Abstract
Although it is well established that women with germ-line mutations in the BRCA1 gene have a greatly increased lifetime incidence of breast and ovarian cancer, the molecular mechanisms responsible for this tissue-specific carcinogenesis remain undefined. The majority of these breast cancers are of the basal-like phenotype characterized by lack of expression of ER, PR, and ERBB2. Because this phenotype has been proposed to resemble that of normal breast stem cells, we examined the role of BRCA1 in human mammary stem cell fate. Using both in vitro systems and a humanized NOD/SCID mouse model, we demonstrate that BRCA1 expression is required for the differentiation of ER-negative stem/progenitor cells to ER-positive luminal cells. Knockdown of BRCA1 in primary breast epithelial cells leads to an increase in cells displaying the stem/progenitor cell marker ALDH1 and a decrease in cells expressing luminal epithelial markers and estrogen receptor. In breast tissues from women with germ-line BRCA1 mutations, but not normal controls, we detect entire lobules that, although histologically normal, are positive for ALDH1 expression but are negative for the expression of ER. Loss of heterozygosity for BRCA1 was documented in these ALDH1-positive lobules but not in adjacent ALDH1-negative lobules. Taken together, these studies demonstrate that BRCA1 plays a critical role in the differentiation of ER-negative stem/progenitor cells to ER-positive luminal cells. Because BRCA1 also plays a role in DNA repair, our work suggests that loss of BRCA1 may result in the accumulation of genetically unstable breast stem cells, providing prime targets for further carcinogenic events.
Keywords: breast cancer, stem cell, hereditary cancer
Accumulating evidence has provided support for the cancer stem cell hypothesis, which holds that cancers originate in tissue stem and/or progenitor cells through the dysregulation of self-renewal processes. The ensuing tumors are driven by a cellular subcomponent that retains stem cell properties (1). Evidence for a cancer stem cell component has been generated in human breast cancers (2), as well as cancers of hematopoietic and solid tumor origin (3–6). A number of developmental pathways such as NOTCH (7), Hedgehog (8, 9), and Wnt (9, 10) have been shown to be involved in the regulation of self-renewal and the differentiation of stem and progenitor cells in a number of cell types. Furthermore, these pathways are frequently dysregulated during carcinogenesis (9–13). It is not known whether genes that contribute to hereditary cancers play a role in regulating stem cell fate. Heterozygous germ-line mutations in the BRCA1 gene predispose women to breast and ovarian cancer with a lifetime risk of breast cancer of up to 85% (14). The vast majority of breast tumors in these patients display a basal-like phenotype characterized by a lack of expression of ER, PR, and ERBB2, but robust expression of markers of myoepithelial differentiation (15). Mouse models using conditional BRCA1 KOs have suggested an important role for BRCA1 in mammary gland development (16). Furuta et al. (17) demonstrated that knockdown of BRCA1 in the MCF10A mammary epithelial cell line abrogated the ability of the cells to form acini in 3D culture, suggesting a possible role of BRCA1 in mammary differentiation. Moreover, in vitro studies on mouse mammary epithelial cells suggested a role of BRCA1 in their differentiation (18). BRCA1 also has been shown to play an important role in DNA repair, activation of cell-cycle checkpoints, and maintenance of chromosome stability (19). Although loss of these functions may contribute to tumorigenesis, the propensity of BRCA1 germ-line mutation carriers to develop breast and ovarian carcinoma remains unexplained.
Results and Discussion
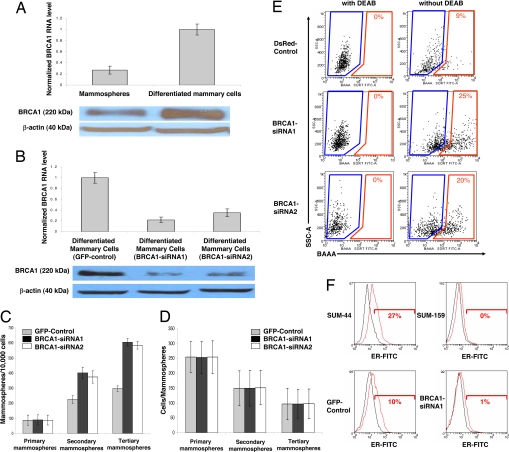
We have proposed that basal-like carcinomas originate from mammary stem cells that exhibit blocks in differentiation (20). Foulkes (21) proposed that the clinical, molecular, and pathological features of breast cancer in BRCA1 mutation carriers fit a model in which BRCA1 functions as a stem cell regulator. To elucidate the role of BRCA1 in mammary stem cell self-renewal and differentiation, we used an in vitro system in which primitive mammary stem/progenitor cells can be propagated in culture as floating spherical colonies termed “mammospheres.” Mammospheres contain a small number of stem cells capable of self-renewal, as well as multipotent progenitor cells capable of differentiation into luminal epithelial cells and myoepithelial cells (22). When mammary epithelial cells are induced to differentiate by attachment to collagen-coated dishes, the level of BRCA1 increases ≈4-fold at both the mRNA and protein levels (Fig. 1A). To determine whether BRCA1 plays a direct role in mammary stem cell self-renewal and differentiation, we used BRCA1 siRNA lentiviruses to knock down BRCA1 expression in primary mammary cells and examined the effect on mammary stem cell self-renewal and differentiation. As shown in Fig. 1B, two independent BRCA1 siRNA lentiviruses reduced BRCA1 expression by >80% at both the mRNA and protein levels compared with a GFP-control lentivirus. Knockdown of BRCA1 had no effect on primary mammosphere formation, but significantly increased secondary mammosphere formation by 70% and tertiary mammosphere formation by 100% (Fig. 1C). Knockdown of BRCA1 had no effect on mammospheres size (Fig. 1D). The number of mammospheres formed upon serial passage at clonal density reflects self-renewal of primitive mammary stem cells, whereas the mammospheres' size is a reflection of progenitor cell proliferation (22).
Fig. 1.
BRCA1 expression increases during mammary differentiation. Knockdown of BRCA1 increases mammary stem/progenitor cells and decreases ER expression. (A) Level of BRCA1 mRNA and protein in mammospheres compared with differentiated cells (cultured for 7 days on collagen) measured by real-time RT-PCR and Western blot analysis, respectively. (B) Level of BRCA1 mRNA and protein measured by real-time RT-PCR and Western blot analysis in BRCA1 knockdown mammary epithelial cells compared with GFP-control-infected mammary epithelial cells. We used two independent BRCA1 siRNA lentiviruses and a GFP-control lentivirus to infect the cells. (C) BRCA1 knockdown increases mammosphere number upon serial passage. (D) BRCA1 knockdown has no effect on mammospheres' size. (E) ALDH1 enzymatic activity as assessed by the ALDEFLUOR assay and flow cytometry demonstrates that BRCA1 knockdown increases the ALDEFLUOR-positive population in vitro. (F) As assessed by flow cytometry, BRCA1 knockdown decreases ER expression in BRCA1 knockdown cells compared with DsRed-control primary mammary epithelial cells. SUM-44 ER-positive and SUM-159 ER-negative breast cancer cell lines serve as positive and negative controls.
We have previously shown that mammary stem/progenitor cells capable of self-renewal and mammosphere formation in vitro and mammary gland reconstruction in NOD/SCID mice express aldehyde dehydrogenase (ALDH) activity as accessed by the Aldefluor assay (23). As shown in Fig. 1E, knockdown of BRCA1 in primary normal mammary cells increased the Aldefluor-positive population compared with DsRed-control lentivirus infected cells from 9–22%. Together these results suggest that the loss of BRCA1 results in an increase in the stem/progenitor cell population.
Studies in murine (24) and human mammary cells (25) suggest that the most primitive stem cells do not express estrogen receptor, but are capable of differentiation into ER-positive luminal epithelial cells. To determine whether knockdown of BRCA1 affects estrogen receptor expression, we used flow cytometry to quantitate ERα expression in primary human mammary cells obtained from reduction mammoplasties. This assay was validated by using ER-positive SUM-44 and ER-negative SUM-159 human mammary carcinoma cell lines. As shown in Fig. 1F, knockdown of BRCA1 in primary human mammary cells reduced the percentage of cells expressing ER from 10% to 1%.
These experiments involving the estrogen receptor suggested that BRCA1 is involved in the differentiation of ER-negative stem/progenitor cells to ER-positive luminal epithelial cells. To further elucidate the role of BRCA1 in mammary differentiation, we used flow cytometry with the lineage-specific markers [epithelial-specific antigen (ESA)] for the luminal epithelial lineage and CD10 for the myoepithelial lineage. Cell differentiation was induced by culturing cells on collagen-coated dishes in the presence of FBS (22). In this system, primitive double-negative (DN) cells (ESA-negative/CD10-negative) give rise to double-positive (DP) cells expressing both luminal epithelial and myoepithelial markers (ESA-positive/CD10-positve) and to single-positive luminal epithelial and myoepithelial cells (ESA-positive or CD10-positive). DP cells are luminal epithelial-committed progenitor cells and give rise to single-positive luminal epithelial cell [supporting information (SI) Fig. 7]. Interestingly, the level of BRCA1 mRNA is ≈5-fold higher in luminal epithelial cells compared with the three other populations (DN, DP, and myoepithelial cells) (Fig. 2A).
Fig. 2.
BRCA1 knockdown blocks epithelial differentiation in vitro. (A) Level of BRCA1 mRNA measured by real-time RT-PCR in the four populations defined by the expression of the luminal marker (ESA) and the myoepithelial marker (CD10). (B) Knockdown of BRCA1 blocks epithelial differentiation in vitro. BRCA1 siRNA or GFP-control-infected cells were induced to differentiate by culturing cells on collagen plates. Expression of lineage-specific markers was determined by flow cytometry at different time points (0, 7, 12, 26, and 35 days). CD10 is a marker of myoepithelial cells, and ESA is a marker of luminal epithelial cells. FACS analysis scatter plots according to ESA and CD10 expression are presented for the two groups, days 0 and 35. Evolution of the four populations for the two groups is plotted as a function of the number of culture days.
BRCA1 knockdown cells cultured on collagen plates showed a significant decrease in the number of luminal epithelial cells (ESA-positive), with a concomitant increase in the number of undifferentiated cells (DN) and myoepithelial cells compared with GFP-control cells (SI Fig. 8). After 35 days in culture, BRCA1 knockdown cells were comprised of 65% undifferentiated cells (DN), 2% luminal epithelial committed progenitor cells (DP), 11% myoepithelial cells, and 22% luminal epithelial cells compared with GFP-control cells that contained 1% undifferentiated cells, 5% luminal epithelial committed progenitor cells, 4% myoepithelial cells, and 85% luminal epithelial cells (Fig. 2B and SI Fig. 9). These data suggest that BRCA1 is required for the luminal differentiation or, alternatively, that BRCA1 is necessary for the survival of differentiated luminal cells.
To determine whether BRCA1 could affect differentiation of mammary stem/progenitor cells in vivo, we used a humanized NOD/SCID mouse model in which primary human mammary epithelial cells are introduced into the cleared fat pads of NOD/SCID mice whose stroma has been humanized by the introduction of mixed irradiated and nonirradiated human mammary fibroblasts (8, 26). Primary mammary cells infected with GFP (GFP-control lentiviruses) or BRCA1 siRNA GFP lentivirus were introduced into these humanized mammary fat pads. After 2 months, mice were killed, and the mammary structures were examined by immunohistochemistry. As shown in Fig. 3, all of the structures produced stained positive with anti-GFP antibody, demonstrating human origin and continued gene expression in these structures. In addition, all cells in these structures stained positive for the pan-cytokeratin AE1/AE3, demonstrating their epithelial origin (data not shown). Ductal structures generated by GFP-control cells were composed of an inner layer of CK18-positive luminal epithelial cells surrounded by a single layer of SMA-positive myoepithelial cells recapitulating the cellular organization found in normal mammary glands (Fig. 3). In contrast, BRCA1 knockdown cells generated abnormal structures composed of a single cell layer (Fig. 3). In some glands, this single layer of cells expressed only the myoepithelial marker SMA, whereas other glands were composed of cells that were negative for the expression of both the epithelial marker CK18 and the myoepithelial marker SMA (Fig. 3). Interestingly, glands produced from the BRCA1 knockdown cells, but not GFP-control lentivirus, uniformly expressed the stem cell marker ALDH1 but were negative for ER expression. Thus, the knockdown of BRCA1 in this animal model produced structures displaying stem cell and differentiation markers similar to those produced by BRCA1 knockdown in vitro. Together these in vitro and mouse model experiments suggest that BRCA1 expression is required for the differentiation of ALDH1-positive/ER-negative mammary stem/progenitor cells into ALDH1-negative/ER-positive epithelial cells and/or the survival of the luminal cells. Loss of BRCA1 function results in the accumulation of ALDH1-positive/ER-negative stem/progenitor cells. This model is depicted graphically in Fig. 4.
Fig. 3.
BRCA1 knockdown blocks epithelial differentiation in NOD/SCID mice xenografts. Mammosphere-initiating cells transduced with GFP-control lentivirus or BRCA1 siRNA lentivirus were introduced into the humanized cleared fat pads of NOD/SCID mice. Mammary structures formed were stained by H&E or examined by immunohistochemistry for expression of GFP, ALDH1, CK18, SMA, and ER. (i, iii, vii, and ix) Fatpads injected with GFP-control-infected cells display mammary epithelial duct structures (i) from human origin (iii, GFP-positive, red staining) that comprised two cell layers with the inner layer expressing the luminal marker CK18 (vii, brown staining), and the outsider layer expressing the myoepithelial marker SMA (ix, red staining). (v and xi) No ALDH1 expression was detected (v), and some cells display ER expression (xi, brown staining). (ii, iv, vi, viii, x) BRCA1 knockdown cells identified by GFP positivity (iv, red staining) produced abnormal structures composed of a single cell layer (ii) that was ALDH1-positive (vi, red staining) and negative for the expression of luminal markers CK18 (viii), and Estrogen Receptor, and with variable expression of the myoepithelial marker SMA (x, red staining). (Scale bars: 100 μm.)
Fig. 4.
Model depicting the proposed role of BRCA1 in mammary, stem, and progenitor cell fate. BRCA1 is required for the differentiation of ALDH1-positive/ER-negative stem/progenitor cells into ER-positive luminal epithelial cells. Loss of BRCA1 function results in aberrant luminal differentiation and also may have an effect on the survival of the luminal cells. Moreover, the loss of BRCA1 function results in the accumulation of ALDH1-positive/ER-negative stem/progenitor cells.
To determine the clinical relevance of the in vitro and mouse model studies, we examined the expression of the stem cell maker ALDH1 and ER in breast tissue obtained from 13 women with documented deleterious BRCA1 germ-line mutations (SI Table 1). These samples were compared with breast tissue obtained from reduction mammoplasties in 22 women with no family history of breast cancer. As shown in Fig. 5, we detected foci of ALDH1-positive cells comprising entire acini in 5 of 13 breast tissue samples obtained from BRCA1 mutation carriers. No such lobules were found in any of the control samples in which only a rare ALDH1-positive cell was found at areas of ductal branching (23). Cells expressing the stem/progenitor marker ALDH1 were morphologically normal, but were negative for expression of the epithelial cell marker CK18 (Fig. 5). These ALDH1-positive clusters also demonstrated reduced expression of CK14 and a markedly reduced expression of estrogen receptor (Fig. 5).
Fig. 5.
Phenotypic characterization of ALDH1-positive lobules in BRCA1 mutation carrier samples. (i) Immunostaining for ALDH1 (red staining) was performed in samples obtained from prophylactic mastectomy specimens of women with confirmed BRCA1 mutations. Foci of ALDH1-positive cells comprising entire acini were detected (red circle) in samples obtained from 5 of 13 patients. Double staining with ALDH1 (i, red staining) and CK18 (ii, brown staining) or CK14 (iii, brown staining) and ER (iv, brown staining) of morphologically normal breast epithelium from BRCA1 carrier patients showed ALDH1-expressing lobules that displayed absence of expression of CK18 and ER (red circles) and a reduced expression of CK14, whereas ALDH1-negative acini (blue circles) were composed of a continuous outer layer of CK14-positive myoepithelial cells surrounding an inner layer of CK18-positive and ER-positive epithelial cells.
We hypothesized that the ALDH1-positive lobules in BRCA1 mutation carriers resulted from a loss of BRCA1 expression in these lobules, producing a block in stem/progenitor cell differentiation. Because it has been shown that tumors in BRCA1 mutation carriers demonstrate loss of heterozygosity (LOH) through loss of the normal BRCA1 allele (27), we predicted loss of the normal allele in ALDH1-positive, but not the surrounding ALDH1-negative, lobules. To test this prediction, we performed laser capture microdissection (LCM) of ALDH1-positive lobules as well as adjacent ALDH1-negative lobules from samples obtained from four BRCA1 mutation carriers. DNA was extracted and analyzed for four microsatellite markers, two within the BRCA1 locus and two immediately telomeric (27). Four of the five BRCA1 mutation carriers presenting ALDH1-positive acini were available for LCM analysis. In each of the four BRCA1 mutation carriers, LOH at the BRCA1 locus was demonstrated in at least one of the BRCA1 polymorphic markers in ALDH1-positive, but not in adjacent ALDH1-negative lobules (Fig. 6A). We used similar methodology to analyze tissue from six of eight BRCA1 mutation carriers who had no detectable ALDH1-positive lobules. We performed LCM of these ALDH1-negative lobules as well as adjacent stroma cells from each of these samples. Analysis of the four microsatellite markers did not reveal the presence of LOH at the BRCA1 locus in any of these samples (Fig. 6B).
Fig. 6.
Stem/progenitor cell expansion in BRCA1 mutation carriers is associated with LOH at the BRCA1 locus. (A) LOH analysis at the BRCA1 locus in ALDH1-positive acini versus ALDH1-negative acini from BRCA1 mutation carriers containing ALDH1-positive acini. ALDH1-positive and adjacent ALDH1-negative lobules were isolated by using laser capture microdissection. Extracted DNA from each microdissected sample was analyzed for four microsatellites in and telemeric to BRCA1 loci (D17S855, D17S1323, D17S1325, and D17S806). In each of the four BRCA1 mutation carrier patients analyzed, LOH at the BRCA1 locus was demonstrated in at least one of the BRCA1 microsatellite markers in ALDH1-positive acini but not in ALDH1-negative acini. (B) LOH analysis at the BRCA1 locus in ALDH1-negative acini versus stromal cells from BRCA1 mutation carriers with no detectable ALDH1-positive lobules. LOH analysis performed as previously comparing ALDH1-negative acini and adjacent stromal cells. In each of the six BRCA1 mutation carrier patients analyzed, no LOH at the BRCA1 locus was observed (LOH, loss of heterozygocity; ROH, retention of heterozygocity; NI, noninformative).
We next determined whether ALDH1 expression is associated with higher risk for the development of invasive breast cancer. Among the 13 BRCA1 mutation carriers, six women developed breast cancer within the following 9 years. Seven women did not develop breast cancer during this follow-up period (SI Table 1). Four of five BRCA1 mutation carriers with ALDH1-positive acini developed cancer, whereas only two of the eight BRCA1 mutation carriers with no detectable ALDH1-positive lobules developed breast cancer (Fisher's test, P = 0.04). Although this dataset represents a limited number of patients, the results are statistically significant and consistent with the hypothesis that expansion of the ALDH1-positive stem/progenitor cell population is an early step in breast carcinogenesis.
In summary, in vitro systems and mouse models suggest an important role for BRCA1 in regulating the differentiation of ALDH1-positive/ER-negative stem/progenitor cells into ER-positive epithelial cells. Alternatively, BRCA1 may be required for the survival of luminal cells, but not stem/progenitor or myoepithelial cells. In BRCA1 mutation carriers, loss of the normal BRCA1 allele is associated with the development of lobules characterized by expression of the stem/progenitor marker ALDH1 with a concomitant lack of expression of epithelial differentiation markers and ER.
Recently using an MMTV-Cre;Brca1f11/f11p53f5&6/f5&6 model, it has been suggested that progesterone plays a role in carcinogenesis in BRCA1 KO mice (28). In this model, unlike in the human situation, BRCA1 KO tumors expressed estrogen and progesterone receptors. In contrast, when BRCA1 was knocked out in more primitive basal cells using a K14cre;Brca1F/+;p53F/F construct, it resulted in the generation of ER-negative/PR-negative tumors more closely recapitulating the phenotype found in woman with BRCA1 germ-line mutations (29). These observations suggest that the BRCA1-associated tumor phenotype depends on the state of differentiation of cells in which BRCA1 function is lost and suggests that in human breast cancer this loss of function occurs in primitive ER-negative stem/progenitor cells. Furthermore, these results suggest that the protective effect of ovariectomy on breast cancer development in BRCA1 carriers may be due to elimination of paracrine signals from differentiated ER/PR-positive luminal cells to more primitive stem/progenitor cells (30). This mechanism would explain the protective effect of endocrine manipulation on the development of ER-negative breast cancers in these women (31).
Taken together these studies suggest that loss of BRCA1 function results in blocked epithelial differentiation with expansion of the undifferentiated stem/progenitor cell compartment. Because BRCA1 also functions in DNA repair and in maintaining chromosome stability, we propose that loss of BRCA1 function may produce genetically unstable stem/progenitor cells that serve as prime targets for further carcinogenic events, including p53 mutation (16, 32). These studies lend support to the cancer stem cell hypothesis by suggesting that dysregulation of stem cell self-renewal and differentiation may initiate hereditary as well as sporadic basal-like breast carcinomas, a portion of which also are characterized by loss of BRCA1 expression (15). Furthermore, the ability to detect expanded stem cell clusters in histologically normal tissue from BRCA1 mutation carriers may identify women within this population at particularly high risk for subsequent development of breast cancer.
Materials and Methods
Dissociation of Mammary Tissue.
One hundred to two hundred grams of normal breast tissue from reduction mammoplasties was minced with scalpels dissociated enzymatically, and single cells were cultured in suspension to produce mammospheres or on a collagen substratum to induce cellular differentiation as described (7, 8).
siRNA Contructs.
Two human BRCA1 siRNA oligos were purchased from Ambion (Silencer predesigned siRNAs) and were used to knock down BRCA1 expression in primary human mammary epithelial cells. The lentiviral siRNAs were constructed and produced, and the cells were infected as described previously (8). In our experiments, >70% of cells in suspension culture were infected with the lentiviruses. The infected cells were sorted based on the GFP- or DsRed-positivity by using a FACStarPLUS (Becton Dickinson) flow cytometer. SI Fig. 10 depicts the experimental design of the lentiviral transfection strategy. Both BRCA1 siRNA lentivirus products (1 and 2) produced similar levels of inhibition of BRCA1 expression and had similar effects in the in vitro systems. We therefore chose to use the BRCA1 siRNA1 construct for the mouse xenograft studies.
Differentiating Culture Conditions.
Single-cell suspensions were plated on collagen-coated plates at a density of 5,000 viable cells per 10-cm-diameter dish. Cells were grown in Ham's F-12 medium (Gibco) with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 10 μg/ml cholera toxin (Sigma–Aldrich), 10 ng/ml epidermal growth factor (BD Biosciences), and 1X Pen/Strep/Fungizone Mix (Gibco). Cells were collected for the lineage analysis by FACS at different time points (days 0, 7, 12, 26, and 35). All experiments were done in triplicate by using single-cell suspensions derived from three different patients.
Real-Time RT-PCR.
After mammospheres were formed in suspension culture or cells reached 85% confluency on the collagen plates (≈7 days), total RNA was isolated by using an RNeasy Mini Kit (Qiagen) and used for real-time quantitative RT-PCR assays in an ABI Prism 7900HT sequence-detection system with 384-well block module and automation accessory (Applied Biosystems). Primers and probes for the Taqman system were selected from the Applied Biosystems web site (www.appliedbiosystems.com). The sequences of the PCR primer pairs and fluorogenic probes used for BRCA1 and RPLPO are available on the Applied Biosystems web site (BRCA1 assay ID: Hs_00173233_mi; RPLPO assay ID: Hs_99999902_mi). The relative expression level of BRCA1 was computed with respect to the internal standard RPLPO gene to normalize for variations in the quality of RNA and the amount of input cDNA.
Western Blotting.
Cells were lysed by using the Pierce Nuclear and Cytoplasmic Extraction Reagent Kit (Pierce). Samples were normalized for protein concentration by using the Pierce BCA protein assay; 50 μg of each nuclear extract sample was analyzed by SDS/5% PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. Immobilized proteins were probed by using antibodies specific for BRCA1 (D9; Santa Cruz Biotechnology) or actin (Santa Cruz Biotechnology) and visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Aldefluor Assay and Flow Cytometry.
Single cells were isolated from lentivirus-infected primary mammospheres and the Aldefluor kit (Stem Cell Technologies) was used to isolate cells with high ALDH enzymatic activity as described previously (23). The 1 μg/ml propidium iodide (PI) (Sigma–Aldrich) was used to access cell viability.
For flow-cytometry analysis, freshly dissociated cells cultured on collagen substrata were stained with PE-conjugated ESA (dilution 1:100; Miltenyi Biotec) and PE-cy5-conjugated CD10 (dilution 1:25; Novocastra) in HBSS (Gibco) with 2% FBS and incubated on ice for 20 min., followed by washing in HBSS with 2% FBS and resuspending in HBSS supplemented with 2% FBS with 1 μg/ml PI (Sigma–Aldrich) for the cell viability. Analysis was performed by using a FACStarPLUS (Becton Dickinson) flow cytometer.
NOD/SCID Mouse Model.
The number 4 inguinal mammary glands of 3-week-old female NOD/SCID mice were cleared and humanized as previously described (9) following a previously established protocol (24). After ≈4 weeks, a 60-day release estrogen pellet (0.72 mg per pellet; Innovative Research of America) was placed s.c. on the back of the mouse's neck by using a trocar, and ≈1.2 × 105 to ≈2.0 × 105 single cells were resuspended in 20–50 μl of 1:1 matrigel 5% serum Ham's F-12 and injected into each of the cleared fatpads. All of the implantation experiments were repeated three times (at least two mice each time) by using single cells obtained from three different patients (SI Table 2). Two to three months after the implantation, the fatpads were removed and fixed in 10% formalin. The tissue was then embedded in paraffin and sectioned.
Immunostaining.
Immunostaining was performed on paraffin-embedded tissue sections as described in ref. 23. The primary antibodies, ALDH1 (BD Transduction Laboratories), cytokeratin 18 (Novocastra), cytokeratin 14 (Novocastra), smooth muscle actin (SMA) (DAKO), estrogen receptor (NeoMarkers), and GFP (NeoMarkers) were used at the dilutions indicated by the manufacturer.
BRCA1 Mutation Carriers Sample Collection.
Thirteen breast tissue samples derived from women carrying a known deleterious BRCA1 heterozygous mutation were collected. Breast tissue samples were obtained from the Surgical Pathology files at the University of Michigan with Institutional Review Board approval. BRCA1 mutation status and clinical information was obtained through the Breast and Ovarian Cancer Risk Evaluation Program at the University of Michigan Comprehensive Cancer Center and are listed in SI Table 1.
LCM.
We used laser capture microdissection to obtain separate and pure cell populations from lobules expressing ALDH1 and lobules negative for ALDH1 expression or from ALDH1-negative lobules and adjacent stromal cells. Then 5-μm paraffin-embedded sections obtained from breast tissue from women with confirmed germ-line BRCA1 mutations were immunostained for ALDH1, and LCM was performed on these stained sections. For each patient, tissue from eight serials sections was collected. The Arcturus PixCell II Laser capture microdissecting system (Arcturus Engineering) utilizes a transparent thermoplastic film applied to the surface of the tissue sections on histopathology slides. The ALDH1-positive or −negative normal epithelial cells and stromal cells to be microdissected were identified and targeted by microscope examination. At least 20 different lobules were microdissected for each group. The ALDH1-negative lobules were randomly selected on each slide. A narrow (≈15 μm) carbon dioxide laser-beam pulse-specificity activated the film above these cells. The resulting strong focal adhesion allowed selective procurement of targeted cells.
DNA Extraction.
DNA from microdissected tissues was extracted by using the Picopure extraction kit (Arcturus Engineering). Microdissected tissue was incubated in 50 μl of lysis buffer at 42°C overnight, followed by heat inactivation at 95°C for 10 min.
Markers for LOH and Amplification.
Four microsatellites from the BRCA1 region on chromosome arm 17q were selected (two intragenic microsatellites, D17S855 and D17S1323, and two telomeric microsatellites, D17S1325 and D17S806). Selection was done based on several criteria, including localization, degree of heterozygosity, previous use in LOH studies, and efficiency of amplification of DNA from microdissected tissue. The order of the markers on the map is known precisely due to the availability of the sequence of the human genome (www.ensembl.org; http://genome.ucsc.edu). Primers were designed according to UniSTS (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = unists). Microsatellite analysis was done after amplification by PCR using a PerkinElmer/Cetus thermal cycler model 9600.
Allelic Profiles and LOH Analysis.
The resulting PCR products were visualized and analyzed with an automated fluorescent sequencing apparatus (ABI 3730 Sequencer, DNA sequencing core; University of Michigan, Ann Arbor, MI). The allelic profiles were read on computer printouts separately by two observers, with one observer reading twice at two intervals. For a given informative marker, LOH was defined by a sharp decrease of either peak of >75%.
Statistical Analysis.
Distributions of molecular markers and other categorical variables were compared by using a standard Fisher exact test. The statistical test was two sided at the 5% level of significance and was done by using the R version 2.3.0 software.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Thomas Giordano (University of Michigan) for tissue procurement and advice with laser capture microdissection; Dr. William Foulkes for reviewing the manuscript; Dr. John Stingl and Dr. Connie Eaves (Terry Fox Laboratory, Vancouver, BC, Canada) for the fibroblast cell line; the University of Michigan Cancer Center Flow Cytometry Core; and the University of Michigan Cancer Center Vector Core. This work was supported by National Institutes of Health Grants CA101860, CA129765, and P 30 C CA46592.
Footnotes
Conflict of interest statement: M.S.W. has financial holdings in and is a scientific advisor for OncoMed Pharmaceuticals.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711613105/DC1.
References
- 1.Wicha MS, Liu S, Dontu G. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 4.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M, et al. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 5.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, Smith BD, Civin CI, Jones RJ. Blood. 2004;103:2332–2336. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 7.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Dontu G, Wicha MS. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reya T, Clevers H. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 11.Brennan K, Brown AMC. J Mammary Gland Biol Neoplasia. 2003;9:119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- 12.Diévart A, Beaulieu N, Jolicoeur P. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 13.Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, Gipp J, Shaw A, Lamm ML, Munoz A, et al. Endocrinology. 2004;145:3961–3970. doi: 10.1210/en.2004-0079. [DOI] [PubMed] [Google Scholar]
- 14.Narod SA, Foulkes WD. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 15.Turner NC, Reis-Filho JS. Oncogene. 2006;25:5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 16.Bradley A, Medina D. J Mamm Gland Biol Neoplasia. 1998;3:363–364. doi: 10.1023/a:1018727813904. [DOI] [PubMed] [Google Scholar]
- 17.Furuta S, Jiang X, Gu B, Cheng E, Chen PL, Lee WH. Proc Natl Acad Sci USA. 2005;102:9176–9181. doi: 10.1073/pnas.0503793102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubista M, Rosner M, Kubista E, Bernaschek G, Hengstschlager M. Oncogene. 2002;21:4747–4756. doi: 10.1038/sj.onc.1205580. [DOI] [PubMed] [Google Scholar]
- 19.Venkitaraman AR. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 20.Dontu G, El-Ashry D, Wicha MS. Trends Endocrinol Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Foulkes WD. J Med Genet. 2004;41:1–5. doi: 10.1136/jmg.2003.013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginestier C, Hur MH, Charafe-Jauffret E, MonvilleJ F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer C, Liu S, et al. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE, The W, et al. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 25.Clayton H, Titley I, Vivanco M. Exp Cell Res. 2004;297:444–460. doi: 10.1016/j.yexcr.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 26.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albertsen HM, Smith SA, Mazoyer S, Fujimoto E, Stevens J, Williams B, Rodriguez P, Cropp CS, Slijepcevic P, Carlson M, et al. Nature Genet. 1994;7:472–479. doi: 10.1038/ng0894-472. [DOI] [PubMed] [Google Scholar]
- 28.Poole AJ, Li Y, Kim Y, Lin SC, Lee WH, Lee EY. Science. 2006;314:1467–1470. doi: 10.1126/science.1130471. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Holstege H, Gulden Hvd, Treur-Mulder M, Zevenhoven J, Velds A, Kerkhoven RM, Vliet MHv, Wessels LFA, Peterse JL, et al. Proc Natl Acad Sci USA. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kauff ND, Satagopan JM, Robson ME, Scheuer L, Hensley M, Hudis CA, Ellis NA, Boyd J, Borgen PI, Barakat RR, et al. N Engl J Med. 2002;346:1609–1615. doi: 10.1056/NEJMoa020119. [DOI] [PubMed] [Google Scholar]
- 31.Narod SA, Brunet JS, Ghadirian P, Robson M, Heimdal K, Neuhausen SL, Stoppa-Lyonnet D, Lerman C, Pasini B, de los Rios P, et al. Lancet. 2000;356:1876–1881. doi: 10.1016/s0140-6736(00)03258-x. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.