic epidermis consisting of a cell layer of 21 ± 2.6 μm (mean ± SD) thickness 18 h after treatment in WT mice compared with 42 ± 3.2 μm in the K14.ATF2f/f mice (Fig. 2 a Middle and b). The difference was even more striking after 48 h, where the thickness of epidermal layers was 45 ± 2.3 μm in WT mice vs. 102 ± 2.8 μm in K14.ATF2f/f mice (Fig. 2 a Bottom and b). These data reveal that the lack of transcriptionally active ATF2 in keratinocytes potentiates epidermal hyperplasia after exposure to a tumor promoter.
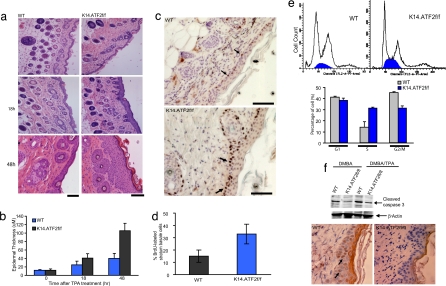
Fig. 2.
Epidermal hyperplasia induced by TPA treatment. (a) Dorsal skin of 8-week-old mice after three topical treatments with 10 μg of TPA was excised and stained with H&E. Histological analysis shown was performed 18 and 48 h after TPA treatment. (Scale bars, 50 μm.) (Top) Acetone-treated control skin. (b) Quantitative analysis of the epidermal thickness (in micrometers) after TPA treatment (P < 0.001). Bars indicate SD (n = 10). The thickness of the epidermis (in micrometers) was calculated as described in Materials and Methods. (c) Immunohistochemical analysis of BrdU incorporation. The dorsal skin of 8-week-old mice received three topical treatments with 10 μg of TPA. Twenty-four hours after TPA treatment, BrdU was injected i.p., and 1 h later the dorsal skin was isolated. Arrows indicate examples of BrdU-positive suprabasal cells. (Scale bars, 50 μm.) (d) Quantitative analysis of the BrdU-positive cells after TPA treatment (P < 0.001). Bars indicate SD (n = 10). (e) (Upper) Cell cycle profile of primary keratinocytes isolated from WT and K14.ATF2f/f pups subjected to FACS analysis. (Lower) Percentages of cells in G1, S, and G2/M phases (mean ± SD of three experiments). (f) Apoptosis was assessed with active caspase 3 antibody of skin from WT and K14.ATF2f/f mice after 5 days of DMBA alone or DMBA and 18 h of TPA treatment. The TPA treatment was done for skin thickening to facilitate visualization of active caspase 3-positive cells in an immunohistochemistry study. (Upper) Western blotting with the active caspase 3 antibody. (Lower) Staining pattern in the skin upon treatment with DMBA after 5 days, followed by TPA treatment. Arrows indicate active caspase 3-positive cells.
Induced DNA Synthesis in Basal Epidermal Cells of TPA-Treated K14.ATF2f/f Mice.
In light of the enhanced hyperplastic responses seen in the epidermis of K14.ATF2f/f mice, we assessed possible changes in the rate of keratinocyte proliferation, which could account for the increased hyperplasia. BrdU labeling was carried out to quantify DNA synthesis in basal keratinocytes. In WT mice, the percentage of BrdU-labeled basal cells was 5% before and 15 ± 3% 24 h after TPA treatment (Fig. 2c). Whereas K14.ATF2f/f mice exhibited BrdU labeling that was similar to the WT animals (5%) before TPA treatment, 24 h after TPA treatment the K14.ATF2f/f mice showed a significant increase in the number of BrdU-labeled cells [(32 ± 3.0%) compared with WT ATF2 mice (15 ± 3%); P < 0.0045 (Fig. 2d)]. Further, primary cultures of K14.ATF2f/f keratinocytes exhibit a marked increase in the S phase of the cell cycle compared with the primary WT keratinocytes (Fig. 2e). These data suggest that the lack of transcriptionally active ATF2 causes increased cell proliferation in primary keratinocytes and their hyperproliferation in the epidermis in response to treatment with tumor promoter.
Reduced Apoptosis in Epidermal Keratinocytes of K14.ATF2f/f Mice.
We next determined whether the higher incidence of papilloma formation could be attributed to altered rate of apoptosis, expected after DNA damage such as DMBA treatment. Reduced levels of active caspase 3 were observed in DMBA-treated K14.ATF2f/f skin compared with the skin of DMBA-treated WT mice, suggesting increased keratinocyte survival in K14.ATF2f/f skin (Fig. 2f). This finding implies that the absence of transcriptionally active ATF2 supports a mutator phenotype, with a consequent increase in tumorigenicity.
Reduced Levels of Presenilin1 (PS1) Coincide with Elevated β-Catenin Expression in the K14.ATF2f/f Papillomas.
To elucidate the molecular pathways that were modified in the absence of transcriptionally potent ATF2, gene expression microarrays were used. A total of 113 probes showed significant differential expression between WT and K14.ATF2 f/f mice (false discovery rate, 10%) [supporting information (SI) Table 1; raw data are available at the National Center for Biotechnology Information Gene Expression Omnibus, accession no. GSE9328]. Genes that were down-regulated in the K14.ATF2f/f papillomas included PS1, which is required for the proteolytic processing of Notch during development and Alzheimer's disease pathogenesis (28). PS1 also serves as a scaffold protein that affects β-catenin phosphorylation and stability independently of the Wnt-regulated axin–CK1α complex (29).
Interestingly, PS1 KO mice that are rescued through neuronal expression of the human PS1 transgene develop spontaneous skin cancers (30). PS1-null keratinocytes have higher cytosolic β-catenin and β-catenin/lymphoid enhancer factor-1/T cell factor (β-catenin/LEF)-mediated signaling. Consistent with these reports, epidermis of K14.ATF2f/f mice not only exhibited reduced levels of PS1, but also had an elevated level of β-catenin expression (Fig. 3a).
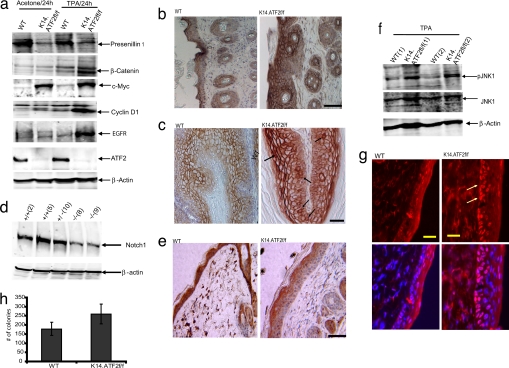
Fig. 3.
Reduced level of PS1 coincides with reduced Notch1 and elevated β-catenin expression in the TPA-treated skin and papillomas of K14.ATF2f/f mice. (a) Epidermis of mice that received three topical applications of acetone or TPA (10 μg) was isolated 24 h after the last treatment. Tissue lysates were subjected to Western blotting with antibodies to PS1, β-catenin, ATF2, c-Myc, cyclin D1, and EGFR. β-Actin was used as loading control. (b) The dorsal skin of 8-week-old mice was subjected to treatment as indicated in a and was excised, and paraffin sections were prepared and stained with β-catenin antibody. (Scale bar, 50 μm.) (c) β-Catenin staining in papilloma sections of the K14.ATF2f/f and WT mice. Cells exhibiting nuclear β-catenin staining are marked with arrows. (Scale bar, 50 μm.) (d) Keratinocytes isolated from K14.ATF2f/f and WT 1-day-old pups were lysed and were subjected to immunoblot analysis with antibodies to Notch1. β-Actin was used as loading control. (e) The dorsal skin of 8-week-old mice treated and prepared as indicated in b was immunostained with antibodies to Notch1. (Scale bar, 50 μm.) (f) Western blotting for phospho-JNK (pJNK1) and total JNK levels was performed as in a. Two mice for each group are shown. β-Actin was used as loading control. (g) Immunohistochemical analysis of the expression of phospho-c-Jun upon TPA treatment in the frozen skin sections of WT and K14.ATF2f/f mice. (Scale bars, 50 μm.) Arrows point to the expression of phospho-c-Jun in the epidermis of WT skin. (h) Increased anchorage-independent cell growth in K14.ATF2f/f keratinocytes. H-RasV12-infected WT and K14.ATF2f/f keratinocytes were seeded in soft agar. Colonies were counted 21 days later and scored microscopically. The data represent an average of three experiments (P < 0.005).
Consistent with the immunoblot analysis (Fig. 3a), β-catenin levels were found to be up-regulated in the epidermis of the K14.ATF2f/f mice (Fig. 3b) and in papilloma samples (Fig. 3c). β-Catenin staining was mostly localized to the membrane of the upper layer of the epidermis in both WT and K14.ATF2f/f papillomas (Fig. 3c). Increased accumulation of cytosolic and nuclear β-catenin was observed in the basal layer of the epidermis of papillomas derived from the K14.ATF2f/f mice. Because cell proliferation takes place within the basal cell layer of the epidermis, increased accumulation of cytosolic and nuclear β-catenin in the epidermis of K14.ATF2f/f mice points to its possible contribution to increased epidermal hyperplasia and papilloma formation.
These data suggest that in the absence of transcriptionally active ATF2, there is a decrease in the expression of PS1 with concomitant increase in the expression of β-catenin.
Elevated Cyclin D1 and c-Myc, and Reduced Notch1 Expression in K14.ATF2f/f Tissues.
Because cyclin D1 and c-Myc are direct target genes for β-catenin/LEF signaling (31, 32), we next assessed changes in their levels of expression. Western blot analysis reveled that cyclin D1 protein was indeed increased in the epidermis of K14.ATF2f/f mice (Fig. 3a) after TPA treatment. Basal levels of c-Myc were also up-regulated in the epidermis of the K14.ATF2f/f mice with a modest increase after TPA treatment (Fig. 3a). The finding that cyclin D1 and c-Myc, two of the major transcriptional targets of β-catenin/LEF signaling, are up-regulated in epidermis of K14.ATF2f/f mice shows that the increased β-catenin protein is functional (see also SI Fig. 5).
Because PS1 is also implicated in the activation of the Notch1-signaling pathway (33) we analyzed changes in the expression of processed (cleaved) Notch1. Lower levels of processed Notch1 expression were found in protein lysates prepared from the epidermis (data not shown) and in keratinocytes derived from the epidermis of K14.ATF2f/f mice compared with the WT mice (Fig. 3d). Interestingly, nuclear localization of Notch1 was detected throughout the WT epidermis, but only in the upper layer of the epidermis of the K14.ATF2f/f mice (Fig. 3e). These results suggest that the epidermis of K14.ATF2f/f mice exhibits lower levels of PS1 expression, which is associated with lower levels of processed Notch1 and higher β-catenin expression.
Elevated EGF Receptor (EGFR), Phospho-JNK, and Phospho-c-Jun Expression in K14.ATF2f/f keratinocytes.
Because PS1 was also implicated in the negative regulation of EGFR signaling and turnover (34, 35), we have assessed changes in EGFR levels in the skin of WT and ATF2 mutant mice. Western blot analysis revealed elevated levels of EGFR in K14.ATF2f/f skin treated with TPA compared with the WT counterpart (Fig. 3a).
Increased EGFR expression is expected to result in activation of respective downstream signaling pathways, including the activation of JNK and c-Jun. Immunohistochemistry of TPA-treated K14.ATF2f/f skin samples revealed increased levels of phospho-c-Jun expression in the basal layers of the epidermis (Fig. 3g), consistent with increased activity of its kinase, JNK (Fig. 3f). Of note, c-Jun was also implicated in positive regulation of EGFR (20). These data identify changes in JNK–Jun signaling pathways and their upstream regulator, EGFR.
Increased Anchorage-Independent Growth of Ras-Infected K14.ATF2f/f Keratinocytes.
A hallmark of malignant transformation is anchorage-independent growth. A soft agar assay was used to measure anchorage-independent growth of keratinocytes derived from the skin of WT and K14.ATF2f/f newborn mice that were infected in culture with a mutant Ras oncogene. A modest, albeit significant, increase was observed in the number (but not the size) of colonies formed in the H-RasV12-infected K14.ATF2f/f keratinocytes compared with the WT counterparts (Fig. 3h; P < 0.005). These findings suggest that lack of ATF2 transcriptional activity increases the tumorigenic potential of keratinocytes in vitro as well as in vivo.
Reduced Level and Altered Subcellular Localization of ATF2 in Skin Cancer Tissue Microarrays.
To characterize the expression of ATF2 in human skin cancer, we used a tissue microarray (TMA) containing normal skin histospots and specimens from 40 patients with SCC or BCC. Significantly, in contrast to the predominant nuclear localization of ATF2 in normal skin (Fig. 4a), samples from malignant tissues (SCC and BCC) exhibited marked reduction of nuclear ATF2 expression (P = 0.0003), whereas the cytoplasmic ATF2 expression was not significantly different (P = 0.13; Fig. 4b and data not shown). Decreased nuclear and variable cytosolic staining was seen in the SCC subset of specimens compared with normal skin (Fig. 4b; P = 0.0005 and P = 0.194, respectively). Similarly, analysis of the BCC subset of samples identified decreased nuclear ATF2 expression compared with normal skin (P < 0.0001) and variable cytoplasmic ATF2 expression (Fig. 4b; P = 0.116). These data indicate that ATF2 expression is either decreased or shifted from the nucleus to the cytosol in the majority of human SCC and BCC samples. Because both changes would result in reduced ATF2 transcriptional activities, these data suggest that ATF2 function is attenuated in these skin tumors.
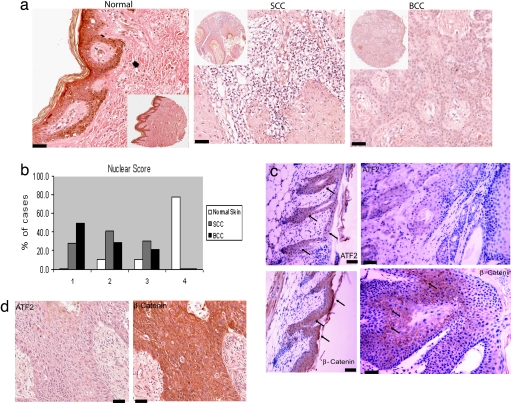
Fig. 4.
Reduced nuclear ATF2 expression in TMA samples from SCC and BCC patients. (a and b) ATF2 protein expression levels were assessed by using a tissue microarray that was stained with an antibody directed against ATF2. (a) Examples of staining in normal skin, SCC, and BCC. (Scale bars, 50 μm.) Scoring of ATF2 nuclear and cytosolic localization was performed as detailed in Materials and Methods. Forty samples from SCC patients, 14 samples of BCC patients, and 10 normal matched and nonmatched skin tissues were analyzed. Scores were divided into four categories: 1, scores ranging from 0 to 75; 2, scores ranging from 76 to 150; 3, scores ranging from 151 to 225; and 4, scores ranging from 226 to 300. (b) Distribution of nuclear ATF2 staining in normal skin, BCC, and SCC. (c) (Left) Staining of the skin derived from the WT mouse that developed papillomas with ATF2 (Upper) and β-catenin (Lower). (Right) Staining of the corresponding papillomas with antibodies to ATF2 (Upper) and β-catenin (Lower). (Scale bars, 50 μm.) (d) Staining of the TMA used in a with ATF2 (Left) and β-catenin antibody (Right). (Scale bars, 50 μm.)
Consistent with the finding in BCC and SCC samples, the level of ATF2 expression was markedly reduced in the papillomas developed in the WT mice compared with the normal-appearing skin (Fig. 4c). Conversely, the expression of β-catenin increased in these papillomas, consistent with our finding of elevated β-catenin expression in the K14.ATF2f/f papillomas. Significantly, and in agreement with our data, an increase in β-catenin staining intensity was seen in samples from SCC and BCC (Fig. 4d). These finding establish that ATF2 transcriptional activity is attenuated because of reduced or altered (cytosolic) localization in papillomas and human skin cancers, which coincides with elevated β-catenin expression.
Discussion
The present work provides genetic evidence for a suppressor role of ATF2 in nonmalignant skin cancer. Using K14.ATF2f/f mice, we demonstrate that the number and incidence of papillomas increases in the absence of a transcriptionally active ATF2 in the basal layer of the epidermis. Consistent with these finding, keratinocytes prepared from K14.ATF2f/f mice that were infected with mutant Ras oncogene exhibited a marked increase in their ability to grow on soft agar compared with their WT counterpart, suggesting a transformed phenotype in vitro. Important support for the finding in the mouse model used here comes from the analysis of human skin tumors; unlike the strong nuclear expression of ATF2 in normal skin, SCC and BCC samples exhibit a significantly reduced nuclear staining. The latter is also consistent with reduced expression of ATF2 found in papillomas developed in the WT animals, supporting the notion that ATF2 need to be inactivated to support skin tumor development. Although the present study did not assess whether a lack of transcriptionally active ATF2 could contribute also to a papilloma-to-carcinoma transition, the data from SCC and BCC, where ATF2 is largely inactivated because of either lower expression or reduced nuclear localization, support such a role. It is of interest to note that loss of ATF2 transcriptional activities per se, or in combination with either DMBA or TPA, is not sufficient to promote papilloma formation, suggesting that ATF2 is contributing to changes elicited by initiating and promoting events in keratinocytes. Among the changes observed are reduced apoptosis of DMBA-treated skin and increased proliferation of TPA-treated skin, as well as of primary keratinocytes. Collectively, these finding reveal that loss of ATF2 transcriptional activities is associated with, and contributes to, skin tumor formation.
The role of the AP1 complex in the promotion of skin carcinogenesis has been demonstrated in mouse models in which the activity of JNK, c-Jun, and other AP1 family members has been inhibited (18–21, 36–40). Surprisingly, unlike the promoting function of c-Jun and JNK2, ATF2 elicits suppressor function in the skin carcinogenesis process. The transcriptional activity of ATF2 is required for this tumor suppressor function because the mouse model used in this work does not produce a transcriptionally active form of ATF2. We show that PS1 and its targets β-catenin, EGFR, cyclin D1, and Notch1 are regulated by ATF2, and we suggest that their deregulation in the mutant mice is associated with the enhanced skin tumor phenotype seen.
Consistent with these findings is the observation that EGFR–JNK–c-Jun–Cyclin D1, which contributes directly to skin tumor progression (34, 35, 41, 42) are up-regulated in the absence of transcriptionally active ATF2. Furthermore, although the latter can be associated with loss of PS1 suppression, PS1 positively regulates Notch1, and reduced PS1 results in lower level of Notch1 expression, seen in the K14.ATF2f/f mice tumors and tissues, as in skin tumor models, where it has been implicated as a tumor suppressor (43, 44).
Consistent with our findings in the skin, ATF2 was recently implicated in eliciting a tumor suppressor function in mammary tumors (45). In contrast, however, previous studies suggested a tumor-promoting role of ATF2 in melanoma (12–17). Similarly, Notch1 functions as a tumor suppressor in mouse skin, as opposed to its function as an oncogene in other organs (44). Thus, tissue-dependent expression of regulatory or accessory factors (i.e., heterodimeric transcription factors) is expected to alter the repertoire of genes that are regulated by ATF2 in keratinocytes vs. other tissue types, including melanoma. Among possible mechanisms that could explain the different functions in the two tissue types is the altered expression and subcellular localization of ATF2 in skin tumors as opposed to melanoma. Whereas in melanoma, nuclear ATF2 expression is associated with poor prognosis (13), its nuclear localization is significantly reduced in BCC and SCC (Fig. 4). Decreased nuclear expression of ATF2 in human skin tumors further implicates loss of ATF2 function in keratinocyte transformation.
Materials and Methods
Additional procedures can be found in SI Methods.
Animal Treatment and Tumor Induction Protocols.
To study the function of ATF2 in the skin, we used the Cre-loxP system for disruption of the ATF2 gene in keratinocytes (40). The K14.ATF2f/f mice and their littermate controls (WT) were of identical FVB/C57BL/6 genetic background. For tumor induction, mice were initiated with a dose of 10 μg of DMBA (Sigma) in 100 μl of acetone applied to the dorsal surface 2 days after shaving. TPA (10 μg in 200 μl of acetone; Sigma) was applied every week for 30 weeks beginning 1 week after initiation. The appearance of lesions in each mouse was monitored and recorded every week. Dorsal skin and/or papillomas were dissected from euthanized mice and fixed in 10% neutral-buffered formalin for 48 h and were embedded in paraffin. Sections (5 μm) were stained with H&E for histopathological analyses. Papilloma incidence and multiplicity were recorded weekly. Papilloma multiplicity was calculated as the average number of skin papillomas per mouse. Papilloma incidence was calculated as the percentage of mice with skin papillomas.
Immunohistochemistry.
Skin specimens were fixed in neutral buffered formalin solution and processed for paraffin embedding. Skin sections (5 μm thick) were prepared and deparaffinized with xylene. For β-catenin, ATF2, and Notch1 immunostaining, tissue sections were incubated in DAKO antigen retrieval solution for 20 min in a boiling bath, followed by treatment with 3% hydrogen peroxide for 20 min. Antibodies against ATF2 (1:100; Santa Cruz Biotechnology), β-catenin (1:500; Abcam), and Notch1 (1:100; Santa Cruz Biotechnology) were allowed to react with tissue sections at 4°C overnight. Biotinylated anti-rabbit IgG was allowed to react for 30 min at room temperature, and diaminobenzidine was used for the color reaction. Hematoxylin was used for counterstaining. The control sections were treated with normal mouse serum or normal rabbit serum instead of each antibody. For the frozen sections, phospho-c-Jun and ATF2 antibodies (1:100; Cell Signaling) were used.
Measurement of Hyperplasia.
Hyperplasia was assessed in six WT and K14.ATF2f/f mice (8 weeks old) after treatment with acetone or TPA at the indicated time points. The thickness of the epidermis (in micrometers) was measured with an imaging system (SlideBook; Intelligent Imaging) in 15 fields per section.
Statistical Analysis.
Data are shown as the means ± SD. Unless indicated otherwise, statistical differences were determined with one-way ANOVA.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Huaxi Xu (Burnham Institute for Medical Research) for presenilin antibody, Scott Lowe (Cold Spring Harbor Laboratory, Cold Spring, NY) for the H-Ras retroviral vector; Maranke Koster and Dennis Roop for providing the protocols for isolating keratinocytes and DMBA/TPA skin carcinogenesis studies; and Jerry Lu for help in isolating mouse epidermis. This work was supported by National Cancer Institute/National Institutes of Health Grant CA099961 (to Z.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.E.A. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database www.ncbi.nlm.nih.gov/geo (accession no. GSE9328).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706057105/DC1.
References
- 1.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta S, Campbell D, Dérijard B, Davis RJ. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 3.van Dam H, Castellazzi M. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 4.Benbrook DM, Jones NC. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 5.Kerppola TK, Curran T. Mol Cell Biol. 1993;13:5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huguier S, Baguet J, Perez S, van Dam H, Castellazzi M. Mol Cell Biol. 1998;18:7020–7029. doi: 10.1128/mcb.18.12.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawasaki H, Song J, Eckner R, Ugai H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama KK. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin AA, Kubik MF, Uittenbogaard MN, Brauweiler A, Utaisincharoen P, Matthews MA, Dynan WS, Hoeffler JP, Nyborg JK. J Biol Chem. 1993;268:21225–21231. [PubMed] [Google Scholar]
- 9.Nakamura T, Okuyama S, Okamoto S, Nakajima T, Sekiya S, Oda K. Exp Cell Res. 1995;216:422–430. doi: 10.1006/excr.1995.1053. [DOI] [PubMed] [Google Scholar]
- 10.Tsai EY, Yie J, Thanos D, Goldfeld AE. Mol Cell Biol. 1996;16:5232–5244. doi: 10.1128/mcb.16.10.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falvo JV, Parekh BS, Lin CH, Fraenkel E, Maniatis T. Mol Cell Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z. Mol Cell. 2005;18:577–587. doi: 10.1016/j.molcel.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger AJ, Kluger HM, Li N, Kielhorn E, Halaban R, Ronai Z, Rimm DL. Cancer Res. 2003;63:8103–8107. [PubMed] [Google Scholar]
- 14.Bhoumik A, Jones N, Ronai Z. Proc Natl Acad Sci USA. 2004;101:4222–4227. doi: 10.1073/pnas.0400195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhoumik A, Ivanov V, Ronai Z. Clin Cancer Res. 2001;7:331–342. [PubMed] [Google Scholar]
- 16.Bhoumik A, Huang TG, Ivanov V, Gangi L, Qiao RF, Woo SL, Chen SH, Ronai Z. J Clin Invest. 2002;110:643–650. doi: 10.1172/JCI16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhoumik A, Gangi L, Ronai Z. Cancer Res. 2004;64:8222–8230. doi: 10.1158/0008-5472.CAN-04-0714. [DOI] [PubMed] [Google Scholar]
- 18.Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R, Colburn N. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JJ, Dong Z, Dawson MI, Colburn NH. Cancer Res. 1996;56:483–489. [PubMed] [Google Scholar]
- 20.Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, Sibilia M, Wagner EF. Dev Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 21.Chen N, Nomura M, She QB, Ma WY, Bode AM, Wang L, Flavell RA, Dong Z. Cancer Res. 2001;61:3908–3912. [PubMed] [Google Scholar]
- 22.Arnott CH, Scott KA, Moore RJ, Hewer A, Phillips DH, Parker P, Balkwill FR, Owens DM. Oncogene. 2002;21:4728–4738. doi: 10.1038/sj.onc.1205588. [DOI] [PubMed] [Google Scholar]
- 23.Matthews CP, Birkholz AM, Baker AR, Perella CM, Beck GR, Jr, Young MR, Colburn NH. Cancer Res. 2007;67:2430–2438. doi: 10.1158/0008-5472.CAN-06-0522. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa T, Bernier F, Sato M, Nomura S, Singh M, Inoue Y, Tokunaga T, Imai H, Yokoyama M, Reimold A, et al. J Biol Chem. 1999;274:17813–17819. doi: 10.1074/jbc.274.25.17813. [DOI] [PubMed] [Google Scholar]
- 25.Yuspa SH. Cancer Res. 1994;54:1178–1189. [PubMed] [Google Scholar]
- 26.Roop DR, Lowy DR, Tambourin PE, Strickland J, Harper JR, Balaschak M, Spangler EF, Yuspa SH. Nature. 1986;323:822–824. doi: 10.1038/323822a0. [DOI] [PubMed] [Google Scholar]
- 27.DiGiovanni J, Bhatt TS, Walker SE. Carcinogenesis. 1993;14:319–321. doi: 10.1093/carcin/14.2.319. [DOI] [PubMed] [Google Scholar]
- 28.De Stooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 29.Kang DE, Soriano S, Xia X, Eberhart CG, De Stooper B, Zheng H, Koo EH. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- 30.Xia X, Qian S, Soriano S, Wu Y, Fletcher AM, Wang XJ, Koo EH, Wu X, Zheng H. Proc Natl Acad Sci USA. 2001;98:10863–10868. doi: 10.1073/pnas.191284198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 33.Song W, Nadeau P, Yuan M, Yang X, Shen J, Yanker BA. Proc Natl Acad Sci USA. 1999;96:59–63. doi: 10.1073/pnas.96.12.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Repetto E, Yoon IS, Zheng H, Kang DE. J Biol Chem. 2007;282:31504–31516. doi: 10.1074/jbc.M704273200. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YW, Wang R, Liu Q, Zhang H, Liao FF, Xu H. Proc Natl Acad Sci USA. 2007;104:10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoumpourlis V, Papassava P, Linardopoulos S, Gillespie D, Balmain A, Pintzas A. Oncogene. 2000;19:4011–4021. doi: 10.1038/sj.onc.1203732. [DOI] [PubMed] [Google Scholar]
- 37.Bowden GT, Schneider B, Domann R, Kulesz-Martin M. Cancer Res. 1994;54:1882S–1885S. [PubMed] [Google Scholar]
- 38.Dong Z, Birrer MJ, Watts RG, Matrisian LM, Colburn NH. Proc Natl Acad Sci USA. 1994;91:609–613. doi: 10.1073/pnas.91.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong Z, Lavrovsky V, Colburn NH. Carcinogenesis. 1995;16:749–756. doi: 10.1093/carcin/16.4.749. [DOI] [PubMed] [Google Scholar]
- 40.Finch JS, Albino HE, Bowden GT. Carcinogenesis. 1996;17:2551–2557. doi: 10.1093/carcin/17.12.2551. [DOI] [PubMed] [Google Scholar]
- 41.Bianchi AB, Fischer SM, Robles AI, Rinchik EM, Conti CJ. Oncogene. 1993;8:1127–1133. [PubMed] [Google Scholar]
- 42.Robles AI, Rodriguez-Puebla ML, Glick AB, Trempus C, Hansen L, Sicinski P, Tennant RW, Weinberg RA, Yuspa SH, Conti CJ. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 44.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, et al. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maekawa T, Shinagawa T, Sano Y, Sakuma T, Nomura S, Nagasaki K, Miki Y, Saito-Ohara F, Inazawa J, Kohno T, et al. Mol Cell Biol. 2007;27:1730–1744. doi: 10.1128/MCB.01579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.