Abstract
Multiple cell-autonomous mechanisms exist in complex metazoans to resist oncogenic transformation, including a variety of tumor- suppressor pathways that control cell proliferation and apoptosis. In vertebrates, additional mechanisms of tumor resistance could potentially rely on cancer cell elimination by specialized cytotoxic leukocytes, such as natural killer (NK) cells. Such mechanisms would require that cancer cells be reliably distinguished from normal cells. The ligands for NKG2D, an activating NK cell receptor, are expressed on many tumor cell lines and at least some primary human tumors. However, it is unknown whether their expression is induced as a direct result of oncogenic transformation in vivo. We provide evidence that NKG2D ligands are induced on spontaneously arising tumors in a murine model of lymphomagenesis and that c-Myc is involved in their regulation. Expression of NKG2D ligands is induced at an early, distinct stage of tumorigenesis upon acquisition of genetic lesions unique to cancer cells, potentially defining a critical step in carcinogenesis. This finding suggests that the regulation of NKG2D ligands depends on a mechanism for intrinsic sensing of oncogenic transformation.
Keywords: Eμ-Myc, innate immunity, tumorigenesis
The susceptibility of target cells to natural killer (NK)-mediated killing is thought to be determined by the relative expression of ligands for NK-activating and -inhibitory receptors (1). NKG2D is an activating receptor expressed on human and mouse NK cells that recognizes a diverse family of ligands, including the Rae-1 family (α, β, γ, δ, and ε), H60, and MULT1 in the mouse and the MICA and ULBP families in humans (2–9). Engagement of NKG2D triggers the cytotoxic activity of NK cells toward target cells expressing ligand(s) (2, 3, 5). Accordingly, NKG2D ligands are not expressed on the surface of normal cells, but their expression is generally induced on unwanted cells, such as virally infected, stressed, and DNA-damaged cells (2–4, 10, 11). Because primary human tumors, many tumor cell lines, and at least some carcinogen-induced tumors can express NKG2D ligands, it is likely that NKG2D–ligand interactions participate in tumor immunosurveillance (9, 12, 13). In tumor cell lines that fail to express NKG2D ligands, ectopic expression of Rae-1 leads to their elimination in vivo (14, 15). Data from murine models have demonstrated an increased incidence of methylcholanthrene (MCA)-induced fibrosarcomas in NKG2D-neutralizing conditions (16). Furthermore, active evasion of immunosurveillance has been observed in individuals with epithelial-derived tumors where soluble, surface-shed NKG2D ligands (MICA) present in the sera result in down-regulation of NKG2D and subsequent impairment of cytotoxic cells (17).
Despite these observations, it is currently unknown whether expression of NKG2D ligands is regulated by pathways implicated in tumorigenesis. Indeed, any “tumor-specific” gene can be up-regulated either as a direct result of oncogenic transformation or as an indirect consequence of tumor growth and evolution. To address this question, we took advantage of a well characterized mouse model of spontaneous B cell lymphoma, whereby the Ig μ enhancer drives continuous expression of the Myc oncogene (Eμ-Myc) (18–20). Eμ-Myc mice develop Burkitt-like lymphoblastic lymphomas with variable kinetics (mean latency of ≈6 months) and faithfully recapitulate several aspects of tumor progression (19).
We first examined whether NKG2D ligands are up-regulated on spontaneously arising cancer cells. Splenocytes were isolated from an Eμ-Myc mouse with lymphoma and a littermate control. B cells were stained with recombinant mNKG2D/Fc, a reagent that detects cell-surface expression of all known NKG2D ligands (8). As expected, normal wild-type B cells did not express any ligands on their surface as determined by negative mNKG2D/Fc staining (Fig. 1A). In contrast, nearly all spontaneously arising lymphomas analyzed (n > 50) expressed ligand(s) for NKG2D (Fig. 1A). This expression was unique to the B cell compartment, defined by B220-positive staining, because T cells within a lymphoma-bearing animal failed to show surface staining (data not shown). We next asked which of the several NKG2D ligands are expressed on cancer cells in this model. H60 and Rae-1α, -1β, and -1γ are not expressed in the C57BL/6 background (21). An antibody specific for Rae-1ε largely blocked mNKG2D/Fc staining (Fig. 1B). In addition, primary lymphomas could be directly stained with a Rae-1ε-specific antibody (Fig. 1B). MULT-1 was not detected on the surface of tumor cells (data not shown). Together, these data suggest that Rae-1ε is the dominant ligand expressed in this system. However, it is possible that other NKG2D ligands may be induced on cancer cells and contribute to NKG2D-mediated immunosurveillance. There are likely to be several different mechanisms that regulate NKG2D ligand expression, permitting cells to engage in “assisted apoptosis” (22). We assayed for transcriptional induction of Rae-1ε (Raet1) in Eμ-Myc preneoplastic B cells and Eμ-Myc tumor B cells. Raet1 mRNA was strongly induced in tumor cells and moderately induced in preneoplastic cells, suggesting transcriptional control as at least one level of regulation (Fig. 1C).
Fig. 1.
Ligands for NKG2D are up-regulated in spontaneously arising lymphomas. (A) Splenocytes from Eμ-Myc mice with lymphoma and littermate controls (LMC) were stained with B220 (B cell marker) and mNKG2D/Fc and analyzed by flow cytometry. The histograms shown, if representing gated populations, are as indicated (e.g., B220+). The shaded histogram represents isotype control for mNKG2D/Fc, and the heavy black line represents mNKG2D/Fc staining. Log fluorescence is plotted on the x axis, and the percentage of maximum events per cells is plotted on the y axis. This scaling is maintained for all subsequent histograms. (B) (Left) Splenocytes from Eμ-Myc mice with lymphoma were stained with B220 and mNKG2D/Fc. The dotted line is mNKG2D/Fc staining after blocking with anti-mouse Rae-1ε, the heavy black line is mNKG2D/Fc staining after blocking with isotype control for anti-mouse Rae-1ε, the thin black line is mNKG2D/Fc staining alone, and the shaded histogram is isotype control for mNKG2D/Fc. (Center) Splenocytes from Eμ-Myc mice with lymphoma were stained with mNKG2D/Fc (heavy black line)/isotype control (shaded histogram) or (Right) anti-mouse Rae-1ε (heavy black line). The shaded histogram here represents anti-mouse Rae-1α/β/γ, and the thin black line represents an additional isotype control. Data are representative of three separate experiments. (C) Quantitative PCR analysis of Raet1 mRNA induction in B cells from wild-type (WT), Eμ-Myc preneoplastic, and Eμ-Myc tumor cells. Values are normalized to Ubiquitin gene expression and reflect fold induction over wild-type B cells (assigned a value of 1.0 and depicted in column 1).
Eμ-Myc-driven lymphomas follow a classic multistep model of tumorigenesis where additional, random, and independent mutations are acquired before the eventual manifestation of a clonal/oligoclonal lymphoblastic lymphoma (19, 23). The kinetics of lymphoma onset suggests that one or two additional genetic alterations are required for the transition from a true polyclonal preneoplastic state to disseminated lymphoma (24). Because of the leukemic component of these cancers, we were able to analyze peripheral blood leukocytes (PBLs) for the onset of Rae-1 expression in a given animal over the entire course of tumorigenesis. A representative example (Fig. 2) shows the analysis of Rae-1 expression in the same animal from the preneoplastic stage (8 weeks of age in the example shown) until the animal is terminally ill 6 weeks later. Importantly, these analyses demonstrate that preneoplastic B cells from Eμ-Myc mice do not express NKG2D ligands (Figs. 2 and 3C). Rae-1-positive cells first emerge among pre-B (B220+/IgM−) cells (in the example shown) after 9 weeks of a preneoplastic stage. This stage will vary from animal to animal because of the stochastic acquisition of mutations required for transformation (19). The pre-B cell population continues to stay Rae-1-positive for the remainder of the analysis, eventually occupying a large fraction of the peripheral blood (Fig. 2). Eμ-Myc mice succumb to both pre-B and mature B cell lymphomas, both of which express Rae-1 (Fig. 2 and data not shown). These observations suggest that Rae-1 expression occurs relatively early during the course of lymphoid tumorigenesis and, unlike most known tumor-specific alterations, is not a late event acquired as a result of tumor evolution. We conclude that Rae-1 expression may be controlled by genetic alterations unique to cancer cells and therefore can reliably distinguish normal cells from transformed cells.
Fig. 2.
Rae-1 induction during the course of lymphomagenesis. (Upper) PBLs from an Eμ-Myc mouse before lymphoma onset were stained as indicated at weekly intervals. Log fluorescence is depicted on the x and y axes. The gated population represents pre-B cells, with a corresponding percentage of total PBLs. Each column represents one time point. (Lower) Histograms are derived from the population gated in Upper. Shaded histogram represents mNKG2D/Fc staining at 8 weeks in all six time points. The heavy black line indicates mNKG2D/Fc staining at subsequent intervals. Data are representative of 10 independent experiments. Individual mouse number is indicated next to genotype in this and subsequent figures.
Fig. 3.
Properties of Rae-1 induction. (A) H2AX phosphorylation levels in B cell lysates from wild-type (WT), Eμ-Myc preneoplastic, and Eμ-Myc tumors was determined by immunoblotting. (B) (Left) CFSE-labeled Ink4a/Arf−/− B cells were stimulated with 3 μM CpG and analyzed 96 h later. Gated population represents actively dividing cells as measured by CFSE dilution. (Right) Rae-1ε staining of the gated population (heavy black line). Shaded histogram represents the isotype control. (C) Splenocytes from preneoplastic Eμ-Myc, Ink4a/Arf−/−, and p53−/− mice were stained as described in Fig. 1B. The gate (B220+) depicted for Eμ-Myc preneoplastic cells was applied to all samples.
It is well established that engagement of NKG2D results in NK cell activation and a perforin-dependent cytotoxic response against target cells (8, 9, 25). We tested whether the expression of Rae-1 in primary lymphomas derived from an Eμ-Myc animal would result in NK-mediated killing of cancer cells. Using an in vivo cytotoxicity assay, we observed significant killing of lymphoma cells that was largely perforin dependent [supporting information (SI) Fig. 6A]. The fact that NK cells are capable of killing cancer cells expressing Rae-1 raised the question: Why do lymphomas still arise in Eμ-Myc mice? It has been previously shown that the NKG2D receptor can be down-regulated on NK cells upon ligand binding, resulting in NK cell inactivation (17, 26). We thus investigated whether persistent NKG2D signaling in Eμ-Myc lymphoma-bearing mice would result in receptor down-regulation. Indeed, we found that in Eμ-Myc mice with lymphoma (Rae-1-positive), the levels of NKG2D on NK cells were down-regulated, compared with lymphoma-free littermate control mice (SI Fig. 6B). In the Eμ-Myc model, the accelerated kinetics of tumorigenesis caused by Myc overexpression may result in an overwhelmingly large population of cancer cells that cannot be adequately dealt with by the NK system.
Having established that Rae-1 is induced during spontaneous lymphomagenesis, we next asked what genetic events are responsible for triggering Rae-1 expression on cancer cells. Oncogenic stress created by sustained Myc expression has been shown to induce DNA damage through a variety of mechanisms (29–31). The detection of DNA damage plays an important role in eliminating nascent tumors (27, 28). In addition, DNA damage can lead to NKG2D ligand expression (10). Because the sensing of DNA damage could be the event that induces surface expression of Rae-1 during lymphomagenesis, we examined its role in Eμ-Myc preneoplastic and tumor cells. In both preneoplastic and tumor B cells, we observed the phosphorylated form of H2AX (γ-H2AX), a mark of DNA double-strand breaks (Fig. 3A). Thus, although the sensing of DNA damage likely plays a role in the Eμ-Myc model of tumorigenesis (32), damage does not appear to be sufficient for the expression of Rae-1 in this model because preneoplastic B cells have an activated damage checkpoint yet remain Rae-1-negative. Additionally, the expression of Rae-1 is not simply a result of B cell proliferation, because CpG-induced B cell proliferation did not result in significant Rae-1ε expression (Fig. 3B).
The vast majority of mutations that have been observed in lymphomas in Eμ-Myc mice affect the p19Arf (Arf)–Mdm2–p53 pathway (33–36). Sustained Myc signaling induces Arf expression, resulting in apoptosis through the inhibition of Mdm2 and stabilization of the p53 tumor suppressor protein (37, 38). Thus, stabilization of p53 during sustained Myc expression provides a built-in mechanism to prevent excessive proliferation (39, 40). The preneoplastic window in Eμ-Myc mice is likely a result of this cell-intrinsic apoptotic program. Indeed, mice expressing the Eμ-Myc transgene on p16Ink4a/p19Arf (Ink4a/Arf), Arf, or p53 heterozygous backgrounds develop lymphomas with significantly faster kinetics (33–36). We therefore tested whether NKG2D ligand expression depends on the acquisition of mutations in the Arf–Mdm2–p53 pathway.
First, we found that B cells from preneoplastic Eμ-Myc, Ink4a/Arf −/−, and p53−/− mice do not express Rae-1 (Fig. 3C). In Eμ-Myc;Ink4a/Arf+/− mice, we observed the rapid and synchronous development of B cell lymphomas as previously reported (35, 36). We found that these lymphomas were consistently Rae-1-positive (Fig. 4A Left), suggesting that disruption of the Arf gene in the context of Myc expression can lead to Rae-1 expression. However, Arf has several functions independent of p53 modulation. To further investigate the specific contribution of the p53 pathway in NKG2D ligand expression, we generated Eμ-Myc;p53+/− mice. These mice also rapidly developed aggressive lymphomas that were Rae-1-positive (Fig. 4A Right).
Fig. 4.
Genetic requirements for NKG2D ligand induction. (A) Staining of lymphomas derived from the indicated genotypes as in Fig. 1A. (B) Staining of lymphoma (Eμ-Myc;Ink4a/Arf+/− 17) and PCR of genomic DNA from lymph node cells derived from same animal or indicated genotypes, assaying for LOH at exon 2. (C) Staining of splenocytes from an early Eμ-Myc;Ink4a/Arf+/− animal. High- and low-FSC gates are constructed within a total live gate as indicated. Numbers reflect the percentage of cells within the live gate that fall within these two populations. Data are representative of more than five experiments. (D) A schematic of the Ink4a/Arf locus (37) with primers specific for exon 1α and exon 2 is shown with quantitative PCR results. Exon 1α was used as a normalizer for genomic copy number among different samples. Each data point represents an individual tumor from a unique mouse. Quantitative PCR is from genomic DNA derived from B cells of the indicated organismal genotype. (E) VDJ usage was analyzed by multiplex PCR and resolved on a sequencer. (Left) Pattern of peaks represents a polyclonal population. (Right) High-FSC B cells from early time points in Eμ-Myc;Ink4a/Arf+/− lymphomagenesis show a largely clonal or oligoclonal profile. Fragment size is plotted on the x axis, and fluorescence intensity is plotted on the y axis.
The Ink4a/Arf and p53 tumor suppressor genes in these animals are initially heterozygous. However, these loci undergo loss of heterozygosity (LOH) during the course of lymphomagenesis, underscoring the importance of the p53 pathway in preventing Myc-induced transformation (33, 35, 36). We tested whether LOH at the Ink4a/Arf locus has occurred in the Eμ-Myc;Ink4a/Arf+/− lymphomas, which, as mentioned above, are Rae-1-positive. In Ink4a/Arf−/− mice, exons 2 and 3 are disrupted, preventing expression of both gene products (41). However, it has been established that mutations in Arf, not Ink4a, play a major role in Eμ-Myc lymphomagenesis (33, 36). For further analyses, we chose to monitor this locus, rather than p53, because the analysis of LOH at Arf is well established (42). We found that in these Rae-1-positive cells, LOH occurred at the Arf locus (Fig. 4B) as previously demonstrated in late-stage lymphomas (33, 35, 36).
We next asked whether LOH at the Arf locus is required for Rae-1 induction on lymphomas. We found that at early stages of lymphomagenesis in Eμ-Myc mice, there are both Rae-1-negative (nontumorigenic) and Rae-1-positive (tumorigenic) cells that can be seen in the same animal (Fig. 2, 10 weeks). Thus, this stage in lymphomagenesis would be predicted to exist in Eμ-Myc;Ink4a/Arf+/− animals where disease progression is dramatically accelerated. Observing such a stage would permit the analysis of cells that have minimal genetic alterations sufficient for Rae-1 expression. To this end, we analyzed splenocytes from Eμ-Myc;Ink4a/Arf+/− mice at a relatively early time point (≈5 weeks) and observed cells that could be morphologically distinguished based on forward scatter (FSC) values (Fig. 4C and SI Fig. 7). We observed high-FSC B cells to be Rae-1ε-positive, whereas low-FSC B cells were Rae-1ε-negative within the same animal. Although we did observe heterogeneity in the expression of Rae-1, this finding is not surprising given that these tumors arise independently and thus are inherently dissimilar at some level (SI Fig. 7). Individual tumors analyzed at precisely the same critical time point, as it relates to tumor progression (i.e., LOH), may stain more uniformly.
Because we observed two distinct populations of B cells in the Eμ-Myc;Ink4a/Arf+/− mice, we assessed the status of the Ink4a/Arf locus at this early stage. We found that the remaining allele of the Ink4a/Arf locus was intact in Rae-1ε-negative B cells but was lost in the Rae-1ε-positive B cells (Fig. 4D). This finding raised the possibility that in vivo Myc overexpression (in Eμ-Myc) and LOH at the Ink4a/Arf locus may be sufficient for Rae-1 expression. To address this possibility further, we assessed the clonality of these Rae-1-positive B cells. Although early Eμ-Myc;Ink4a/Arf+/− B cells, before the appearance of two FSC discernible populations, were polyclonal (Rae-1ε-negative), the high-FSC Rae-1ε-positive cells were mono- or oligoclonal (Fig. 4E), presumably reflecting a selection stage that accompanies LOH at the Ink4a/Arf locus. Thus, both dysregulated Myc expression and LOH appear to be necessary for Rae-1 expression on lymphoma cells. However, based on the mono- or oligoclonal nature of the early Rae-1-positive tumor cells, we cannot exclude the possibility that an additional genetic, epigenetic, or developmental event also is necessary for Rae-1 induction. This event would be presumably acquired after LOH at the Ink4a/Arf locus and could formally cooperate with Myc alone to induce Rae-1. Thus, loss of Arf activity would be permissive for acquiring this property but not necessary per se for Rae-1 surface expression. Importantly, however, a combination of deregulated activity of a dominant oncogene product (Myc) and loss of a tumor suppressor protein (either Arf or p53) is involved in the induction of Rae-1 in the B cell lymphoma model studied here. It is likely that different combinations of oncogenes and tumor suppressors might be involved in Rae-1 induction in other cell types.
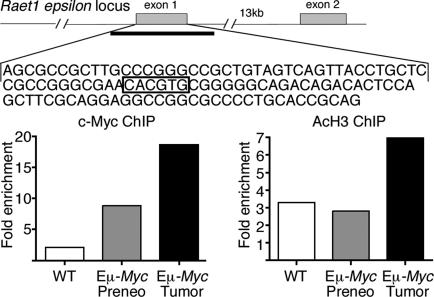
We next examined whether c-Myc plays a direct role in Raet1 induction. Within exon 1 of Raet1 epsilon exists an E-box (CACGTG), the canonical Myc-binding sequence. In addition, this region is within a CpG island, a highly permissive environment for Myc binding (43). We isolated B cells from wild-type, preneoplastic, and tumor-bearing animals and performed ChIP for both c-Myc and acetylated histone H3, a mark of both chromatin accessibility and transcriptional activation (43). We observed enhanced binding (8-fold enrichment over wild type) of Myc to the locus in tumor cells as well as increased histone H3 acetylation relative to wild-type B cells (Fig. 5). This observation suggests that Myc is directly involved in regulating Raet1 epsilon and correlates well with the transcriptional induction we observed previously (Fig. 1C). Raet1 epsilon was modestly induced in the preneoplastic stage, which likely reflects Myc binding to the locus and inducing transcription. The oncogenic state, created in part by LOH at the Ink4a/Arf locus, may further derepress the Raet1 epsilon locus, directly promote transcriptional induction, or support stabilization of the message for surface expression.
Fig. 5.
Myc directly binds within the Raet1 epsilon locus. (Upper) Schematic of Raet1 epsilon locus is shown with indicated E-box (CACGTG) in exon 1. Heavy line under exon 1 demarcates a CpG island of ≈1.5 kb spanning exon 1. (Lower) ChIP analysis of B cells by using antibodies against c-Myc and acetylated histone H3. Precipitated DNA was amplified by using primers flanking the indicated E-box. Data are representative of three independent experiments and are expressed as fold enrichment at the Rae-1 epsilon locus over an amplicon within a control (Chrnb4) locus.
Oncogenic transformation is a multistage process of successive acquisition of genetic and epigenetic alterations affecting cell proliferation and survival (23, 44). With a few notable exceptions, different stages of cancer progression are not easily distinguishable and, therefore, not clearly defined. The results presented here reveal a critical stage of oncogenic transformation that is marked by cell-surface expression of a signal that triggers a cytotoxic response against a nascent cancer cell. This early stage is defined by a combination of genetic events unique to cancer cells, a combined alteration of an oncogene and a tumor suppressor gene. A putative intrinsic sensor of oncogenic transformation that activates the expression of Rae-1 detects this abnormal state. The nature of this sensor is puzzling. Myc alone can alter expression of hundreds of genes, yet even sustained activity of Myc is not sufficient for triggering Rae-1 surface expression (Figs. 2 and 3C). Deficiency in either Arf or p53 also is necessary, yet even complete loss in either tumor suppressor gene by itself is insufficient (Fig. 3C). Thus, the putative sensor appears to detect a unique characteristic of tumor cells: The presence of a dominant oncogene and the loss of a tumor suppressor. Triggering the sensor allows tumor cells to “report” their transformed status to the host immune system. Although we have detected transcriptional induction of Raet1 in lymphomas (Fig. 1C), precisely how the oncogenic state is sensed and relayed to this locus is unclear. At a minimum, our data suggest that Myc is directly involved in this switch. It also appears that posttranscriptional regulation contributes to cell-surface expression of NKG2D ligands (A.M.U., T.B., and R.M., unpublished data).
Although the present analysis was done exclusively on B lymphomas, tumor cells derived from other compartments may phenocopy the stage marked by NKG2D ligand expression. Indeed, we have found that NKG2D ligands also are induced on spontaneous T cell lymphomas (data not shown). The nature of the genetic abnormalities detected in different cancer types may be varied, although the features observed here are likely to be involved in many of them. Tumor immunosurveillance has been studied primarily in the context of adaptive immune responses to tumor-associated or tumor-specific antigens. The innate immune system, however, is not antigen-based, but rather detects signals characteristic of infectious non-self or abnormal self and, in so doing, can reliably identify the origin of the signal. Here, we provide evidence for an innate immunosurveillance mechanism that is causally linked to cancer-specific genetic lesions.
Methods
Mice.
All mice were bred and maintained at the animal facility of the Yale University School of Medicine. Eμ-Myc hemizygous mice were kindly provided by Scott Lowe (Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). Ink4a/Arf−/− mice were obtained from the Mouse Models of Human Cancers Consortium Repository at the National Cancer Institute. p53−/−, CD45.1, and C57BL/6 mice were obtained from The Jackson Laboratory. All mice were on a C57BL/6 background. All mouse experiments were performed after approval by and in accordance with regulatory guidelines and standards set by the Institutional Animal Care and Use Committee of Yale University.
Reagents.
Recombinant mouse NKG2D/Fc chimera (mNKG2D/Fc), recombinant human IgG1 Fc (isotype control for mNKG2D/Fc), monoclonal anti-mouse Rae-1ε, and monoclonal anti-mouse Rae-1α/β/γ antibody were purchased from R&D Systems. Biotinylation of these antibodies was performed by using EZ-Link Biotin from Pierce. Purified anti-CD16/CD32, phycoerythrin (PE) anti-CD45R/B220, anti-CD45.1, anti-NK1.1, fluorescein isothiocyanate (FITC) anti-H-2Kb, anti-CD3ε, anti-IgM, and streptavidin-PE-Cy5 (Cy-Chrome) were all purchased from BD Biosciences. Biotin anti-human IgG (H+L) was purchased from Jackson ImmunoResearch. Biotin anti-NKG2D (CX5) and purified rat IgG2a isotype control were purchased from eBioscience. Anti-CD19 microbeads were purchased from Miltenyi Biotech. Poly(I·C) was purchased from Sigma–Aldrich. Carboxyfluorescein diacetate succinimidyl ester (CFSE) was purchased from Molecular Probes.
Flow Cytometry.
Erythrocyte-depleted splenocytes were stained with relevant antibodies for 30 min on ice and analyzed on a FACScan or FACSCalibur flow cytometer (BD Biosciences). Cells were first incubated with purified anti-CD16/CD32 to block Fc-mediated binding of antibodies. For mNKG2D/Fc staining, cells were stained with biotin anti-human IgG (preadsorbed with 3% normal mouse serum before usage), followed by streptavidin-Cy-Chrome. Data were analyzed by using FlowJo software (Tree Star).
In Vivo Cytotoxicity Assay.
Splenocytes were prepared from CD45.1 and lymphoma-bearing mice (Eμ-Myc;Ink4a/Arf+/−). B cells were isolated from CD45.1 mice by using anti-CD19 microbeads, followed by purification with an AutoMACS sorter (Miltenyi Biotech). Then, >95% of cells were B220+ after purification. Lymphomas were labeled with CFSE. Twenty-four hours before injection, mice were treated with 100 μg of poly(I·C) or PBS (vehicle) i.p. (100 μl total volume). Fifteen million total cells were injected intravenously at a ratio of 2:1 (lymphoma:CD45.1). Eighteen hours after injection, animals were euthanized and splenocytes stained with anti-CD45.1.
PCR.
Genomic DNA was isolated from lymph nodes or spleen with DNAzol (Invitrogen). For LOH assays, primer sequences and PCR conditions were kindly provided by Scott Lowe (42). For quantitative LOH analysis, primers for exon 1α and exon 2 (same as above) were separately used with QuantiTect SYBR Green reagents (Qiagen) and run on an Mx3000 real-time PCR system (Stratagene). Values among separate samples were normalized to exon 1α. In populations with differential Rae-1/FSC properties, B cells were isolated by cell sorting (DAKO). Total RNA was isolated from wild-type, preneoplastic Eμ-Myc, and tumor Eμ-Myc B cells with RNA-Bee reagent (Tel-Test). Total RNA was reverse transcribed with oligo(dT) and SuperScript III (Invitrogen). cDNAs were analyzed by quantitative PCR amplification by using SYBR Green Reagents on an Mx3000 real-time PCR System. The abundance of Raet1 message was normalized to Ubiquitin and computed by using the comparative quantitation module of MXPro Software. Primers for Ubiquitin were as described previously (45). Raet1 primers are as follows: forward, CCCCAATGCAGACAGAAAAT; reverse, GAAGCGGGGAAGTTGATGTA.
Clonality.
Genomic DNA was prepared from sorted B cells (same as above), and PCR analysis was performed as described in ref. 46.
B Cell Proliferation.
B cell enrichment of splenocytes was performed by complement-mediated T cell depletion with anti-Thy-1/anti-CD4, followed by rabbit complement (Cedarlane Laboratories). Purity was >90% after depletion as assessed by B220+ staining. Cells were stimulated with 3 μM CpG DNA1826 (Keck Facility, Yale University, New Haven, CT). Cells were labeled for 10 min at 37° with 5 μM CFSE. Cells were cultured at 1 × 106 cells per ml in complete media (RPMI medium 1640 supplemented with 10% FCS, 10 mM Hepes, 10 mM sodium pyruvate, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin) (Invitrogen).
Western Blotting.
Total splenocytes were used fresh. Cells were spun on a Ficoll gradient to remove dead cells, followed by B cell purification by positive selection performed as described above. Purified B cells were lysed in TNG buffer [50 mM Tris·HCl (pH 7.5), 200 mM NaCl, 50 mM β-glycerol phosphate, 1% Tween-20, and 0.2% Nonidet P-40] supplemented with protease and phosphatase inhibitors. Lysates were cleared by centrifugation at 13,000 × g for 10 min at 4°C, and protein concentration was assayed in the supernatants. Pellets were further extracted with Laemmli SDS loading buffer. Samples were normalized by the protein concentration, and equal amounts of supernatant and pellet fractions were combined and resolved on a 4–12% gradient Tris-Glycine NuPAGE gel (Invitrogen). After standard transfer, Western blotting was performed with mouse anti-γ-H2AX (1:2,000; Upstate Biotechnology) and mouse anti-GAPDH (1:10,000; Sigma–Aldrich) antibodies.
ChIP.
B cells were isolated from wild-type and preneoplastic spleens by T cell depletion as described above. Tumor samples were not depleted because T cells represented a minor fraction of total splenocytes. Purity was >80% as assessed by B220+ staining for all samples. ChIP was performed essentially as described previously (43, 47). Anti-c-Myc (N262) was purchased from Santa Cruz Biotechnology and anti-acetylated H3 from Upstate Biotech. Raet1 epsilon locus primers are as follows: forward, GGCCGCTGTAGTCAGTTACC; reverse, CCCCCTACACCCCATTACTC. Chrnb4 (control) locus primers are as follows: forward, TTGGGTAAGCCAGGCTAAGA; reverse, TCTCATTCGTCTTGGGGACT. Immunoprecipitated DNA and input DNA were amplified with locus-specific primers by quantitative PCR by using input DNA to establish a standard curve. Data are represented as % input (Raet1 epsilon locus)/% input (Chrnb4 locus) and expressed as fold enrichment.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Scott Lowe for providing Eμ-Myc mice and a protocol for carrying out LOH analysis; Greg Barton, Chandrashekhar Pasare, and Noah Palm for critical and insightful discussions; and Charles Annicelli and Sophie Holley for expert assistance with animal care. This work was supported in part by the Training Program in Genetics National Institutes of Health Grant 5T32GM07499 (A.M.U.), a postdoctoral fellowship from the Leukemia and Lymphoma Society (T.B.), and the Howard Hughes Medical Institute (R.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701675105/DC1.
References
- 1.Vivier E, Nunes JA, Vely F. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 2.Raulet DH. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 3.Cerwenka A, Lanier LL. Nat Rev Immunol. 2001;1:41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama WM, Plougastel BF. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 5.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 6.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Science. 1999;285:727–729. [PubMed] [Google Scholar]
- 7.Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 8.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 9.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Nat Immunol. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 10.Gasser S, Orsulic S, Brown EJ, Raulet DH. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Routes JM, Ryan S, Morris K, Takaki R, Cerwenka A, Lanier LL. J Exp Med. 2005;202:1477–1482. doi: 10.1084/jem.20050240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Science. 2001;294:605–609. [PubMed] [Google Scholar]
- 13.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerwenka A, Baron JL, Lanier LL. Proc Natl Acad Sci USA. 2001;98:11521–11526. doi: 10.1073/pnas.201238598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. J Exp Med. 2005;202:583–588. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groh V, Wu J, Yee C, Spies T. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 18.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 19.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langdon WY, Harris AW, Cory S, Adams JM. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 21.Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. J Exp Med. 2003;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medzhitov R, Janeway CA., Jr Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 23.Fearon ER, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 24.Cory S, Vaux DL, Strasser A, Harris AW, Adams JM. Cancer Res. 1999;59:1685s–1692s. [PubMed] [Google Scholar]
- 25.Hayakawa Y, Kelly JM, Westwood JA, Darcy PK, Diefenbach A, Raulet D, Smyth MJ. J Immunol. 2002;169:5377–5381. doi: 10.4049/jimmunol.169.10.5377. [DOI] [PubMed] [Google Scholar]
- 26.Ogasawara K, Hamerman JA, Hsin H, Chikuma S, Bour-Jordan H, Chen T, Pertel T, Carnaud C, Bluestone JA, Lanier LL. Immunity. 2003;18:41–51. doi: 10.1016/s1074-7613(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 27.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 28.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 29.Pusapati RV, Rounbehler RJ, Hong S, Powers JT, Yan M, Kiguchi K, McArthur MJ, Wong PK, Johnson DG. Proc Natl Acad Sci USA. 2006;103:1446–1451. doi: 10.1073/pnas.0507367103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW. Cancer Res. 2006;66:6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 31.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 32.Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ, Jr, Anderson CW, Appella E, Bulavin DV. J Exp Med. 2006;203:2793–2799. doi: 10.1084/jem.20061563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu B, Marin MC, el-Naggar AK, Stephens LC, Brisbay S, McDonnell TJ. Oncogene. 1995;11:175–179. [PubMed] [Google Scholar]
- 35.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr CJ. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 38.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evan GI, Vousden KH. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 40.Lowe SW, Cepero E, Evan G. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 41.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 43.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 45.Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, Cleveland JL. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 46.Egle A, Harris AW, Bath ML, O'Reilly L, Cory S. Blood. 2004;103:2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- 47.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.