Abstract
Although central to pathogenesis, the molecular mechanisms used by microbes to regulate virulence factor production in specific environments during host–pathogen interaction are poorly defined. Several recent ex vivo and in vivo studies have found that the level of group A Streptococcus (GAS) virulence factor gene transcripts is temporally related to altered expression of genes encoding carbohydrate utilization proteins. These findings stimulated us to analyze the role in pathogenesis of catabolite control protein A (CcpA), a GAS ortholog of a key global regulator of carbohydrate metabolism in Bacillus subtilis. Inasmuch as the genomewide effects of CcpA in a human pathogen are unknown, we analyzed the transcriptome of a ΔccpA isogenic mutant strain grown in nutrient-rich medium. CcpA influences the transcript levels of many carbohydrate utilization genes and several well characterized GAS virulence factors, including the potent cytolysin streptolysin S. Compared with the wild-type parental strain, the ΔccpA isogenic mutant strain was significantly less virulent in a mouse model of invasive infection. Moreover, the isogenic mutant strain was significantly impaired in ability to colonize the mouse oropharynx. When grown in human saliva, a nutrient-limited environment, CcpA influenced production of several key virulence factors not influenced during growth in nutrient-rich medium. Purified recombinant CcpA bound to the promoter region of the gene encoding streptolysin S. Our discovery that GAS virulence and complex carbohydrate utilization are directly linked through CcpA provides enhanced understanding of a mechanism used by a Gram-positive pathogen to modulate virulence factor production in specific environments.
Keywords: ccpA, pharyngitis, regulation, streptolysin, transcriptome
Investigations in bacterial pathogens have suggested close links between basic metabolic processes and microbial pathogenesis (1, 2). For example, bacteria alter transcription of carbohydrate utilization genes and virulence factor production in response to changes in environmental conditions encountered during infection in humans (3, 4). Therefore, it is reasonable to speculate that pathogenic bacteria have developed molecular strategies to directly link regulation of carbohydrate utilization and virulence factor production. However, the mechanisms underlying such relationships are largely undefined.
In Bacillus subtilis, alterations in gene transcription in response to environmental carbohydrate concentrations are controlled in part by catabolite control protein A (CcpA), which binds to DNA at catabolite response element (cre) sites (5). Binding of CcpA to cre sites is enhanced by interaction of CcpA with the phosphoprotein HPr-Ser-46-P, the phosphorlyation state of which in turn is affected by uptake of glucose and other readily metabolized carbohydrates by phosphotransferase (PTS) systems (6). Thus, in Bacillus spp., CcpA directly links environmental carbohydrate levels with transcriptional regulation of carbohydrate utilization genes. Most studies of CcpA have been conducted in Bacillus spp. (5, 7). Several Gram-positive pathogens encode proteins with significant homology to B. subtilis CcpA and HPr suggesting that similar molecular processes may occur in other microbes (8–11).
Group A Streptococcus (GAS) causes diverse infections in humans ranging from colonization and uncomplicated pharyngeal and skin infections to necrotizing fasciitis and toxic shock syndrome (12). The diversity in routes and manifestations suggests that GAS colonization and infection involve complex regulatory networks that are differentially regulated in distinct environments (13). In fact, recent genomewide investigations of GAS gene expression have demonstrated that GAS responds to different environments by altering the transcription of genes involved in meeting basic metabolic demands and differential transcription of genes encoding major virulence factors (14–17). These studies have resulted in a new understanding of the relationship between metabolism and virulence in GAS. Here, we report the results of studies that extend this understanding to the molecular level.
Results
Comparison of GAS Gene Transcript Levels in Saliva and a Nutrient-Rich Medium.
Genomewide transcriptome analyses have suggested that differences exist in GAS gene expression during interaction with saliva and the oropharynx compared with growth in laboratory media, but no direct comparison has been done (15, 17). We used real-time TaqMan quantitative reverse transcription (QRT) PCR to test the hypothesis that GAS gene transcript levels differ significantly during growth in human saliva, a major component of innate and acquired immunity in the oropharynx, compared with growth in Todd–Hewitt broth with yeast extract (THY). We measured the transcript levels of 78 GAS genes encoding transcription regulators or proteins with either a known or putative extracellular location because of the likelihood such genes are involved in host–pathogen interaction [see supporting information (SI) Table 1]. Fifty-nine of the 78 genes had at least a twofold significantly different transcript level between the two media for at least one of the time points measured (select genes are shown in SI Fig. 7 with gene functions available in SI Table 1). Differences in gene transcript levels between saliva and THY media were especially prominent in the early-exponential growth phase, as 50 genes had significantly different transcript levels at the time point studied. A key finding was that, at the early time point, we observed ≈10-fold increase in the ccpA transcript level in saliva compared with THY. Together, the data show that GAS markedly alters its transcript profile in response to human saliva and suggest that CcpA mediates some of the observed differences.
Inactivation of the ccpA Gene Results in Medium-Specific Growth Defects.
To test the hypothesis that CcpA directly mediates some of the observed transcript differences we created isogenic mutant strain ΔccpA from wild-type serotype M1 strain MGAS5005 (confirmatory Southern blot shown in SI Fig. 8). We genetically complemented the ΔccpA isogenic mutant strain by using a plasmid capable of replicating in GAS to make strain compΔccpA. The growth curves for the three strains in THY were superimposable (SI Table 2 and SI Fig. 9). Conversely, compared with wild-type strain MGAS5005, the ΔccpA mutant strain had a prolonged lag phase in a glucose medium and an increased (slower) doubling time in a maltose medium (Fig. 9 B and C). Moreover, the ΔccpA strain did not reach as high a cell density or maintain as many viable colony-forming units (CFUs) in human saliva as wild-type strain MGAS5005 (Fig. 9D). Gram stain of the ΔccpA isogenic mutant strain grown in human saliva did not reveal enhanced clumping or increased chain length, two possibilities to explain the observed CFU differences (data not shown). There was no significant difference in growth between strain MGAS5005 and the compΔccpA strain in any of the media tested.
The ΔccpA Isogenic Mutant Strain Has an Altered Colony Morphology Associated with Differential Expression of pel/sagA and hasA Transcripts.
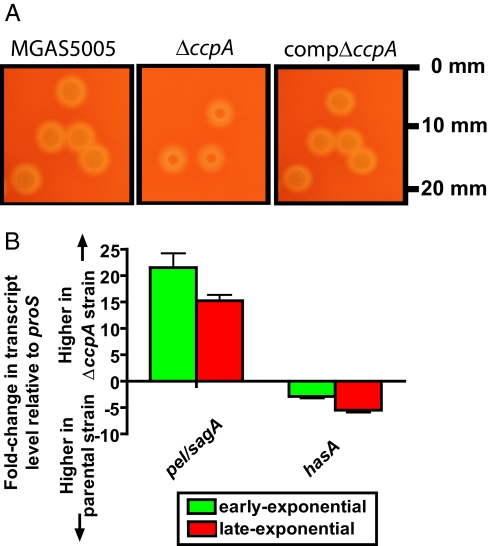
The ΔccpA mutant strain had altered colony morphology and hemolytic phenotype compared with wild-type strain MGAS5005 when grown on blood agar plates (Fig. 1A). The colony diameter (1.7 ± 0.1 mm) of the ΔccpA strain was significantly less than that of wild-type strain MGAS5005 (2.6 ± 0.3 mm) or the compΔccpA strain (2.4 ± 0.4 mm, P < 0.001). The observed difference in colony diameter was not caused by a reduced number of bacterial cells, because there was no significant difference in CFUs within colonies of the three strains (1.45 ± 0.3 × 109, 1.21 ± 0.51 × 109, 1.33 ± 0.49 × 109 for strain MGAS5005, the ΔccpA strain, and compΔccpA strains, respectively, P = 0.461). In addition, the radius of the zone of β-hemolysis surrounding the ΔccpA mutant strain (1.7 ± 0.1 mm) was significantly larger than the zone around wild-type strain MGAS5005 (0.4 ± 0.1 mm) and the compΔccpA strain (0.6 mm ± 0.2 mm, P = 0.005).
Fig. 1.
CcpA influences streptolysins and hyalouronic acid capsule expression. (A) Colony phenotype of strain MGAS5005, its ΔccpA mutant derivative strain, and compΔccpA strain on sheep blood agar plates. The ΔccpA strain has significantly increased hemolysis and decreased colony size compared with strain MGAS5005 and strain compΔccpA. All three images are of same magnification with size indications at right. (B) Changes in colony hemolysis and size were associated with altered pel/sagA and hasA gene transcript levels. Strain MGAS5005 and ΔccpA were grown in THY to early- and late-logarithmic growth phase, and pel/sagA and hasA transcript levels were measured by real-time TaqMan QRT-PCR with the ΔΔCT method. Values above the x axis indicate higher gene transcript levels in the ΔccpA strain, whereas values below the x axis indicate higher gene transcript levels in strain MGAS5005. Error bars indicate standard deviation among quadruplicate samples done on two separate occasions.
The β-hemolytic properties and colony morphology of GAS are determined in part by streptolysin S activity and capsule size, respectively (18, 19). Thus, we used real-time TaqMan QRT-PCR to test the hypothesis that the ΔccpA strain had higher levels of sagA transcript, the first gene in a nine-gene operon responsible for streptolysin S production, and lower transcript levels of hasA, the first gene in a three-gene operon of capsule synthesis genes. Consistent with the hypothesis, the level of sagA transcript was increased ≈20-fold in the ΔccpA mutant strain compared with wild-type strain MGAS5005 (Fig. 1B). Moreover, the hasA transcript was significantly decreased in the ΔccpA mutant strain compared with its parental strain at both time points. Although not studied in strain MGAS5005, in another GAS strain the sagA gene is transcribed as part of an RNA molecule with regulatory activity known as the pleiotropic effect locus (pel) (20). Based on QRT-PCR analysis, we found no significant difference in the transcript level of pel and sagA, consistent with the hypothesis that sagA is part of the larger pel transcript in strain MGAS5005 (data not shown). There was no significant difference in pel/sagA or hasA transcript level between the wild-type and compΔccpA strains (data not shown). Therefore, we conclude that CcpA directly or indirectly influences the transcript levels of GAS genes encoding critical virulence factors.
Analysis of the CcpA Transcriptome.
To test the hypothesis that CcpA influences a broader array of GAS genes, we next compared the genomewide transcriptome of wild-type strain MGAS5005 and the ΔccpA isogenic mutant strain grown in THY. After accounting for multiple comparisons, in the early-exponential growth phase the level of transcripts of ≈20% of all ORFs differed between the wild-type and ΔccpA isogenic mutant strain (Fig. 2A, SI Table 3). In the late-exponential phase ≈10% of all ORFs were differentially expressed.
Fig. 2.
Analysis of the CcpA transcriptome. Strain MGAS5005 and ΔccpA were grown to early- and late-logarithmic growth phase in THY, and expression microarray analysis was performed as described in Materials and Methods. Values above the x axis indicate higher gene transcript levels in the ΔccpA strain and values below the x axis indicate higher gene transcript levels in wild-type strain MGAS5005. (A) Summary of genes with altered transcript levels grouped by functional category. Percentage numbers refer to genes affected by CcpA inactivation/total number of genes in that particular category in the entire GAS genome. (B) Genes encoding carbohydrate utilization proteins. Shown are the mean fold changes in transcript levels of genes in an operon known or putatively involved in the transport and/or metabolism of the indicated carbohydrate. The transcript levels of all genes in the indicated operons were similarly affected. (C) Mean transcript levels of genes encoding select virulence factors. For B and C, error bars show standard deviation among four samples.
The largest number of transcripts differentially expressed between the two isogenic strains encode proteins known to be or putatively involved in carbohydrate transport and metabolism (Fig. 2B, SI Table 3). The differentially expressed genes included ATP-binding cassette (ABC) transporters or PTS operons responsible for glucose, lactose, maltodextrin, mannose, fructose, cellobiose, lactose, galactose, tagatose, and sialic acid transport. The transcript levels of genes in all of the aforementioned operons were increased in the ΔccpA strain except for the maltodextrin operon. The decreased level of the maltodextrin operon in the ΔccpA strain is in accord with the decreased growth of the ΔccpA strain in a maltodextrin medium (SI Fig. 9C). We also observed significant differences in transcript levels of 21 genes encoding known and putative transcriptional regulators. Six of these genes comprise three two-component (TCS) gene regulatory systems, including M5005_spy0784/5, M5005_spy1305/6, and sptR/sptS. M5005_spy0784/5 positively influences a putative mannose/fructose phosphotransferase system, whereas sptR/sptS affects carbohydrate metabolism and virulence factor production in human saliva (15, 21).
A third major category of genes affected in the ΔccpA mutant strain included those encoding proteins putatively or known to be involved in GAS virulence. As predicted from QRT-PCR data, the transcript levels of the nine-gene operon encoding streptolysin S were significantly elevated in the ΔccpA strain, whereas the entire hasABC operon involved in capsule synthesis was significantly down-regulated in the ΔccpA strain (Fig. 2C). Other virulence factors affected by ccpA inactivation included spd, which encodes an extracellular DNase, and endoS, which encodes a protein that cleaves human Ig (22, 23). No significant difference in transcript levels was observed for emm, which encodes the anti-opsonophagocytic M protein, or mga, which encodes a transcriptional regulator involved in up-regulation of several virulence factors (Fig. 2C). Taken together, we conclude that CcpA is a global regulator of carbohydrate metabolism in GAS and has important effects on regulation of genes encoding transcriptional regulators and major virulence factors.
CcpA Contributes to GAS Virulence and Ability to Colonize the Mouse Oropharynx.
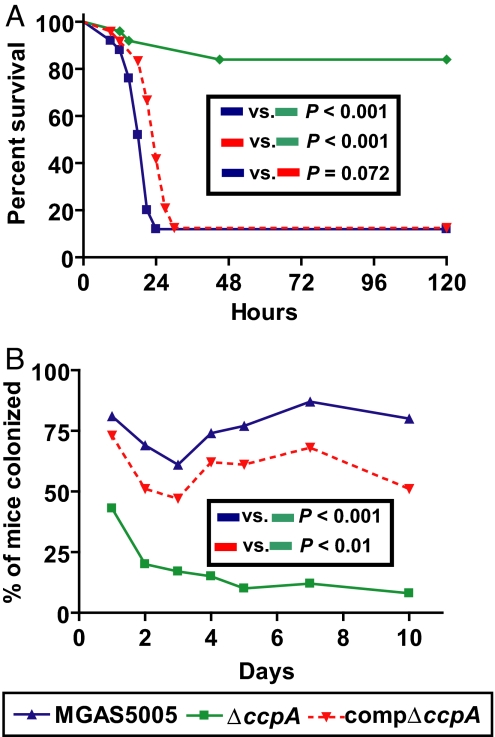
In light of our findings that CcpA affects the transcript levels of multiple GAS virulence factors, we next tested the hypothesis that the ΔccpA isogenic mutant strain was less virulent for mice than for the wild-type parental strain. Consistent with the hypothesis, significantly more mice inoculated i.p. with wild-type strain MGAS5005 died than with the ΔccpA isogenic mutant strain (P < 0.001; Fig. 3A). Similarly, the compΔccpA strain was significantly more virulent than the ΔccpA strain (P < 0.001) and as virulent as strain MGAS5005 (P = 0.072).
Fig. 3.
Inactivation of CcpA significantly decreases GAS virulence. (A) Invasive disease model. Twenty-five adult outbred CD-1 mice per group were inoculated i.p. with ≈1 × 107 CFU of the indicated strains. Percent survival is graphed with P values for Kaplan–Meier survival analysis. (B) Oropharyngeal colonization model. Adult outbred CD-1 mice (35 per group) were inoculated intranasally with ≈1.0 × 107 CFU of the indicated strains. Mice oropharynxes were swabbed daily. Percentage of mice with GAS isolated by day with P values shown for repeated measures analysis.
Next, we tested the hypothesis that ccpA contributes to the ability of GAS to colonize the mouse oropharynx. As hypothesized, after intranasal inoculation, the percentage of mice colonized over time with strain MGAS5005 was significantly greater than the ΔccpA mutant strain (P < 0.001; Fig. 3B). Similarly, the average number of CFUs recovered from mice inoculated with strain MGAS5005 was significantly greater than with the ΔccpA mutant strain (P < 0.001; data not shown). The compΔccpA strain was recovered at significantly higher CFUs and from more mice compared with the ΔccpA strain (P < 0.01 for both). Taken together, these data indicate that CcpA contributes to GAS virulence and ability to colonize the mouse oropharynx.
CcpA Affects the Transcript Levels of GAS Virulence Factors During Growth in Human Saliva.
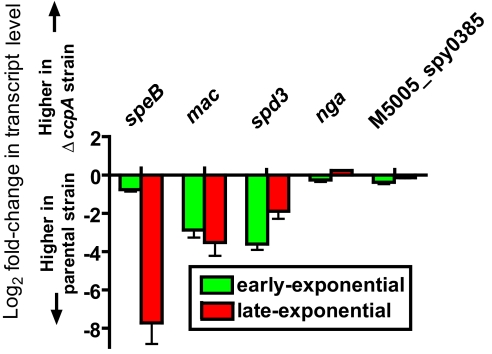
We next sought to determine whether, under glucose-limiting conditions, CcpA influences the transcript levels of genes encoding virulence factors not influenced by CcpA in glucose-rich conditions (e.g., THY). To this end, we tested the transcript levels of several GAS virulence factors in strain MGAS5005 and the ΔccpA strain during growth in human saliva, a glucose-poor medium. The transcript levels of speB (encoding a cysteine protease), mac (encoding an Ig-degrading enzyme), and spd3 (encoding a DNase) were significantly higher in strain MGAS5005 than in the ΔccpA strain during growth in human saliva (Fig. 4). These data demonstrate that CcpA is needed for GAS to respond to human saliva by increasing transcript levels of virulence factors shown to affect the ability of GAS to persist in human saliva (speB) and cause pharyngitis in a non-human primate model (spd3) (23, 24).
Fig. 4.
CcpA affects GAS virulence factor production in glucose-limited medium. Gene transcript levels were measured by TaqMan real-time QRT-PCR in strains MGAS5005 and ΔccpA grown to early- and late-logarithmic growth phase in human saliva. Bars indicate log2 differences in gene transcript levels between the two strains with error bars showing standard deviation for quadruplicate samples done on two separate occasions. Values above the x axis indicate higher gene transcript levels in the ΔccpA strain and values below the x axis indicate higher gene transcript levels in strain MGAS5005.
Recombinant GAS CcpA Binds to the Streptolysin S Promoter Region.
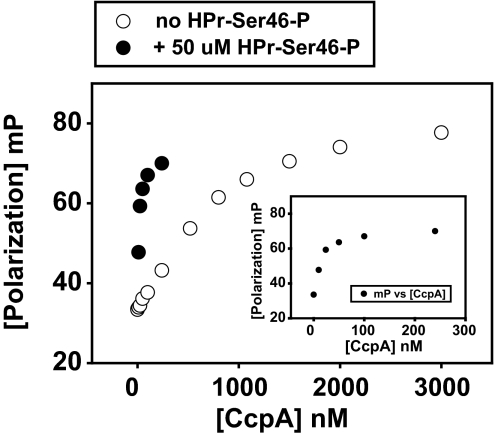
When complexed with its coeffector HPr-Ser-46-P, CcpA functions by binding to cognate DNA sites (cre) in the promoter region or within target genes in B. subtilis (25, 26). A bioinformatic analysis of the genome of strain MGAS5005 identified 37 sites that contain a B. subtilis consensus cre sequence (TGWAANCGNTNWCA; see SI Text and SI Table 4). To elucidate whether CcpA exerts its regulatory effect on virulence factor production by direct DNA interaction, we analyzed the binding of purified recombinant GAS CcpA (SI Fig. 10) to DNA sequences in the promoter region of three different genes by fluorescence polarization: (i) pel/sagA, a virulence gene whose expression was repressed by CcpA in our transcriptome analysis; (ii) lctO, which contains a consensus cre sequence and was differentially transcribed between strain MGAS5005 and the ΔccpA mutant strain and thus served as a positive control; and (iii) ftsX, a gene that does not contain a consensus cre sequence, was not differentially expressed in the transcriptome analysis, and therefore was chosen as a negative control (SI Table 5). GAS CcpA bound specifically to the pel/sagA DNA (Fig. 5). The binding affinity was specific (Kd = 950 ± 98 nM, assuming a 100% active CcpA) and enhanced ≈65-fold by the presence of 50 μM HPr-Ser-46-P (Kd = 14.5 ± 2.2 nM), further supporting the notion that CcpA binds to the pel/sagA promoter in vivo and controls its transcription. Nonphosphorylated HPr did not increase the CcpA–DNA interaction (data not shown). Similar results were found for lctO, and, as expected, specific binding was not observed for the promoter region of ftsX (SI Fig. 11). Thus, binding of CcpA to the promoter region of pel/sagA demonstrates a direct mechanism of CcpA regulation of a key GAS virulence factor.
Fig. 5.
The CcpA–(HPr-Ser-46-P) complex interacts specifically with the promoter region of pel/sagA. Purified GAS CcpA was titrated to 1 nM pel/sagA cre in the absence (open circles) and presence (closed circles) of 50 μM HPr-Ser-46-P. Millipolarization units (mP) are plotted against the CcpA concentration. The shift to the left in the binding curve demonstrates stimulation of DNA–CcpA complex formation by HPr-Ser-46P (KDs for DNA interaction are 948 ± 89 nM and 14.5 ± 2.2 nM with and without HPr-Ser-46P, respectively). (Inset) Shown is relationship between CcpA concentration and mP units in the presence of HPr-Ser-46-P to clarify that saturation is already reached at a CcpA concentration of 240 nM.
Discussion
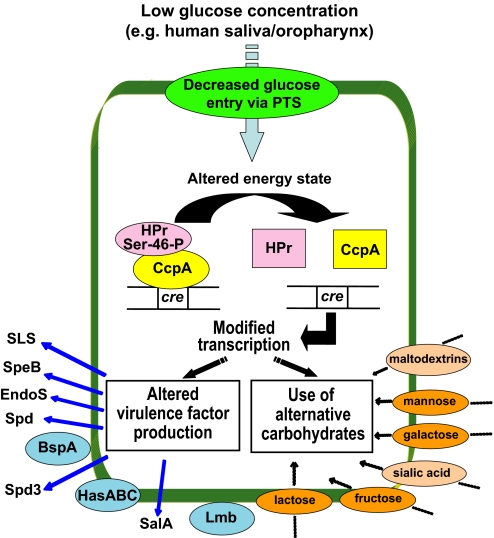
Although CcpA orthologs have been investigated in other Gram-positive organisms, before this study the CcpA transcriptome had not been determined in a human pathogen (8, 9, 27, 28). A key discovery was that CcpA influenced the transcript levels of several GAS virulence factors, including the potent cytolysin streptolysin S, the extracellular DNase Spd, and the Ig-degrading EndoS. In Bacillus spp., transport of glucose or other readily metabolized carbohydrates through the PTS system leads to phosphorylation of the phosphocarrier protein HPr at residue Ser-46, resulting in HPr-Ser-46-P (6). HPr-Ser-46-P serves as a protein coeffector for CcpA that mediates CcpA binding to cre sites, which ultimately results in altered gene expression (26). Strain MGAS5005 contains a gene (M5005_spy1121) with 66% identity and 82% similarity at the amino acid level to HPr in Bacillus subtilis subsp. subtilis strain 168, suggesting that similar CcpA pathways are likely to function in GAS. Our GAS data suggest a model that, when environmental glucose levels are low, such as in the human oropharynx and saliva, CcpA-(HPr-Ser-46-P)-cre interaction is relieved, thus allowing transcription of carbohydrate utilization genes and virulence factors vital to host–pathogen interaction (Fig. 6).
Fig. 6.
Hypothesis explaining how CcpA affects GAS carbohydrate utilization and virulence factor production. Decreased extracellular glucose levels result in dephosphorylation of HPr-Ser-46-P leading to dissociation of HPr and CcpA. CcpA/HPr dissociation results in changes in CcpA binding to DNA catabolite response element sites (cre) and altered transcription. Systems involved in the uptake and utilization of specific carbohydrates are shown on the right side of the figure. Phosphotransferase systems are shown in orange and ATP-binding cassette transport systems are in tan. On the left side of the figure, in blue, are virulence factors with the relationship of the protein names to the cell surface corresponding to their inferred location (e.g., actively secreted, anchored to the cell wall, or embedded in the cell membrane). Putative or known functions of affected virulence factors: SLS, cytolysin; SpeB, cysteine protease; EndoS, Ig cleavage; Spd and Spd3, DNases; BspA, epithelial cell-binding protein; HasABC, hyaluronic acid capsule production; SalA, salivaricin production; Lmb, laminin binding protein.
Although CcpA has been shown to influence virulence factor production in other Gram-positive pathogens, there is no information available regarding the mechanism by which this occurs (8, 27, 28). Our finding that CcpA binds to the pel/sagA promoter region demonstrates a direct link in GAS between environmental carbohydrate concentrations and virulence factor production. In light of the known lytic effect of streptolysin S on neutrophils, the data suggest that the nutritional status of GAS may serve as a mechanism for sensing when key virulence factors are needed to inhibit host defenses (29). Alternatively, streptolysin S may have a unappreciated role in nutrient acquisition. Because sagA is transcribed as part of the larger regulatory RNA pel, our data demonstrating that CcpA affects transcript levels of an RNA regulatory system are in accordance with a study in Staphylococcus aureus in which CcpA influenced transcript levels of RNAIII, the RNA effector molecule of the agr system (8, 20). Taken together, our findings substantiate the idea that in Gram-positive pathogens CcpA interacts with RNA regulatory systems, the importance of which are being increasingly appreciated in both humans and microbes.
Finally, we also discovered that CcpA positively influences the transcript level of several virulence factors, including speB, during growth in the nutrient-limited medium of human saliva. Given that human saliva is the first substance with which GAS interacts in the oropharynx, these data strongly suggest that GAS CcpA is required for the up-regulation of key virulence factors during the initial stages of host–pathogen interaction (24). The absence of increased production of key virulence factors early during pharyngeal infection may explain the relative inability of the ΔccpA mutant strain to colonize the oropharynx. Thus, our data demonstrate the GAS CcpA directly represses production of virulence factors, such as streptolysin S, under nutrient-rich conditions and augments production of other virulence factors, such as SpeB, under nutrient-limited conditions, thereby providing a key mechanism by which GAS responds to changing environments (Fig. 6).
Human pathogenic microbes differentially regulate production of key virulence factors in vivo, a hallmark of pathogen–host interaction. We have discovered that the major human pathogen GAS modulates virulence factor production required for survival and infectivity by a CcpA-mediated pathway. Given the highly conserved nature of CcpA, similar genomewide studies of other Gram-positive pathogens may yield enhanced understanding of links between basic metabolic processes and pathogenesis.
Materials and Methods
Bacterial Strains and Culture Media.
Serotype M1 strain MGAS5005 is genetically representative of the clone responsible for most contemporary (post-1987) human infections; its genome has been sequenced (30). The ΔccpA isogenic mutant strain was created from parental serotype M1 strain MGAS5005 by nonpolar insertional mutagenesis. We used pDC123, a plasmid capable of replicating in GAS, to genetically complement the ΔccpA strain in trans to create the compΔccpA strain (SI Text). Ultrapure carbohydrates (Sigma) were added at a concentration of 0.5% (wt/vol) to a carbohydrate-free preparation of a commercially available, chemically defined medium (CDM; SAFC Biosciences) to create glucose medium, maltose medium, etc. Growth in human saliva was performed with specimens collected from healthy adult volunteers under a protocol approved by the Baylor College of Medicine Institutional Review Board (24).
RNA Isolation, TaqMan Transcript Level Analysis, and Expression Microarray Analysis.
RNA was purified from a minimum of four replicate cultures by using an RNeasy kit (Qiagen). The concentration and quality of RNA were assessed with an Agilent 2100 Bioanalyzer and analysis of the A260/A280 ratio. Select gene transcript level analysis was performed with TaqMan real-time QRT-PCR (primers and probes listed in SI Table 6). A custom-made Affymetrix GeneChip that contains 100% of the ORFs of strain MGAS5005 was used for expression microarray (transcriptome) studies. Principal component analysis (PCA) indicated that the data were of high quality and that the two time points provided distinct information regarding GAS gene transcripts differentially expressed between these two strains (SI Fig. 12). To compare gene transcript levels between the wild-type and mutant strain, a two-sample t test (unequal variance) was applied, followed by a false discovery rate correction (Q < 0.05) to account for multiple testing. Genes were considered differentially transcribed if the t test had a corrected P value of <0.05 and the difference in mean gene transcript level was at least 2-fold. For further information on assessment of transcript levels see SI Text.
Mouse Virulence Studies.
Mouse experiments were performed according to protocols approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. For the invasive disease model, 25 female outbred CD-1 Swiss mice (Harlan–Sprague–Dawley) were injected i.p. with ≈2.5 × 107 GAS CFU (31). Throat colonization studies were performed by inoculating 35 mice in each group with 1 × 107 CFU of GAS (32). The throat of each mouse was swabbed before inoculation and then daily thereafter.
Purification and Binding Characteristics of CcpA.
CcpA was purified to homogeneity from Escherichia coli (SI Fig. 10, SI Text). Fluorescence anisotropy was used to determine the binding characteristics of CcpA to 5′-fluorescein-labeled oligonucleotides in the presence and absence of 50 μM HPr-Ser-46-P (33, 34; SI Text).
Supplementary Material
ACKNOWLEDGMENTS.
We thank B. Lei for providing the vector used to overexpress CcpA. This work was supported by American Heart Association Grant 0565133Y (to S.A.S), National Institute of Allergy and Infectious Diseases K08 Career Development Award AI-064564 (to S.A.S.), and funds from the Robert A Welch Foundation G-0040 (R.G.B).
Footnotes
The authors declare no conflict of interest.
Data deposition: Expression microarray data have been deposited in the Gene Expression Omnibus (GEO) database at NCBI, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE10156).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711767105/DC1.
References
- 1.Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006;25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 2007;64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- 3.Larocque RC, et al. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect Immun. 2005;73:4488–4493. doi: 10.1128/IAI.73.8.4488-4493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH., Jr Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 6.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 8.Seidl K, et al. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother. 2006;50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 12.Tart AH, Walker MJ, Musser JM. New understanding of the group A Streptococcus pathogenesis cycle. Trends Microbiol. 2007;15:318–325. doi: 10.1016/j.tim.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 14.Malke H, Steiner K, McShan WM, Ferretti JJ. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: The action of CodY and RelA. Int J Med Microbiol. 2006;296:259–275. doi: 10.1016/j.ijmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Shelburne SA, et al. Central role of a two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA. 2005;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho KH, Caparon MG. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol. 2005;57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- 17.Virtaneva K, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nizet V, et al. Genetic locus for streptolysin S production by group A Streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold M, et al. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol. 2004;53:1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- 21.Sitkiewicz I, Musser JM. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group A Streptococcus. Infect Immun. 2006;74:1339–1351. doi: 10.1128/IAI.74.2.1339-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collin M, Olsen A. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 2001;20:3046–3055. doi: 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumby P, et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shelburne SA, III, et al. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 2005;73:4723–4731. doi: 10.1128/IAI.73.8.4723-4731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev. 2003;67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher MA, et al. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell. 2004;118:731–741. doi: 10.1016/j.cell.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 27.Giammarinaro P, Paton JC. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect Immun. 2002;70:5454–5461. doi: 10.1128/IAI.70.10.5454-5461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varga J, Stirewalt VL, Melville SB. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J Bacteriol. 2004;186:5221–5229. doi: 10.1128/JB.186.16.5221-5229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyoshi-Akiyama T, et al. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J Infect Dis. 2005;192:107–116. doi: 10.1086/430617. [DOI] [PubMed] [Google Scholar]
- 30.Sumby P, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 31.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelburne SA, III, et al. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect Immun. 2006;74:4605–4614. doi: 10.1128/IAI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kliegman JI, Griner SL, Helmann JD, Brennan RG, Glasfeld A. Structural basis for the metal-selective activation of the manganese transport regulator of Bacillus subtilis. Biochemistry. 2006;45:3493–3505. doi: 10.1021/bi0524215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.