Abstract
The maize Hm1 gene provides protection against a lethal leaf blight and ear mold disease caused by Cochliobolus carbonum race 1 (CCR1). Although it was the first disease-resistance (DR) gene to be cloned, it remains a novelty because, instead of participating in the plant recognition and response system as most DR genes do, Hm1 disarms the pathogen directly. It does so by encoding an NADPH-dependent reductase, whose function is to inactivate Helminthosporium carbonum (HC) toxin, an epoxide-containing cyclic tetrapeptide, which the pathogen produces as a key virulence factor to colonize maize. Although CCR1 is strictly a pathogen of maize, orthologs of Hm1 and the HC-toxin reductase activity are present in the grass family, suggesting an ancient and evolutionarily conserved role of this DR trait in plants. Here, we provide proof for such a role by demonstrating its involvement in nonhost resistance of barley to CCR1. Barley leaves in which expression of the Hm1 homologue was silenced became susceptible to infection by CCR1, but only if the pathogen was able to produce HC toxin. Phylogenetic analysis indicated that Hm1 evolved exclusively and early in the grass lineage. Given the devastating ability of CCR1 to kill maize, these findings imply that the evolution and/or geographical distribution of grasses may have been constrained if Hm1 did not emerge.
Keywords: host–pathogen specificity, nonhost resistance, host-specific toxin, Cochliobolus, Helminthosporium carbonum toxin reductase
An important goal of research on disease is to establish how specificity, a hallmark of most infectious diseases, is established. A given pathogen normally can cause disease in only a single or limited number of species, while the species outside of its host range are immune to it. Although it remains largely unknown how such specificity is established in most plant diseases, one exception is provided by fungal pathogens that use host-specific (host-selective) toxins (HSTs) as key mediators of pathogenicity (1, 2). Most HSTs are low-molecular-weight fungal metabolites that inflict damage on only those plants that are susceptible to the producing pathogen. HSTs facilitate disease development in a genotype-specific fashion, thus determining both the host range of the pathogen and the specificity of the disease. It is generally assumed that the presence of a corresponding molecular target in the host and not in nonhost species underlies all cases of HST selectivity (1, 2).
A classic example of a plant disease involving an HST is that of the maize leaf spot and ear mold (3). This lethal disease is caused by Cochliobolus carbonum race 1 (CCR1), a fungal ascomycete whose asexual form (anamorph) is known as Helminthosporium carbonum (HC) (synonym Bipolaris zeicola). CCR1 is among the most destructive pathogens of maize. It can kill susceptible maize plants at any stage of development (Fig. 1) (4). Unlike most other plant pathogens, CCR1 can invade every part of the host, causing blight of the leaves, rot of the roots and the stalk, and mold of the ear (Fig. 1 A–H).
Fig. 1.
Exceptional ability of CCR1 to invade and devastate susceptible maize. (a) A field of maize plants killed after infection with CCR1. (b) The same field ≈4 weeks earlier at the time of inoculation. (c–g) CCR1 can invade every part of the maize plant, causing spots on leaves (c), blight of the foliage (d), and rot of the roots (e), the stalk (f), and the ear (g). (h) Aerial hyphae are often produced on infected tissues if climate is warm and humid. (i) Structure of HC toxin. (j) Typical size and appearance of leaf lesions incited by CCR1 if it lacks the ability to produce HC toxin or the host harbors HCTR to degrade HC toxin.
The ability of CCR1 to cause disease depends on two conditions. The first requires the host to lack Hm1, a disease-resistance (DR) gene present almost ubiquitously in maize (5, 6). Second, CCR1 must produce HC toxin (3), an archetypal HST of the structure cyclo(d-Pro-l-Ala-d-Ala-l-2-amino-8-oxo-9,10-epoxy decanoic acid) (Fig. 1I) (7–10). Genetic variants of CCR1 that lack the ability to produce HC toxin are nonpathogenic and unable to invade beyond the penetration site (Fig. 1J). However, they do resume growth and colonization of the host tissue if HC toxin is administrated exogenously to the infection site (11). Maize lines that are resistant to CCR1 are tolerant of HC toxin compared with susceptible lines (11). On these resistant lines, CCR1 is contained at the infection site in the same fashion as the HC-toxin-deficient CCR1 does in a susceptible host (Fig. 1J).
Interestingly, the Hm1-based resistance mechanism that evolved naturally in maize targeted HC toxin for detoxification (12). It is mediated by an enzyme encoded by Hm1 (13). Named HC-toxin reductase (HCTR), this enzyme is an NADPH-dependent reductase that bears significant homology to dihydroflavonol reductase (DFR) involved in the biosynthesis of flavonoids and anthocyanins throughout the plant kingdom. In addition to the Hm1 gene, which confers complete protection in every part of the plant, certain lines of maize contain a second DR gene Hm2, which confers effective resistance only in adult plants (5, 6). The cloning of Hm2 has demonstrated that it encodes a structural, albeit truncated, duplicate of HM1 [supporting information (SI) Fig. 5] (36).
Like most pathogens of plants, CCR1 exhibits a high degree of host specificity and can cause disease only in maize. All other plant species, including those that are closely related to maize, are virtually immune to CCR1. Notwithstanding the absolute requirement of maize as a host for CCR1, all grass species tested possess candidate genes or sequences with high homology to Hm1. These include sorghum, rice, barley, wheat, rye, oats, millet, fescue, bluegrass, reed canarygrass, and bamboo (5, 14). In barley, rice, and sorghum, these homologs are syntenic with that of the maize Hm genes (5, 14), indicating that they are truly orthologous and derived from a common ancestor. HCTR activity has also been detected in all grasses tested, including barley, wheat, sorghum, rice, and oats, implying that these genes are not relics of the past but still maintained functionally in nonmaize cereals (10, 15). In contrast, no HCTR activity, nor the sequences that could be considered truly homologous to Hm1, has been detected outside of the grass lineage. This includes the model dicot Arabidopsis, whose genome has been sequenced fully. These results and observations suggest that the need to detoxify HC toxin probably arose only in grasses.
The widespread presence of HCTR-encoding sequences in the grass family raises the question: Why are functional homologs of Hm1 present and maintained in plant species that are outside of the host range of CCR1? One possibility is that HCTR performs another essential function, in addition to reducing HC toxin. In this scenario, the ability of HCTR to inactivate HC toxin would only be incidental, happening entirely by serendipity. No evidence exists yet to support such a role for HCTR in plants, although it was reported recently that a rice homolog of Hm1 was able to protect plants from multiple stresses when overexpressed ectopically as a transgene (16, 17). An alternative hypothesis for the ubiquitous presence of Hm1 in all grasses is that they all perform the same function as the maize Hm1 gene and serve to guard their hosts against a pathogen such as CCR1. The results presented here support such a role for Hm1 and show that that the HCTR function evolved exclusively to contend with HC toxin.
Results and Discussion
Conserved Role of Hm1 in Barley.
To address the possibility that the threat imposed by CCR1 is responsible for the conserved maintenance of HCTR across all cereals, we took advantage of barley as an experimental system. This decision was dictated by two criteria. First, the barley Hm1 homologs were already cloned and characterized, at least from the line Morex, which was found to harbor a tandemly duplicated pair of transcriptionally active Hm1 genes (14). Second, virus-induced gene silencing (VIGS) has been developed as a reverse genetics tool for functional characterization of genes in this cereal (18), including those involved in DR (19). The tripartite genome of the barley stripe mosaic virus (BSMV) has been harnessed as a vector to implement VIGS in this cereal (18). The line that responds the best to VIGS in barley is Black Hulless. To examine whether it contains one or more copies of Hm1, sequences corresponding to this gene were PCR-amplified and sequenced. The results obtained showed that Hm1 exists as a single copy gene in Black Hulless, which, like the Morex homologs, encodes a peptide that is 72% identical and 85% similar to that of maize HM1 (14) (SI Fig. 6).
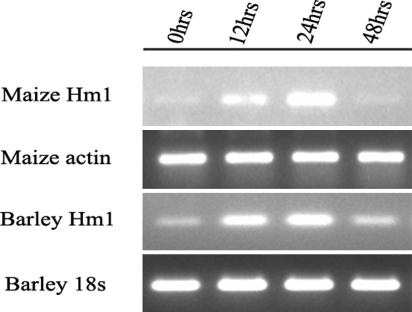
The transcriptional behavior of the barley Hm1 gene was examined in response to inoculation with CCR1 and compared with that of the maize Hm1 gene (Fig. 2). Interestingly, both the maize and barley genes behave in a similar manner in response to infection by CCR1. Although both are constitutively expressed at very low levels in seedling leaves, their transcript levels rapidly and transiently increase many fold in response to infection. This increase is detectable by 12 h after infection (hai), reaches peak levels ≈36 hai, and begins to decline by 48 hai. Because the transcriptional activity of a gene often corresponds to its biological function, these results strengthened the hypothesis that the function of the barley Hm1 was the same as that of the maize Hm1, i.e., to negate the disease-inducing ability of HC toxin.
Fig. 2.
Expression of the maize and barley Hm1 genes in response to CCR1 infection. RT-PCR amplification of total RNA isolated from maize and barley seedlings was conducted at different times after inoculation with CCR1. The controls (maize actin and barley 18S genes) show equal amplification in all samples.
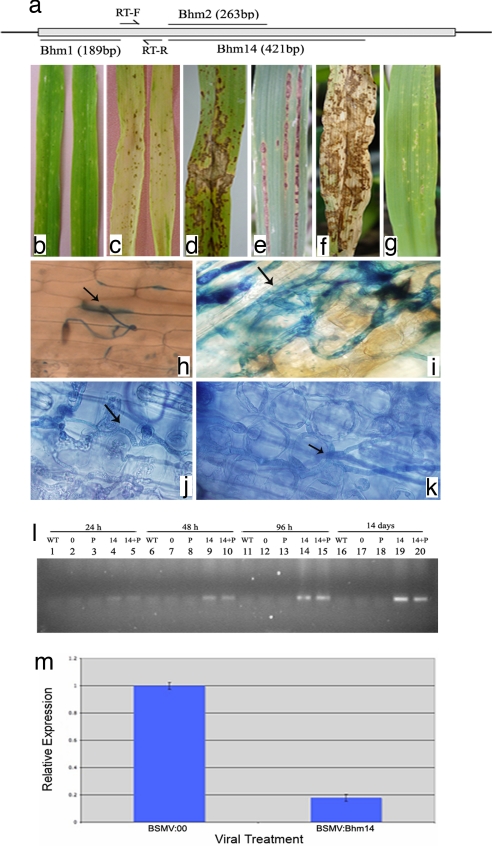
To silence the barley Hm1 by VIGS, three separate constructs were made containing different parts of the barley Hm1 gene in the BSMV vector (Fig. 3a). These constructs were used individually and in combination with the phytoene desaturase (Pds) gene to infect leaves of 7-day-old barley seedlings. Silencing of Pds, which results in photobleaching of the affected tissue (18, 20), was used as a visible marker for VIGS. Plants infected with the Pds construct alone (BSMV:Pds) or an empty vector (BSMV:00) served as controls.
Fig. 3.
VIGS-mediated suppression of Hm1 results in barley susceptibility to CCR1. (a) Schematic representation showing the size and relative locations of barley Hm1 fragments designed to produce VIGS vectors. (b and c) Typical appearance of lesions caused by CCR1 on normal barley leaves (b) or leaves in which the Pds gene is suppressed via VIGS (c). (d–f) CCR1-caused expanding lesions on barley leaves in which Hm1 was suppressed alone (d and e) or in combination with Pds (f). (g) Resistant lesions of Hm1-suppressed barley to an HC toxin-deficient isolate of CCR1. (h–k) Microscopic demonstration showing that while CCR1 fails to grow beyond the site of penetration in normal barley leaves (h), it is able to grow and invade substantially inside the Hm1-suppressed barley leaves (i–k). Arrows indicate an appressorium (h) and fungal hyphae (i–k) ramifying through the barley leaf tissue. (l) Increase in CCR1 biomass over time in Hm1-suppressed barley leaves as demonstrated by PCR amplification of its 5.8S rRNA ITS sequence. WT indicates no viral infection; others denote infection with BSMV:00 (0); BSMV:Pds (P); BSMV:Bhm14 (14); and BSMV:Pds plus BSMV:Bhm14 (14+P). (m) Real-time RT-PCR of barley Hm1 transcripts after infection of WT and Hm1-suppressed leaves with CCR1. (Magnification: h, ×20; i–k, ×100.)
A week after infection with BSMV vectors, the plants were challenged with either the wild-type (HC toxin-producing) or the HC toxin-nonproducing isolate of CCR1. Similar to the reaction of resistant maize, BSMV:00 plants inoculated with either isolate of CCR1 responded by producing minute chlorotic flecks or lesions at the site of infection (Fig. 3b). Microscopic examination of the interaction revealed that pathogen behavior and growth were similar. For instance, the fungus germinated and produced appressoria within 8–10 h after inoculation, allowing the pathogen to directly penetrate epidermal cells (Fig. 3h). However, subsequent growth of the pathogen ceased, and the pathogen was contained within the chlorotic fleck. The BSMV:Pds plants responded similarly to pathogen inoculation, as did the empty vector controls, and restricted the pathogen to the infection site (Fig. 3c).
By contrast, plants whose Hm1 gene was silenced exhibited spreading disease lesions when their leaves were sprayed with the HC toxin-producing isolate of CCR1 (Fig. 3 d–f). These lesions continued to enlarge over time and coalesced to cause extensive tissue damage typical of symptoms associated with maize leaf blight (Figs. 3 d–f and 1c). Microscopic analysis indicated that the pathogen continued to invade after penetration and grew through barley epidermal and mesophyll cells as it does in maize (Fig. 3 i–k). This resulted in substantial growth of the pathogen, as is also evidenced by a PCR-based fungal mass analysis of CCR1 (Fig. 3l) (21). We were able to reisolate CCR1 from surface-sterilized barley leaves containing disease lesions, and these isolates retained full pathogenicity toward susceptible maize.
All three BSMV:Hm1 constructs were equally effective in suppressing barley resistance to CCR1, indicating that the VIGS studies are reproducible and that susceptibility to CCR1 is not a result of off target silencing. Thus, CCR1 has the ability to colonize barley, provided its Hm1 gene is silenced. But this happens only if the Hm1-silenced plants are challenged with an HC toxin-producing isolate of CCR1; HC toxin-deficient isolates fail to extend beyond the site of penetration regardless of whether Hm1 is silenced or not (Fig. 3g). These results are consistent with a specific role of the barley Hm1 homolog against HC toxin. To address whether the loss of resistance to CCR1 was, in fact, the result of down-regulation of the Hm1 gene, the expression of this gene was examined by real-time PCR in the silenced tissue after infection of barley leaves with BSMV:Bhm14. As expected, the expression of Hm1 significantly decreased in silenced leaves (Fig. 3m), providing a cause and effect relationship between the suppression of Hm1 and the induction of susceptibility to CCR1.
These results provide compelling evidence for the role of Hm1 in nonhost resistance of barley to CCR1. When the function of this gene is blocked, barley becomes susceptible to CCR1. This is essentially what happened in maize naturally. Like all plants, maize initially was not a host of CCR1. However, when mutant alleles of both Hm1 and Hm2 assorted together during inbred development, the resulting lines were susceptible to CCR1 (5). This led to the genesis of a new disease of maize by a pathogen that was not previously known to exist (4, 5, 22). The present study sheds light on this subject and implies that the breakdown of HCTR-based resistance in maize unveiled an ancient case of parasitism in which HC toxin played a decisive role. Although we may never know the identity of the pathogen that caused this disease of the past and forced the grass lineage to acquire Hm1, it is feasible that it was either CCR1 itself or its immediate ancestor. This notion is based on the fact that CCR1 is the only pathogen known to produce HC toxin (10), and it also exhibits the ability to colonize dead or senescing tissues of a wide range of grasses (23, 24).
Hm1 Evolved Exclusively in Grasses.
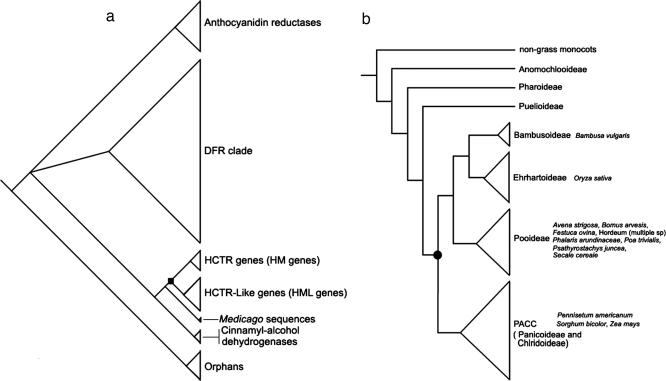
The conserved structure and function of Hm1 in maize and barley predict the existence of this DR gene in the grass family before its radiation into major cereals 50 million to 60 million years ago (25, 26). To gain an insight into the origin and subsequent evolution of Hm1, we constructed a large data set consisting of representatives of HCTR (Hm1 and Hm2), Hml, Dfr, Cad, and other related flavonoid biosynthesis genes from a range of land plant species (SI Fig. 7). A Bayesian phylogenetic analysis revealed the following about the evolutionary history of Hm1 and its paralogs or homeologs. First, Hm1 is an ancient gene preserved in all major grasses. It had a single (monophyletic) origin rather early in the evolution of the grass lineage (Fig. 4). Second, the HCTR gene family represents a distinct lineage separate from the Hml clade, both of which are present exclusively in grasses. This clearly contrasts with all other genes included in the analysis, which are represented widely throughout the plant kingdom. Third, Hm2 emerged specifically in the maize lineage probably as a result of a whole genome duplication event that occurred in this lineage after divergence from the sorghum lineage (Fig. 4).
Fig. 4.
Evolutionary origin of Hm1. (a) Summary phylogenetic tree, based on a Bayesian analysis of DNA sequences. This tree shows relationships between Dfr, Anthocyanidin reductases, Cad, HCTR (Hm1 and Hm2), and HCTR-like (Hml) genes. Both Monocot and eudicot sequences are found in the Dfr, Anthocyanidin reductases, and Cad clades. Two sequences from Medicago are sister to the HCTR and HCTR-like clades. The black square indicates gene duplication event resulting in the grass-specific HCTR and HCTR-like gene sequences. (b) Grass phylogeny [modified from Barker et al. (26)]. Listed are species for which HCTR genes have been sequenced or detected by Southern blot hybridization (14). The black dot indicates theb minimum point at which HCTR gene function may have originated, although the function may be found in all grasses.
Fourth, this analysis led to the identification of an additional Hm1-like (Hml) gene whose encoded peptide bears >70% identity with HM1 and HM2 (SI Fig. 5). Hm1 and Hml appear to have resulted from a duplication event before the radiation of major clades of grasses (Fig. 4) (26). The exact placement of this duplication event remains unresolved, as does the question of which evolved first, Hm1 or Hml. Interestingly, the Hml clade has undergone significant expansion in rice (SI Fig. 7). Hml maps to an area of the genome (chromosome 7S) that has never been shown to be associated with resistance to CCR1, making it unlikely to encode HCTR. The function of the Hml gene remains to be addressed.
Implications.
The maintenance of HCTR gene function in maize and barley, coupled with the unique phylogenetic position of the Hm1 gene (with no closely related orthologs in eudicots), suggests that Hm1 may have played a critical role in the evolution of most of our cereal crops. Given the devastating potential of CCR1 to kill susceptible corn, it is likely that this fungus or its ancestral form would have threatened the existence of grasses, or at least severely constrained their geographical distribution, had Hm1 not evolved to detoxify HC toxin. Thus, it seems likely that Hm1 served as a guardian of the grass family, allowing it to survive, thrive, and evolve into crops that feed the world.
In addition to revealing the antiquity and ubiquity of the Hm1-encoded DR function in grasses, this study has broad implications for the field of plant pathology. The first concerns the definition of HC toxin as an HST. Our results show that HC toxin is not a HST, as is traditionally assumed. Rather, it is the malfunction of Hm1 that renders HC toxin host-specific. Consistent with this idea are the findings that all plant histone deacetylases, which appear to be the targets of HC toxin's action, are sensitive to HC toxin (10, 27). However, this raises another question: Why are dicots that do not possess HCTR activity resistant to CCR1? Second, this study helps establish a paradigm for nonhost resistance in plants and shows that this kind of resistance, which often exists at the species level, could result from the operation of an active, evolutionarily conserved DR mechanism. In this regard, Hm1 can be viewed as a nonhost resistance gene that protects the entire grass family from CCR1. Finally, the success of Hm1 in keeping a deadly pathogen like CCR1 in check since the dawn of grasses provides a good example for designing similar protection strategies against other diseases.
Materials and Methods
Plant and Fungal Materials.
Maize and barley seedlings were grown and maintained in the greenhouse as described (5, 20). The maize line used for expression analysis was B73. Fungal strains were cultured and maintained as described (5).
VIGS Experiments.
Three separate fragments, Bhm1 (189 bp), Bhm2 (263 bp), and Bhm14 (431 bp), were used to silence the barley Hm1 gene. These were generated by PCR amplification from barley genomic DNA as follows: Bhm1 was amplified with the forward primer 5′-TCG AGG CAG GCT ACA CCG TCC-3′ and reverse primer 5′-TGG CGA CGA GGA AGA CGA AGT GG-3′. Bhm2 was amplified with the forward primer 5′-TCA TCT CTG AAT CTT GTT GGA CTC-3′ and reverse primer 5′-GAT CCT CGG CAG CAT GAA G-3′. Amplification of Bhm14 was accomplished by using the same forward primer as for Bhm2 but the reverse primer was 5′-TAC TTG CTG CCG TAG TGG TCC AAG-3′. All of these fragments were cloned into the BSMV vector as described (20). The silencing control construct (BSMV:Pds) contained an 185-bp fragment of the Pds gene (18, 20). In vitro transcription of viral RNAs and subsequent inoculation of 7-day-old barley seedlings was done as described (20). Challenge inoculations were carried out by spraying seedlings with either an HC toxin-producing or -nonproducing isolate of CCR1. Inoculated leaves were sampled daily for microscopic observations and stained with trypan blue in lactophenol before visualization (11). Pictures were taken with an Olympus BX41 with DP70 digital camera. For fungal biomass analysis, CCR1-specific primers and PCR protocols were used as described (21). All VIGS experiments involved a minimum of eight plants, and each experiment was performed at least three times.
RNA Analysis.
Total RNA from maize and barley leaves was extracted by using TRIZOL (Invitrogen) and treated with RNase-free DNase I (Promega). For analysis by RT-PCR, 0.2 μg of total RNA of each of maize and barley was reverse-transcribed to generate first-strand cDNA by using the one-step RT-PCR kit (Qiagen) in accordance with the manufacturer's instructions. PCR conditions were 94°C for 60 s, 60°C for 60 s, and 72°C for 60 s (34 cycles for the Hm1 genes and 28 cycles for the control genes). The primers Hm1F (5′-CGATCGCTGGGTGCCAGTTC-3′) and Hm1R (5′-TGAAGTCTCTGTACCCGACG-3′) were used to amplify the maize Hm1 transcript. The primers ActinF (5′-GCATCCTGACACTGAAGTAC-3′) and ActinR (5′-GATAGCAACATACATTGCTGG-3′) were used to amplify the maize Actin gene transcript as a control. The barley primers used to amplify the Hm1 transcript were RT-F (5′-TTC GTC TTC CTC GTC GCC AAC-3′) and RT-R (5′-CGT CCT AGA CTC CGC GCA TA-3′), and the 18S control primers were the same as before (20). Real-time RT-PCR quantification of the Barley Hm1 expression in VIGS-silenced leaves was performed at 36 h after CCR1 inoculation as described (20). The experiment was replicated three times, and each replicate contained an RNA pool of four plants infected with either BSM:00 or BSMV:Bhm14.
Molecular Cloning and Analysis.
DNA was extracted from maize seedlings as described (5). The maize Hm2 gene was cloned as overlapping PCR fragments from a maize tester homozygous for this allele by using the following primer sets: Hm2F1, 5′-TCGCAGAAACCGGATTAGTGGGTA-3′ and Hm2R1, 5′-TTTGTACCCATCGCCGGAACC-3′; Hm2F1, 5′-AGTGCCCTAGTCCATCGAGTAGCA-3′ and Hm2R2, 5′-TAGTGGTCCGTGATGTCGTGGATG-3′; Hm2F3, 5′-CCTTCGCAGCGCACACTTCGA-3′ and Hm2R3 5′-AGTGGTTCTGCTTGGTTGAAAGGA-3′. PCR conditions were as described (5). The PCR amplicons were subcloned into the pGem-T easy vector system (Invitrogen), and three separate clones were sequenced at the Purdue Genomic Facility. Hml was isolated from a genomic library of B73 as described (28). Hml was mapped to the maize chromosome 7 by using oat-maize addition lines DNA (29). The barley Hm1 homolog was cloned by RT-PCR from total RNA isolated from Black Hulless seedling as described above. The primers were: BHm1F (5′-CGATCGCTGGGTGCCAGTTC-3′) and BHm1R (5′-TGAAGTCTCTGTACCCGACG-3′).
Phylogenetic Analysis.
Sequences for phylogenetic analysis were obtained from GenBank and PlantGDB (30, 31). Bayesian phylogenetic analyses were conducted by using MrBayes version 3.1.2 (32, 33). MrModeltest v. 2.2, with Akaike Information Criterion and hierarchical likelihood ratio testing, was used to determine the best-fitting model of molecular evolution, GTR + I + Γ (34, 35). Two independent runs were done with MrBayes. Using random starting trees, Markov Chain Monte Carlo using three heated chains and one cold chain and a flat Dirichlet prior on nucleotide frequencies and relative rate parameters, the Bayesian analysis was run for 1,000,000 generations sampling every 100 generations, resulting in 10,000 trees. Chain stationarity was achieved after 154,500 generations. A total of 8,455 trees were used to create a 50% majority rule consensus tree; the percentage of times clades occurring in this sample of trees reflects clade posterior probability values.
Supplementary Material
ACKNOWLEDGMENTS.
We thank H. Ruess for the images for Fig. 1 a and b; R. Latin for access to his microscope; and P. Balint-Kurti, L. Dunkle, L. Johal, R. Nicholson, P. Virk, and C. Woloshuk for editorial and scientific comments on the manuscript. This work was supported by start-up funds from Purdue University and a National Science Foundation grant (to G.S.J.) and the U.S. Department of Agriculture-Agricultural Research Service Current Research Information System (Project 3602-21220-010-00) (S.R.S.). This is Purdue University Agricultural Research Program paper 2008-18270.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. EU367521).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711406105/DC1.
References
- 1.Wolpert TJ, Dunkle LD, Ciuffetti LM. Host-selective toxins and avirulence determinants: What's in a name? Annu Rev Phytopathol. 2002;40:251–285. doi: 10.1146/annurev.phyto.40.011402.114210. [DOI] [PubMed] [Google Scholar]
- 2.Walton JD. Host-selective toxins: Agents of compatibility. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheffer RP, Ullstrup AJ. Host-specific toxic metabolite from Helminthosporium carbonum. Phytopathology. 1965;55:1037–1038. [Google Scholar]
- 4.Ullstrup A. Two physiologic races of Helminthosporium maydis in the corn belt. Phytopathology. 1941;31:508–521. [Google Scholar]
- 5.Multani DS, et al. Plant-pathogen microevolution: Molecular basis for the origin of a fungal disease in maize. Proc Natl Acad Sci USA. 1998;95:1686–1691. doi: 10.1073/pnas.95.4.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson OE, Ullstrup AJ. Resistance to leaf spot in maize. J Hered. 1964;55:195–200. [Google Scholar]
- 7.Gross ML, et al. The structure of the toxin from Helminthosporium carbonum. Tetrahedron Lett. 1982;23:5381–5384. [Google Scholar]
- 8.Kawai M, Rich DH, Walton JD. The structure and conformation of HC-toxin. Biochem Biophys Res Commun. 1983;111:398–403. doi: 10.1016/0006-291x(83)90319-4. [DOI] [PubMed] [Google Scholar]
- 9.Liesch JMC, et al. Structure of HC-toxin, a cyclic tetrapeptide from Helminthosporium carbonum. Tetrahedron. 1982;38:45–48. [Google Scholar]
- 10.Walton JD. HC-toxin. Phytochemistry. 2006;67:1406–1413. doi: 10.1016/j.phytochem.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Comstock JC, Scheffer RP. Role of host-selective toxin in colonization of corn leaves by Helminthosporium carbonum. Phytopathology. 1973;63:24–29. [Google Scholar]
- 12.Johal GS, Briggs SP. Reductase activity encoded by the HM1 disease resistance gene in maize. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 13.Meeley RB, Johal GS, Briggs SP, Walton JD. A biochemical phenotype for a disease resistance gene of maize. Plant Cell. 1992;4:71–77. doi: 10.1105/tpc.4.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han F, Kleinhofs A, Kilian A, Ullrich SE. Cloning and mapping of a putative barley NADPH-dependent HC-toxin reductase. Mol Plant–Microbe Interact. 1997;10:234–239. doi: 10.1094/MPMI.1997.10.2.234. [DOI] [PubMed] [Google Scholar]
- 15.Meeley R, Walton J. In: Advances in Molecular Genetics of Plant Microbe Interactions. Nester EW, Verma DPS, editors. Dordrecht, The Netherlands: Kluwer; 1993. pp. 463–475. [Google Scholar]
- 16.Uchimiya H, et al. Transgenic rice plants conferring increased tolerance to rice blast and multiple environmental stresses. Mol Breeding. 2002;9:25–31. [Google Scholar]
- 17.Hayashi M, et al. Enhanced dihydroflavonol-4-reductase activity and NAD homeostasis leading to cell death tolerance in transgenic rice. Proc Natl Acad Sci USA. 2005;102:7020–7025. doi: 10.1073/pnas.0502556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holzberg S, Brosio P, Gross C, Pogue GP. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002;30:315–327. doi: 10.1046/j.1365-313x.2002.01291.x. [DOI] [PubMed] [Google Scholar]
- 19.Hein I, et al. Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 2005;138:2155–2164. doi: 10.1104/pp.105.062810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scofield SR, Huang L, Brandt AS, Gill BS. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005;138:2165–2173. doi: 10.1104/pp.105.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berbee ML, Pirseyedi M, Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS, glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. [Google Scholar]
- 22.Ullstrup AJ. Further studies on a species of Helminthosporium parasitizing corn. Phytopathology. 1944;34:214–222. [Google Scholar]
- 23.Nelson RR. Genes for pathogenicity in Cochliobolus carbonum. Phytopathology. 1970;60:1335–1337. [Google Scholar]
- 24.Scheffer RP. Role of toxins in evolution and ecology of plant pathogenic fungi. Experientia. 1991;47:804–811. [Google Scholar]
- 25.Bremer K. Gondwanan evolution of the grass alliance of families (Poales). Evol Int J Org Evol. 2002;56:1374–1387. doi: 10.1111/j.0014-3820.2002.tb01451.x. [DOI] [PubMed] [Google Scholar]
- 26.Barker NP, et al. Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Missouri Botanical Garden. 2001;88:373–457. [Google Scholar]
- 27.Brosch G, Ransom R, Lechner T, Walton JD, Loidl P. Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell. 1995;7:1941–1950. doi: 10.1105/tpc.7.11.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johal GS, Multani DS. U.S. Patent 6. 2004;784:341. [Google Scholar]
- 29.Okagaki RJ, et al. Mapping maize sequences to chromosomes using oat-maize chromosome addition materials. Plant Physiol. 2001;125:1228–1235. doi: 10.1104/pp.125.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Q, Schlueter SD, Brendel V. PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 2004;32:D354–D359. doi: 10.1093/nar/gkh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 33.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 34.Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- 35.Nylander JA. MrModeltest. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. Version 2.0. [Google Scholar]
- 36.Chintamanani S, Multani DS, Ruess H, Johal GS. Distinct mechanisms govern the dosage-dependent and developmentally regulated resistance conferred by the maize Hm2 gene. Mol Plant-Microbe Interact. 2008;21:79–86. doi: 10.1094/MPMI-21-1-0079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.