Summary
The Mixed Lineage Leukemia (Mll) gene is essential for embryonic hematopoietic stem cell (HSC) development but its role during adult hematopoiesis is unknown. Using an inducible knockout model, we demonstrate that Mll is essential for the maintenance of adult HSCs and progenitors, with fatal bone marrow failure occurring within 3 weeks of Mll deletion. Mll-deficient cells are selectively lost from mixed bone marrow chimeras demonstrating their failure to self-renew even in an intact bone marrow environment. Surprisingly, HSCs lacking Mll exhibit ectopic cell cycle entry resulting in the depletion of quiescent HSCs. In contrast, Mll deletion in myelo-erythroid progenitors results in reduced proliferation and reduced response to cytokine-induced cell cycle entry. Committed lymphoid and myeloid cells no longer require Mll, defining the early multipotent stages of hematopoiesis as Mll-dependent. These studies demonstrate that Mll plays selective and independent roles within the hematopoietic system, maintaining quiescence in HSCs and promoting proliferation in progenitors.
Keywords: hematopoiesis, MLL, histone methyltransferase, leukemia, hematopoietic stem cell
Introduction
To maintain the adult hematopoietic system, an ongoing and flexible interplay between lineage-instructive transcription factors and epigenetic regulators is likely to play a central role in determining appropriate numbers of each cell type. Several sequence-specific transcriptional regulators have been shown to play important roles in hematopoiesis, but the role of chromatin-modifying epigenetic regulators in this process is less clear. Polycomb (PcG) and trithorax (trxG) family genes first identified in Drosophila melanogaster influence target gene expression in a manner that is heritable through multiple daughter cell divisions (Cavalli and Paro, 1998; Cavalli and Paro, 1999), making these proteins uniquely qualified to influence cell identity and plasticity during development or in regenerating tissues such as the hematopoietic system.
The Mixed Lineage Leukemia (MLL) gene was the first trxG gene identified as a proto-oncogene (Djabali et al., 1993; Domer et al., 1993; Gu et al., 1992; Tkachuk et al., 1992). Chromosomal translocations disrupting the MLL gene are primarily associated with acute lymphocytic (ALL) or myelogenous leukemia (AML) of infants, as well as secondary AML, accounting for more than 70% of each these two groups (reviewed in (Biondi et al., 2000; Pui and Relling, 2000). MLL fusion genes produced by chromosomal translocation encode proteins composed of the N-terminal ~1/3 of MLL fused to one of over 50 identified translocation partners. The expression of MLL fusion proteins can influence lineage fidelity within the hematopoietic system (Drynan et al., 2005), and can produce cells with mixed lineage identity in human leukemia (Armstrong et al., 2002).
Retroviral expression of MLL fusion proteins in bone marrow cells is sufficient to produce AML or ALL in the mouse, as is the expression of MLL fusions by knock-in or cre-mediated de novo chromosomal translocation (Ayton and Cleary, 2001; Daser and Rabbitts, 2005). The retroviral transduction model enabled structure-function assays that identified motifs within the MLL N-terminus and the fusion partner that are essential for transformation. Collectively, such experiments indicate that MLL fusion proteins act via a gain-of-function mechanism through the acquisition of ectopic transactivation or dimerization capacity (reviewed in Ayton and Cleary, 2001). The N-terminal portion of MLL is likely to play a large role in targeting the fusion proteins to many of the same loci regulated by endogenous MLL, as fusion proteins have been shown by chromatin immunoprecipitation (ChIP) to localize to at least some of the same regulatory sequences as full-length wild-type MLL (Milne et al., 2005; Xia et al., 2005) and increase expression of those target genes (Horton et al., 2005).
Recent advances in purifying native MLL and complexes from cell lines have yielded important insights into the mechanism by which MLL acts on its target genes. Such complexes harbor histone methylation, acetylation, and chromatin remodeling activities (Milne et al., 2002; Nakamura et al., 2002; Wysocka et al., 2005; Yokoyama et al., 2004). The Su(var)3-9/Enhancer of zeste/Trithorax (SET) domain at the extreme C-terminus of MLL methylates histone H3 at lysine 4 (H3 K4), a modification associated with active transcription (Santos-Rosa et al., 2002). Consistent components of these complexes include WDR5, ASH2L and menin. WDR5 and ASH2L interact with the C-terminal SET domain acting as cofactors for the histone methyltransferase activity (Dou et al., 2006; Steward et al., 2006). Menin interacts with the MLL N-terminus and bridges MLL to the basal transcriptional machinery (Yokoyama et al., 2004). The interaction of menin with the N-terminus of MLL is important for maintaining expression of an endogenous MLL target gene in HeLa cells, and is essential for transformation by MLL fusion proteins (Yokoyama et al., 2005; Yokoyama et al., 2004). Other proteins have been described as part of the MLL complex, or as transient interactors, but scant in vivo data is available to validate the biological importance of these interactions.
To understand the mechanisms by which MLL fusion proteins are leukemogenic, it is critical to determine the normal function of MLL in vivo, particularly in the cell types that may represent the target for transformation by MLL oncogenes. Several groups have disrupted the Mll gene using different strategies, and in most cases homozygous Mll mutants die during embryogenesis (Ayton et al., 2001; Yagi et al., 1998; Yu et al., 1995). Interestingly, the deletion of the SET domain alone is not lethal but homozygous mutants exhibit mild loss-of-function phenotypes including homeotic transformation and reduced expression of several Hox genes (Terranova et al., 2006).
Due to the embryonic lethality of most Mll mutants, studies aimed at identifying the role of Mll in the hematopoietic system have focused on embryonic hematopoiesis. Experiments using Mll-deficient embryonic stem (ES) cells demonstrated that these cells were incapable of differentiating into any hematopoietic cell types in adult animals or in the fetal liver. This multi-lineage block was due to the lack of HSCs in Mll-deficient embryos as measured functionally (Ernst et al., 2004a). Using an in vitro system we showed that the block in hematopoietic development was accompanied by global reduction in Hox gene expression and could be rescued by the reintroduction of individual Hox genes (Ernst et al., 2004b). These experiments indicate that Mll plays an essential role in the development of definitive HSCs during embryogenesis, and suggest that this is due to global Hox gene misexpression. However, the early and severe nature of the block in hematopoietic development precluded a detailed analysis of the mechanisms underlying the lack of HSCs or the assessment of Mll function in more differentiated hematopoietic lineages. To address these questions, we developed a mouse model in which Mll can be inducibly inactivated in the bone marrow and in specific lineages to provide a model system in which such functions could be studied in more detail.
Results
Excision of Mll exons 3-4 generates a loss-of-function allele
To create an inducible loss-of-function model for Mll, we designed an in-frame deletion to remove the AT-hooks, nuclear targeting sequences (NTS) and subnuclear localization (SNL) sequences by flanking exons 3 and 4 with loxP sites (“floxed”, MllF). Thus, cre recombinase-mediated excision produces an intragenic deletion (MllΔN) resulting in an MLL mutant protein lacking several NTS and other interaction motifs (Fig. 1A). We recently introduced this allele into embryonic stem (ES) cells in combination with a strong loss-of-function allele (Mll-lacZ) and found that these cells exhibited a block in hematopoietic development (Ernst et al., 2004a) and a block in embryoid body hematopoiesis (Ernst et al., 2004b). Embryos homozygous for the MllΔN allele died in utero at embryonic day 12.5, one day later in embryo development than MllΔN/Mll-lacZ and two days later than Mll-lacZ homozygotes (Supplemental Fig. 1). MllΔN/+ animals exhibit normal viability and fertility and can be back-crossed to the C57Bl/6 strain without loss of heterozygote viability, in contrast to the exon 3 insertion Mll-lacZ strain (Yu et al., 1995). Thus, both the age of embryonic lethality and the hematopoietic phenotypes indicate that the MllΔN allele is a strong hypomorphic allele of Mll, slightly less severe than the Mll-lacZ allele.
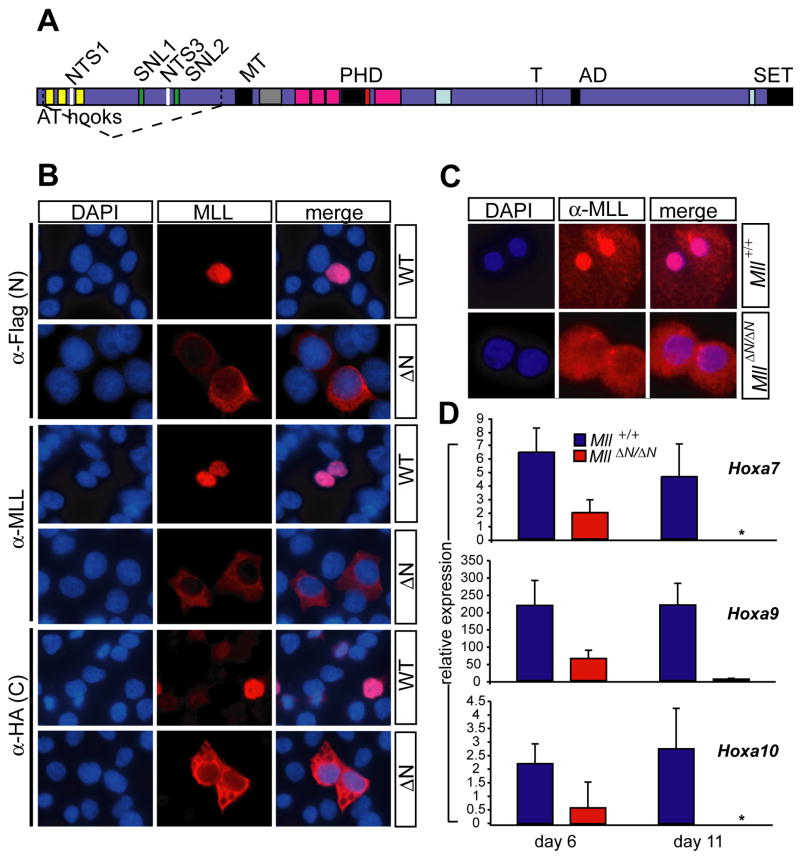
Figure 1.
The MllΔN allele encodes a cytoplasmic protein with reduced activity. A) Diagram of wild-type MLL with the deletion of sequences encoded by exons 3-4 indicated by dashed lines. Homology motifs are shown as colored bars with the following indicated: AT hooks, minor groove DNA binding motif; SNL1 and 2, subnuclear targeting motif; NTS1 and 3, nuclear targeting signals; MT, DNA methyltransferase homology and CpG binding motif; PHD, plant homeodomain homology regions; T, taspase cleavage sites; AD, activation domain; SET, Su(var)3-9/Enhancer of zeste/Trithorax homology region. B) Localization of tagged transfected wild-type MLL versus MLLΔN. 293T cells were transfected with the construct labeled on the right of the panels, and stained with antibodies indicated at the left. Examples shown are representative of three independent experiments. C) Subcellular localization of endogenous MLL in wild-type (upper panels) and MllΔN/ΔN macrophages (lower panels) derived in vitro from bone marrow cells. D) Relative Hox gene expression levels were determined by quantitative PCR using linneg/low bone marrow cells isolated the days indicated after Mll deletion. Each bar represents the average expression level from at least three animals. Error bars represent the standard deviation of the average relative expression among animals. The asterisk denotes an undetectable Ct.
To confirm the predicted subcellular distribution of the MLLΔN protein, we expressed an N- and C-terminally tagged version of the predicted protein encoded by MllΔN in 293T cells. MLL is processed by proteolytic cleavage to produce N- and C-terminal polypeptides that associate with each other within a large protein complex (Hsieh et al., 2003; Yokoyama et al., 2002). To determine the localization of both protein fragments, cells transfected with tagged wild-type and MLLΔN constructs were visualized by fluorescence microscopy using antibodies to the N- and C-terminal tags, as well as an internal MLL antibody that recognizes an epitope in the N-terminal fragment. MLLΔN was clearly excluded from the nucleus as determined by the N-terminal anti-Flag or anti-MLL antibodies, as well as the C-terminal anti-HA antibody (Fig. 1B). In contrast, the full-length protein was exclusively nuclear as expected (Fig. 1B, C). This subcellular localization was also observed using antibodies that detect the endogenous MLLΔN protein produced by cre-mediated Mll excision in MllF/F bone marrow-derived macrophages (Fig. 1C). Western blot analysis demonstrated similar levels of the MLLΔN protein compared to wild-type (not shown).
To independently assess the function of the MLLΔN protein, we examined the expression of Hoxa7, Hoxa9 and Hoxa10, well-characterized MLL target genes, in MllΔN/ΔN hematopoietic cells. As shown in Figure 1D, we observed a progressive reduction (2 to ~50 fold) in Hoxa7, Hoxa9 and Hoxa10 expression between days 6 and 11 after cre-mediated excision of Mll. Other Hox genes, such as Hoxb3 were not affected by Mll loss (Supplemental Fig. 2). Furthermore, highly purified c-Kit+/lineageneg/low/Sca-1+ (KLS) cells exhibited a similar fold reduction in Hoxa gene expression (Supplemental Fig. 3). Thus, excision of Mll exons 3-4 results in the production of a stable protein that fails to become imported into the nucleus, and based on the genetic, biochemical and gene expression data above, results in a non-functional protein and a loss-of-function phenotype.
Acute Mll loss results in rapid bone marrow failure
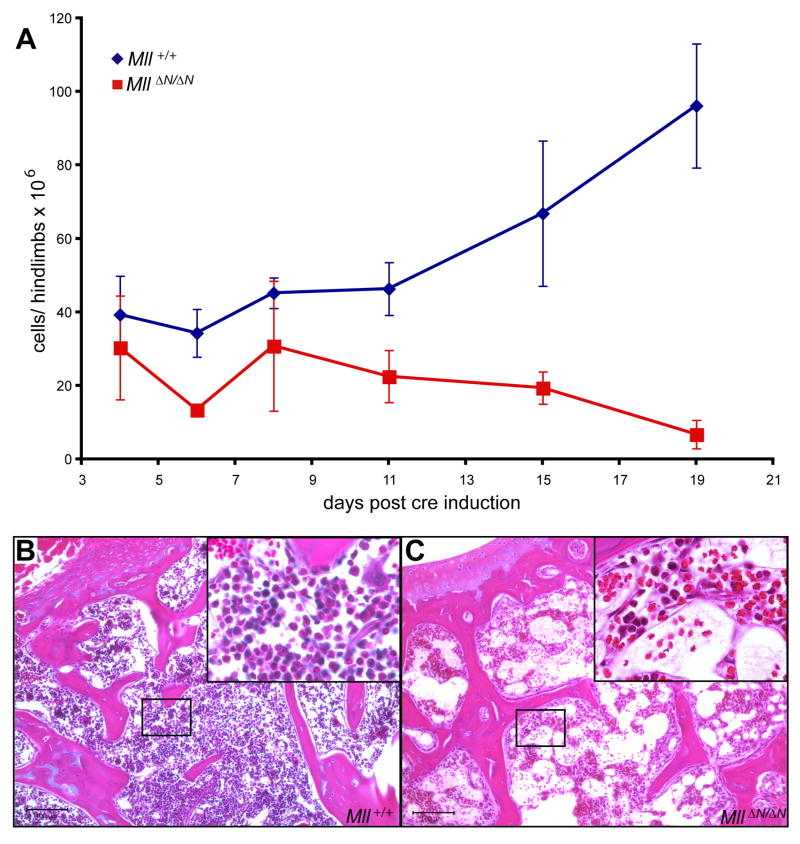
To determine the role of Mll during adult steady-state hematopoiesis, we employed an inducible transgenic cre mouse strain (Mx1-cre, Kuhn et al., 1995). Control and Mx1-cre;MllF/F animals were subjected to a brief series of polyinosinic:polycytidylic acid (pI:pC) injections to induce cre-mediated excision of Mll in the bone marrow. Prior to pI:pC injection bone marrow populations did not differ between control and Mx1-cre;MllF/F animals (Supplemental Fig. 4). As early as 4 days after the first pI:pC injection, nearly all bone marrow cells had undergone cre-mediated excision (Supplemental Fig. 5). Approximately 19 days after inducing cre expression, animals in which 80% or more of the bone marrow was converted to the excised MllΔN allele exhibited bone marrow cytopenia and died or had to be sacrificed (Fig. 2). The bone marrow of the few Mx1-cre; MllF/F animals surviving the pI:pC injections was invariably composed of cells that had not undergone cre-mediated Mll excision (data not shown ). Mx1-cre; MllF/F pI:pC-injected animals that were also injected with wild-type bone marrow cells were healthy beyond 6 months (data not shown), demonstrating that the cause of death was from bone marrow failure.
Figure 2.
Mll maintains bone marrow homeostasis. A) Average bone marrow cellularity per 2 hindlimbs as a function of time after the first pI:pC injection. Control animals (blue, Mll+/+) represent pI:pC injected Mx1-cre;Mll+/+ and MllF/F animals (red) represent pI:pC injected Mx1-cre;MllF/F animals. B–C) Hematoxylin and eosin stained sections from the humerus of control (B) or Mx1-cre;MllF/F (C) animals 15 days after pI:pC injection. Slides were imaged at 100X and insets at 400X magnification.
The maintenance of adult HSCs requires continuous MLL function
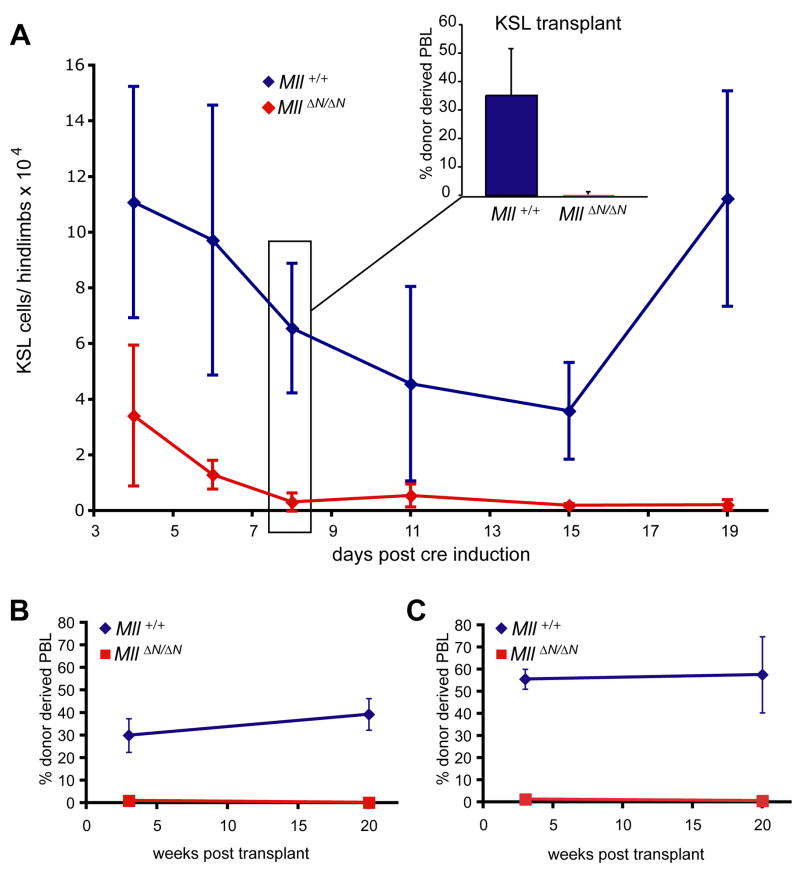
We previously demonstrated that MLL was essential for the development of HSCs in the embryo, so we first determined whether acute loss of Mll affected the HSC-enriched KSL population in the bone marrow. Phenotypic and functional analyses were performed from day 4 after cre-mediated Mll deletion until the point at which Mx1-cre; MllF/F animals died. Notably, the number of KSL cells dropped precipitously and failed to recover (Fig. 3A). To determine whether the reduction in KSL cells reflected a cell-intrinsic defect, we performed competitive transplantation experiments using sorted pooled cells from the day 8 time point. We injected up to 10,000 viable KSL cells purified by fluorescence-activated cell sorting (FACS) from pI:pC treated Mx1-cre;MllF/F or control animals into lethally irradiated recipients. Animals injected with wild-type KSL cells all exhibited donor-derived cells in the peripheral blood, however MllΔN/ΔN KSL cells failed to yield any significant donor contribution as early as 3 weeks after transplantation (Fig. 3A, inset), or as late as 20 weeks (not shown).
Figure 3.
Loss of HSCs upon Mll deletion. A) Total KSL cell number per 2 hindlimbs of control (Mx1-cre;Mll+/+ or MllF/F, blue) or Mx1-cre;MllF/F (red) animals over a 2 week time course. Inset graph shows the results of competitive transplantation assays using 10,000 purified KSL cells to engraft lethally irradiated recipients. The donor cells were purified from Mll+/+ (blue) or MllΔN/ΔN (red) bone marrow from animals as described in (A) 8 days after pI:pC treatment. Peripheral blood contribution was determined 3 weeks after transplantation. For sorting details see Supplemental Fig. 6. B, C) Percentage of Ly5.1+ (donor) peripheral blood leukocytes (PBLs) in lethally irradiated recipients transplanted with control (blue) or MllΔN/ΔN (red) unfractionated bone marrow cells from animals as described in (A). Donor bone marrow was harvested 4 days after pI:pC injection. In both experiments, 3 × 105 wild-type carrier cells were used. These cells were mixed with either an equal number or 3 × 106 donor cells per lethally irradiated recipient for the 1:1 or the 10:1 transplant, respectively. For each set of experiments, 2–4 individual donors and 4–12 recipients were analyzed.
To determine whether HSC activity was present independent of the cell surface phenotype, unfractionated MllΔN/ΔN bone marrow cells were tested in competitive transplantation experiments. We tested either equal numbers (1:1) or a 10-fold excess (10:1) of MllΔN/ΔN bone marrow mixed with wild-type bone marrow cells (Fig. 3B, C). Remarkably, even a 10-fold excess of MllΔN/ΔN cells yielded recipients that were engrafted exclusively by wild-type cells (Fig 3C). Thus, Mll deletion results in the rapid decline in a phenotypically-defined HSC-enriched population, loss of HSC activity from this population, and in fact, from the bone marrow in general.
Consequences of Mll deletion on HSC viability and proliferation
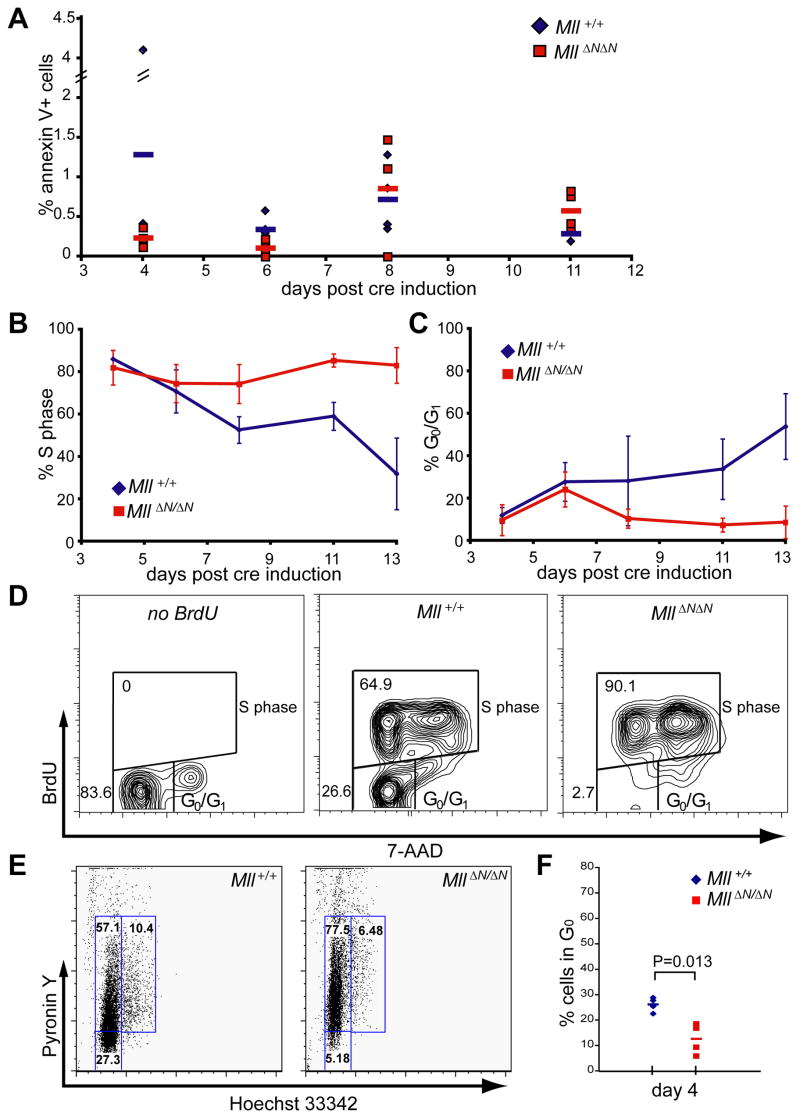
One explanation for the inability of MllΔN/ΔN KSL to compete with wild-type cells in transplantation experiments, as well as their disappearance from conditional knockout bone marrow, is that cells undergo programmed cell death in response to Mll deletion. In fact, an Mx1-cre induced knockout of the anti-apoptotic Mcl-1 gene exhibits very similar kinetics of bone marrow cytopenia and HSC loss (Opferman et al., 2005). To determine whether cell death occurs in response to Mll loss, we assessed annexin V and propidium iodide (PI) staining of KSL cells throughout the time course after Mll deletion. As shown in Figure 4A, we found no significant difference in the proportion of annexin V+ KSL cells at any time point. Annexin V+ cells were detectable in other populations in the bone marrow, but there were no significant differences between the two genotypes in any population tested, nor was there a difference in the overall viability (PI+) at early time points (day 0–11, data not shown).
Figure 4.
Cell death and proliferation within the MllΔN/ΔN KSL population. A) Control (blue), or MllΔN/ΔN (red) cells were stained with lineage, c-Kit, and Sca-1 antibodies and annexin V to assess cell death within the KSL population over time. The percentage of annexin V+ cells within the KSL gated population is shown from individual animals with the average shown as a bar. For gating examples see Supplemental Fig. 6. B–C) Control (blue), or MllΔN/ΔN (red) cells were stained as above, fixed, then stained with anti-BrdU and 7-AAD. Data in (B) indicates percent BrdU positive KLS cells from at least 3 individual animals per time point with the error bars representing 95% confidence intervals. Data in (C) represents the percentage BrdU negative KSL cells (G0/G1 gate, panel D). D) Representative FACS plots illustrating BrdU and 7-AAD staining of KSL cells 11 days post cre induction. E) Representative Hoechst/Pyronin staining of sorted KLS/CD48neg cells at day 4 post cre induction, and F) compiled Hoechst/Pyronin results from 4 individual animals per genotype based on the gates shown in E. Similar results were obtained in at least 3 independent experiments.
Since programmed cell death did not account for the engraftment and homeostasis defects identified in Mll-deficient KSL cells, we considered growth arrest as an alternative hypothesis. For these experiments we assessed the proportion of KSL cells in cycle using 12-hour bromodeoxyuridine (BrdU) pulses (Figure 4B, C). Cell surface staining was performed to identify KSL cells followed by intracellular staining for BrdU detection (identifying cells in S phase during the prior 12 hours). Surprisingly, we found a significant increase in BrdU+ KSL cells in MllΔN/ΔN bone marrow starting at day 8 after Mll deletion, becoming more severe by day 13 (Fig. 4B). Conversely, there were almost no BrdU-negative cells in the MllΔN/ΔN KSL population from day 11 (Fig. 4C, D). We further enriched HSCs from this population to examine the proportion of quiescent (G0) cells by sorting CD48neg/KSL cells (Kiel et al., 2005). Four days after cre induction (prior to the increase in S phase cells) we found that MllΔN/ΔN CD48neg/KSL cells consistently exhibited a shift from G0 to G1 relative to control cells (Fig. 4E, F). Therefore the first defect observed in MllΔN/ΔN HSCs is the failure to maintain quiescence, followed shortly thereafter by an increased number of cycling KSL cells. These data suggest that the depletion of HSCs upon Mll loss is due to their failure to self-renew and maintain a non-cycling population of HSCs.
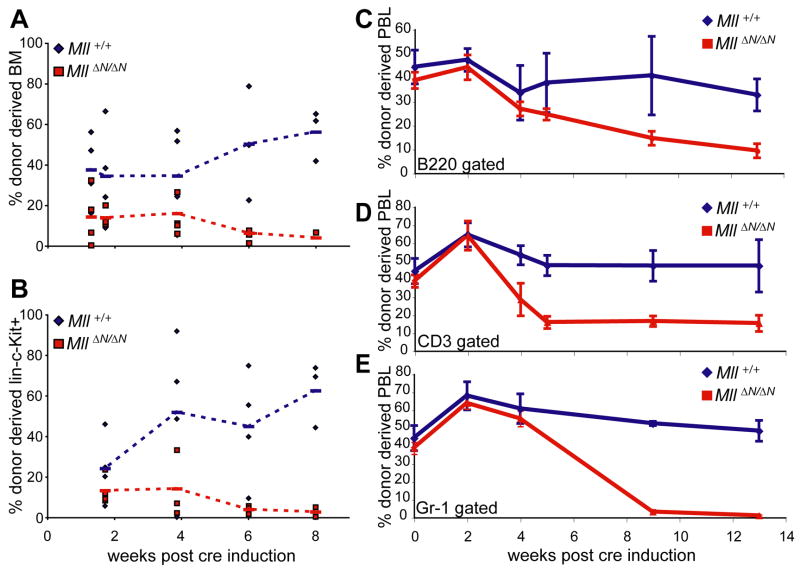
To determine whether MllΔN/ΔN HSCs and their progeny are ultimately depleted in vivo due to a failure to self-renew, we generated chimeric animals in which co-transplanted wild-type bone marrow supports long-term animal viability. Bone marrow chimeras were generated using Ly5.2+ wild-type cells mixed with either Ly5.1+ Mx1-cre;MllF/F or Ly5.1+ transgene negative MllF/F cells. After pI:pC treatment, the Mx1-cre;MllF/F (now MllΔN/ΔN) cells were selectively lost from chimeras (Fig. 5). This loss occurred first in the bone marrow, with the linneg/low/c-Kit+ pool depleted of MllΔN/ΔN cells by 6 weeks (Fig. 5A, B), followed shortly after by a decline in MllΔN/ΔN neutrophils, B- and T-cells in the peripheral blood (Fig. 5C–E). Since whole bone marrow was transferred, the fact that peripheral blood T- and B-cells were not completely depleted (Fig. 5C, D) likely reflects the long half-life of these cells (>12 weeks, (Forster and Rajewsky, 1990; Kondo et al., 1997)) whereas the more rapid and complete loss of MllΔN/ΔN neutrophils (half-life of 1–2 days) reflects the complete exhaustion of MllΔN/ΔN HSCs. These data demonstrate that Mll-deficient HSCs are completely depleted, even in a bone marrow environment that includes wild-type neighboring hematopoietic cells and does not become cytopenic. However, these data do not account for the rapid cell loss in pI:pC injected Mx1-cre;MllF/F animals (Fig. 2A). To investigate this further, we focused on the consequences of Mll deletion on progenitor populations.
Figure 5.
Selective attrition of MllΔN/ΔN cells from mixed bone marrow chimeras. A) Percentage of donor-type cells (Ly5.1+) within total bone marrow of chimeras. Bone marrow chimeras were established by co-injecting equal numbers of Ly5.1 donor bone marrow cells (from either Mx1-cre;Mll+/+ or Mx1-cre;MllF/F donors) plus wild-type bone marrow and waiting >6 weeks until stable engraftment. Chimeras were then injected 4 times with pI:pC and complete excision of Mll was confirmed 2 weeks after the first injection. B) Percentage of donor cells (Ly5.1+) within the linneg/low/c-Kit+ gated population. C–E) Percentage of donor-derived PBLs gated on C) B220 for B-cells, D) CD3 for T-cells, and E) Gr-1 for neutrophils. Blue diamonds, chimeras generated with Mx1-cre;Mll+/+ cells and red squares, chimeras generated with Mx1-cre;MllF/F cells.
Mll plays an independent role in maintaining progenitor pools
To determine whether lineage-committed progenitors were also affected by the loss of MLL function, we analyzed representative myeloid and lymphoid progenitor populations by cell surface phenotype. We found that the absolute number of common lymphocyte progenitors (CLP), common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP) were reduced 1.4- to 4-fold by day 11 after Mll deletion (Fig. 6A). This was predominantly due to an overall reduction in linneg/low/c-Kit+ cells (3.8-fold, data not shown).
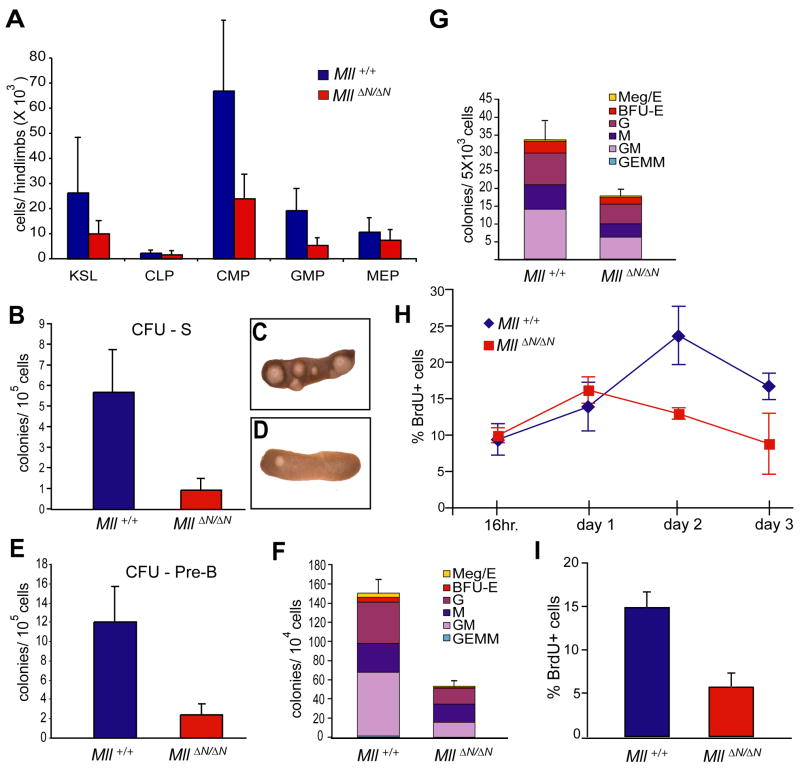
Figure 6.
Reduction in lymphoid and myeloid progenitors upon Mll deletion. A) Average cell number per 2 hind limbs at day 11 post cre induction for the indicated cell types: c-Kit+/lineageneg/low/Sca-1+ (KLS), common lymphocyte progenitors (CLP), common myeloid progenitors, granulocyte-megakaryocyte progenitors (GMP), megakaryocyte-erythroid progenitors (MEP). Averages are from 3–4 animals and error bars represent 95% confidence intervals. B) Colonies per spleen (CFU-S8) produced from pI:pC injected control (MllF/F, blue) or Mx1-cre;MllF/F donors (red). Donor cells (1 × 105) were harvested from animals 4 days after cre induction. At least 4 donor animals and 16 irradiated recipients were used per genotype. C, D) Representative spleens for Mll+/+ (C) or MllΔN/ΔN (D) donor cells. E) CFU-preB frequency in linneg/low bone marrow. Control (MllF/F, blue) or MllΔN/ΔN cells (red) were generated as in (B) but were enriched in linneg/low cells, plated and scored (see Experimental Procedures). F) CFU-C assay using bone marrow cells prepared as in (E) but anti-IL7Rα and -Sca-1 were included in the lineage mix. Cells from at least 3 donors per genotype were plated in triplicate and colonies were scored 7 days later. Represented are the averages from all replicates with error bars indicating the 95% confidence interval of the averages. Meg/E, megakaryocyte/erythroid; BFU-E, burst formation unit-erythroid; G, granulocyte; M, macrophage; GM, granulocyte/macrophage; GEMM, granulocyte, erythroid, macrophage, megakaryocyte colony. G) CFU-C assay using in vitro produced MllΔN/ΔN bone marrow cells. Linneg/low cells from Mll+/+ or MllF/F bone marrow were infected with a cre-IRES-GFP retrovirus and 5,000 GFP+ cells were plated in methylcellulose in triplicate as described in (E). H) Linneg/low/IL7Rα−/Sca-1− cells were enriched from control (MllF/F, blue) or MllΔN/ΔN (red) samples prepared as in (F) from animals 11 days after cre induction. Samples were cultured in liquid medium containing SCF, IL-3, and IL-6. Cells were pulsed for 1 hour with BrdU, harvested and analyzed by flow cytometry at the indicated time points, reflecting days of in vitro culture. I) Cells as described above were incubated with low serum and cytokine-free medium overnight, followed by re-addition of serum and cytokines for 1 hour followed by BrdU pulses as in (H).
To examine progenitor frequencies prior to their phenotypic reduction, we subjected bone marrow from the day 6 time point to in vivo (colony forming units-spleen day 8, [CFU-S8]) and in vitro (colony forming unit-culture [CFU-C]) clonal progenitor assays. Reduced myeloid, erythroid and lymphoid colony frequencies were observed using MllΔN/ΔN bone marrow samples (2–5 fold, Fig. 6B–F). The reduction in all colony types suggests a uniform requirement for Mll in the primitive hematopoietic compartment.
The maintenance of progenitors in vivo may reflect an intrinsic requirement for Mll within the progenitor pool or a prior effect in generating progenitors from HSCs. To distinguish between these possibilities, we excised Mll in myelo-erythroid progenitors (linneg/low/Sca-1−/IL-7Rα− cells) purified from MllF/F or Mll +/+ bone marrow and determined CFU-C frequencies using MllΔN/ΔN cells generated in vitro through retroviral introduction of cre recombinase. This approach also revealed ~2 fold reduced CFU-C from MllΔN/ΔN myelo-erythroid progenitors, with all colony types similarly reduced (Fig. 6G). Together, these data indicate that Mll is required for the maintenance of myelo-erythroid progenitors through a cell-intrinsic mechanism.
Proliferation defects in MllΔN/ΔN myelo-erythroid progenitors
To determine the basis for the functional and phenotypic reduction in MllΔN/ΔN progenitors, we assessed cell death and proliferation. We found no significant difference between the viability of MllΔN/ΔN cells in vivo or using the in vitro cre excision strategy described above (data not shown). We assessed proliferation in vitro using a pool of myelo-erythroid progenitors (linneg/low/IL7Rα−/c-Kit+/Sca-1− cells encompassing the CMP, GMP and MEP populations). Using a time course of in vitro BrdU pulses, we found that MllΔN/ΔN progenitors exhibited reduced proliferation relative to controls (Fig. 6H). In addition, serum/cytokine starvation experiments demonstrated that MllΔN/ΔN progenitors exhibited a 2.6-fold reduction in cells that could re-enter the cell cycle upon serum and cytokine restoration (Fig. 6I). Importantly, the composition of the cell populations before or after in vitro culture was not significantly different between wild-type and MllΔN/ΔN populations (Supplemental Fig. 7 and data not shown). Based on these results, we conclude that Mll is required to sustain cytokine-driven proliferation of myelo-erythroid progenitors. Thus, the combination of ectopic, non-renewing HSC proliferation in conjunction with the reduced proliferation within the larger progenitor pool (linneg/low/c-Kit+) are likely to play a major role in the rapid attrition of cells in the pI:pC injected Mx1-cre;MllF/F animals.
Differentiating T-, B-, and myeloid cells no longer require Mll for homeostasis
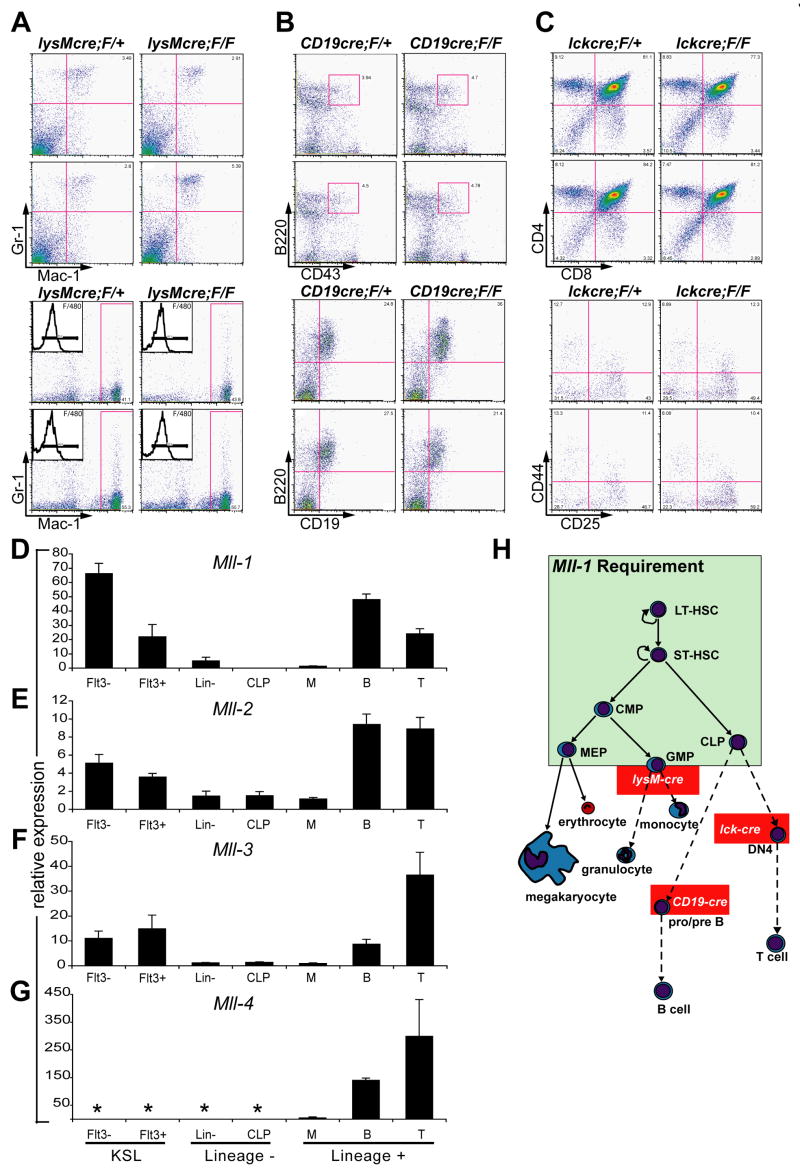
To determine whether Mll is continuously required throughout the differentiation of myeloid and lymphoid cells, we used lineage-specific cre transgenic or knock-in strains to delete Mll in cells subsequent to lineage commitment. In contrast to the results with more primitive progenitors, we found no difference in the steady-state number or function of peripheral B-cells, T-cells or macrophages and neutrophils in CD19-cre, lck-cre or lysozyme-M cre;MllF/F animals respectively (data not shown). We carefully examined the populations representing the beginning of maximal cre expression in each strain and found that there were no differences in cell number or phenotype despite efficient gene deletion within the appropriate population (Fig. 7A–C and data not shown). Thus, these data delineate an early window of hematopoietic development as Mll-dependent, encompassing the stem and progenitor pools. These data also demonstrate that our failure to detect mature MllΔN/ΔN leukocytes in transplanted animals (Figs. 3, 5) was not due to a late block in T-cell, B-cell or granulocyte differentiation, and therefore accurately reflect a defect in stem/progenitor function.
Figure 7.
Mll is dispensable for homeostasis of lineage-committed cells and is not compensated by close homologs in the primitive hematopoietic compartment. A) Analysis of bone marrow (top 4 panels) and peritoneal cells (bottom 4 panels) in lysozyme M-cre;MllF/+ (left column) and lysozyme M-cre;MllF/F animals (right column). Inset in the lower 4 panels indicates F/480 expression on Mac-1/Gr-1 gated cells. B) Analysis of bone marrow cells of CD19-cre;MllF/+ (left column) and CD19-cre;MllF/F (right column) animals. C) Analysis of thymocytes in lck-cre;MllF/+ (left column) and lck-cre;MllF/F (right column) animals. The lower 4 panels represent CD4−/CD8− gated cells stained with the indicated antibodies. Two representative animals are shown for each strain. D–G) The expression of Mll homologs within purified hematopoietic populations. Quantitative real-time PCR results are shown for Mll (Mll-1) (D), Mll-2 (E), Mll-3 (F) and Mll-4 (G). Populations are as described in Fig. 6, with KSL cells further fractionated to enrich HSCs with short- and long-term engrafting potential (Flt3+ and Flt3−, respectively). Also tested are the populations predicted to be affected in the lineage-specific cre strains: myelomonocytic precursors (M, Mac-1+/Gr-1+), pro/pre-B cells (B, B220+/CD19+), and double positive T-cells (T, CD4+/CD8+). Results represent the mean of triplicate reactions with all expression levels normalized to Gapdh. Error bars indicate standard deviation and asterisks indicate an undetectable Ct. Similar results were obtained in three independent experiments. H) Model illustrating the essential, non-redundant role of MLL in maintaining stem and progenitor cells. The green box encompasses cells affected by MLL loss, with those outside the box unaffected at the level of steady-state cell numbers. Red boxes indicate point at which lineage-specific cre expression occurs using the strains shown.
Due to the contrast of these results with the severe, multi-lineage phenotype observed in early progenitors, we considered compensation by Mll homologs as an explanation for the differential effect of Mll deletion on primitive versus more mature, lineage-committed cell types. The Mll homologs Mll-2 through -4 exhibit similar domain organization and encode SET domains predicted to methylate histone substrates with the same specificity (FitzGerald and Diaz, 1999; Prasad et al., 1997; Ruault et al., 2002). Mll-2, the most closely related homolog is expressed in a similar manner to Mll in the hematopoietic system (see below). MLL-2 has been isolated in similar protein complexes as MLL and has also been localized to the same target gene by ChIP (Dou et al., 2005; Hughes et al., 2004; Nakamura et al., 2002; Yokoyama et al., 2004). Mll-1, -2, -3 and -4 are expressed in double positive (DP) thymocytes and CD19+B220+ pro/pre B-cells as were analyzed in the lck-cre and CD19-cre strains (Fig. 7A–G, Supplemental Fig. 8). The overlap in expression of Mll homologs illustrates the potential for compensatory functions within in these cell types, however, Mll-2 and -3 are also expressed significantly within the KSL population and thus would be equally capable to compensate for Mll-1 loss in stem and progenitor populations, which is not the case.
These data demonstrate that the selective expression of Mll homologs within the hematopoietic system cannot account for the selective effects of Mll-1 loss shown in this study. Despite the expression of Mll-2 and -3 within stem and progenitor populations, Mll-1 performs an essential and non-redundant role in maintaining HSCs and progenitors (Fig. 7H). In the more mature populations, it is possible that the relatively high levels of Mll-2, -3, and -4 compensate for Mll-1 loss, but a definitive test of this hypothesis will require the analysis of compound mutants.
Discussion
The central finding of this study is that adult steady-state hematopoiesis depends on Mll, based on essential functions performed within hematopoietic stem and progenitor cells. In the absence of Mll, HSCs exit from a quiescent state and precociously undergo cell cycle entry. Ectopic cycling of HSCs occurs without an increase in this population and coincident with functional deficits, suggesting that cell division in this compartment is accompanied by differentiation and not self-renewal. In contrast, the role of Mll within primitive myelo-erythroid progenitors appears to be quite distinct, in that progenitor frequency is reduced upon Mll deletion through a mechanism involving reduced proliferation. In conjunction with germline loss-of-function analyses (Ernst et al., 2004a), the studies presented here demonstrate that Mll is necessary for both the development and maintenance of HSC. Further characterization will reveal whether all of these MLL dependent processes utilize common or distinct molecular pathways.
Our data generally support the concept that MLL acts on a selective set of target genes in different cell types. Based on a genome-wide ChIP study in the U937 cell line, it has been proposed that MLL functions as a transcription start-site specific global transcriptional regulator (Guenther et al., 2005). This conclusion is partly based on the large overlap in Pol II and MLL localization at active promoters. If this co-localization indicated a biologically important association, one would expect a severe consequence for all cells in which MLL activity is removed. This prediction is difficult to reconcile with the selective phenotypes observed in vivo upon Mll loss. For example, embryonic stem cells and many differentiating tissues in chimeric animals are unaffected by Mll deficiency (Ernst et al., 2004a; Ernst et al., 2004b). These observations, along with the results presented in this study suggest that selective and distinct target genes are regulated by MLL and its homologs in different tissues.
The reduced proliferation of myelo-erythroid progenitors shown in this study is consistent with previous observations using embryoid body-derived hematopoietic progenitors. In these studies, the reduced expression of multiple Hox genes appeared to be the primary cause for the reduced colony frequency in Mll mutant embryoid bodies (Ernst et al., 2004b). Thus, we predict that the reduced Hox gene expression in progenitors is similarly likely to play a major role in reduced progenitor frequency and proliferation upon Mll deletion. Conversely, Hoxa7 and Hoxa9 are consistently overexpressed in MLL fusion-initiated leukemia in humans and in mouse models, and are rate-limiting for leukemogenesis or influential in determining leukemic cell phenotype (Armstrong et al., 2002; Ayton and Cleary, 2003; Kumar et al., 2004; So et al., 2004). Based on the results of the current study and results of Hox expression analyses in Mll loss- and gain-of-function experiments (Ernst et al., 2004b; Horton et al., 2005), we propose that altered Hox gene expression may be responsible for the proliferation component of the leukemogenic program induced by MLL fusion proteins.
The regulation of quiescence in HSCs is less likely to be explained by Hox gene maintenance alone, based on analyses of individual and compound Hox gene knockouts in which such a phenotype has not been reported (Bijl et al., 2006; Brun et al., 2004; Lawrence et al., 2005). This suggests that there are other HSC-specific Mll target genes involved in this process. Such HSC-specific target genes may remain activated by MLL fusion oncoproteins as HSCs differentiate, resulting in the maintenance of a self-renewal program coincident with a proliferation-promoting program mediated by Hox gene overexpression. Candidate genes for this self-renewal program include those reported as re-activated in an MLL-AF9 leukemia model (Krivtsov et al., 2006). Leukemia can be initiated by introducing Mll fusion oncogenes into committed progenitors (Cozzio et al., 2003; Krivtsov et al., 2006; Somervaille and Cleary, 2006), suggesting that the reactivation of such a self-renewal program by Mll fusion oncogenes can occur in progenitors. However, the introduction of Mll fusion oncogenes into HSCs rather than progenitors may facilitate fusion protein access to a quiescence-promoting gene expression program. One prediction of this hypothesis is that the introduction of Mll fusions into HSCs (rather than progenitors) would result in a more persistent, chemotherapy-resistant pool of leukemic cells, a concept that has not been tested in mouse models.
Importantly, our studies suggest that targeting MLL fusion oncogenes through mechanisms that generally interfere with MLL function may cause bone marrow failure. Thus it will be essential to identify strategies that selectively target MLL fusion oncoproteins without compromising wild-type MLL function. This effort will be aided by a comprehensive assessment of target genes affected by MLL fusions versus those affected by wild type MLL.
Experimental Procedures
Mice, competitive transplantation and CFU-S assays
Animals were maintained in accord with the Dartmouth Animal Resources Center and I.A.C.U.C. policies. Mll floxed alleles and ES targeting have been described (Ernst et al., 2004a). To generate MllΔN animals, MllF/F animals were crossed to the EIIa-cre strain (Jackson Labs). MllΔN/ΔN embryos were generated by crossing heterozygotes. Eight a.m. of the day of vaginal plug observation was considered E 0.5. The MllF/F strain was backcrossed to C57Bl/6 for more than 9 generations then back-crossed to B6.SJL (Jackson Labs) for 5 generations. Animals were crossed to the Mx1-cre, lysozyme M-cre, CD19-cre (obtained from Drs. Rajewski and Orkin, Harvard Medical School) and lck-cre strains (obtained from Dr. Wilson, University of Washington) to produce homozygous Mll floxed (MllF/F), cre-expressing strains. Mx1-cre;MllF/F, Mx1-cre;Mll+/+ or MllF/F mice were treated with 2–4 injections of pI:pC (GE Healthcare) every other day. Two, three, or four injections were performed for the day 4, 6, or 8–19 time points respectively. Increasing the number of injections resulted in a higher penetrance of the phenotypes described, without altering the kinetics of their appearance. All animals were 6–12 weeks old for analyses in this study. The following formula was used for the pI:pC dose: (10 × animal mass (g) + 50) μg pI:pC.
For competitive transplantation experiments C57BL/6J recipients were irradiated with a split dose of 1100 Rads 3 hours apart. Bone marrow cells were prepared by crushing with a mortar and pestle in ice-cold Hank’s buffered saline solution (HBSS) supplemented with 2% fetal bovine serum (FBS). Red cells were removed using ACK lysis buffer (0.15M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA pH 7.2). Bone marrow chimeras were generated by injecting a mixture of 1.25 × 106 donor cells (Mx1-cre;MllF/F, MllF/F, or Mx1-cre; Mll+/+, Ly5.1) plus 2.5 × 105 carrier cells (wild-type Ly5.2 or Ly5.1/5.2) and waiting > 6 weeks for stable engraftment. Animals were maintained on sterile food and water (containing 100 μg/mL Baytril) for 3 weeks after irradiation. For the CFU-S8 assay 1 × 105 bone marrow cells from animals receiving 2 pI:pC doses were injected into lethally irradiated recipients. Eight days later spleens were transferred to Tellesniczky’s fixative. Colonies were counted and photographed using a Leica MZ7.5 dissecting microscope fitted with a Cannon Powershot S3 IS digital camera.
Flow cytometry, annexin V binding assays and G0/G1 analysis
Lineage staining was accomplished using unconjugated rat anti-CD4, -CD3, -CD8, -Gr-1, -B220, -CD19, -Ter119, -Mac-1 and for some experiments anti-Sca-1 and -IL-7Rα (all antibodies were from BD Biosciences except the IL7Rα [eBiosciences]). Lineage antibodies were detected with goat ant-rat F(ab′)2 tricolor (Caltag), blocked with rat serum (Sigma) and stained with anti-Sca-1 and -c-Kit antibodies (BD Biosciences). Stained cells were incubated with annexin V FITC as described by the manufacturer and analyzed immediately using a FACSCalibur (BD Biosciences) and FlowJo software (Treestar, Inc). We first determined that Mx1-cre;Mll+/+ and MllF/F animals exhibited indistinguishable levels of annexin V and PI staining after pI:pC injection (data not shown) then used either genotype as controls for the following experiments. Annexin V positive controls were generated for each analysis by incubating cells with 1 mM H2O2 at 37°C for 5 minutes. G0/G1 analysis was performed by first FACS purifying KLS/CD48neg cells then incubating 10,000 sorted cells in 10 μg/mL Hoechst 33342 and 100 μM verapamil (both from Molecular Probes) for 45 minutes at 37°C. Samples were then supplemented to 2 μg/mL Pyronin Y (Sigma), incubated for an additional 15 minutes, washed and analyzed immediately on a FACSAria (BD Biosciences). Anti-CD48 was from BD Biosciences. The unpaired t test was used to evaluate the statistical significance of differences in G0 populations.
In vivo proliferation assays
Mx1-cre;MllF/F, Mx1-cre;Mll+/+ or MllF/F mice were pI:pC injected as described above. At 12 and 6 hours prior to sacrifice animals were injected with 1mg BrdU. Cells were harvested and stained as described above. Stained cells were then fixed, permeabilized, and incubated with FITC-labeled anti-BrdU and 7-AAD following the manufacturer’s suggestions (BD Biosciences).
Ex vivo introduction of cre recombinase and in vitro progenitor assays
Bone marrow cells were prepared from 6–8 week old Mll+/+ and MllF/F mice. Cells were stained with unlabeled antibodies as described above and depleted with sheep anti-rat magnetic beads (Dynal). Linneg/low ells were then resuspended in growth medium (IMDM containing 20% FBS, 200 mM nonessential amino acids [NEAA], 200 mM L-glutamine [L-gln], 10 U/ml penicillin G, μg/ml streptomycin [P/S], 50 μM 2-ME) supplemented with 40 ng/ml IL-6, 20 ng/ml IL-3, 40 U/ml LIF, 80 ng/ml SCF and 10 μg/ml polybrene. Spin-infection was performed for 30 minutes at 25°C in 96-well plates. FACS-purified GFP+ cells were subjected to colony assays as described (Ernst et al., 2004b). For the proliferation assays, linneg/low cells were enriched as described above from Mx1-cre;MllF/F or MllF/F animals at day 11 post cre-induction. Cell densities were adjusted to 5 × 105 cells/mL in growth medium supplemented with 1XBIT (StemCell Technologies), 50 ng/mL SCF, 10 ng/mL IL-3 and 10 ng/mL IL-6. Cells were incubated with BrdU at 10 μM for 1 hour. For the starvation experiment, cells were grown for 2 days, washed and incubated cytokine-free, 0.5% FBS growth medium overnight, shifted to complete medium then pulsed with BrdU as described above. For the pre-B colony assay, 10,000 linneg/low cells were plated in Methocult M3630 (StemCell Technologies) supplemented with 100 ng/mL SCF, 20 ng/mL murine Flt3L, and 10 ng/mL IL-7. Pre-B colonies were scored after 7 days. Cytokines were from R&D Systems.
Real-time, quantitative PCR
Total RNA from the sorted populations indicated in Fig. 7 was purified using the RNeasy Mini kit (Qiagen). After extraction, 1 μg MS2 bacteriophage RNA (Roche) was added as a carrier, and mRNA was amplified using the RiboAmp RNA Amplification Kit (Arcturus) following manufacturer’s recommendations for one round of amplification. Hox gene detection was performed as described (Ernst et al., 2004b) and Mll family primer and probes are shown in Supplemental Figure 5. Relative expression levels were determined using the ΔΔCt method (Livak and Schmittgen, 2001) with data from triplicate multiplexed reactions normalized to Gapdh. Quantitative real-time PCR was performed on a 7900HT machine using the 9600 emulation setting (Applied Biosystems).
Histopathology
For bone marrow sections, the humerus was removed, cleaned and transferred to Bouin’s fixative overnight at 4°C. Fixed samples were sectioned and stained by the Harvard Histopathology Core Facility. Samples were imaged on a Nikon Optiphot-2 microscope using 10X and 40X objectives.
Plasmids
The region spanning the exon 3-4 deletion (ΔN) was amplified by PCR from MllΔN/ΔN fibroblast RNA. This PCR product was inserted into the FseI and PvuI sites of the human MLL cDNA. The resulting MLLΔN cDNA was modified to encode the Flag epitope at the 5′ end and the HA epitope at the 3′ end by inserting annealed oligonucleotides encoding the epitope sequence. The tagged cDNAs (wild-type and MLLΔN) were then subcloned into pCDNA3.1(−) (Invitrogen) at the XhoI and EcoRV sites.
Cell culture, transfection and immunofluorescence
293T cells were maintained in DMEM supplemented with 10% FBS, 200 mM L-gln, P/S, and 100 nM NEAA and transfected using FuGENE 6 (Roche). Thirty-six hours post-transfection, cells were plated on coverslips and grown overnight before fixing in 4% paraformaldehyde in PBS for 5 minutes. Cells were then blocked with PBS containing 2% goat serum and 0.2% Triton X-100 overnight. Anti-Flag M2 (1:100, Sigma), anti-MLL1 (BL1290) and anti-HA (both 1:100, Bethyl Labs) and were incubated with coverslips for 30 minutes at room temperature, then washed 3 times in PBS. Detection was performed using Alexa Fluor-568 goat anti-mouse or Alexa Fluor-488 goat anti-rabbit (both at 1:1000, Molecular Probes). Stained cells were then washed 3 times in PBS, with the final wash containing 1μg/mL 4′, 6-diamidino-2-phenylindole (DAPI). Coverslips were mounted onto slides with Prolong Gold antifade medium (Molecular Probes). Images were captured with a Zeiss AxioVisionZ1 microscope and analyzed in Adobe Photoshop. Macrophages were cytospun on to slides and stained as described above.
Supplementary Material
Acknowledgments
This study is dedicated to Stanley Korsmeyer, in whose lab the generation of the mouse model described here was initiated. This work was supported by funds from the Sydney Kimmel and V Foundations, N.C.R.R. C.O.B.R.E. grant #2P20RR016437, N.I.H. #DK067119 and ACS IRG#82-003-21. We acknowledge the insightful comments of David Traver, Malek Djabali and Iannis Aifantis. We are grateful for the gifts of cre mice from Drs. Rajewsky and Orkin. We thank Hanno Hock for essential advice on pI:pC vendors and Amy Wagers for advice on the Hoechst/Pyronin staining. We thank Gary Ward for expert cell sorting, Nancy Speck and her laboratory members for helpful comments, R. Mako Saito for help with microscopy and Victor Ambros and laboratory members for help with real-time PCR. The authors declare that they have no competing conflict of interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Ayton P, Sneddon SF, Palmer DB, Rosewell IR, Owen MJ, Young B, Presley R, Subramanian V. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis. 2001;30:201–212. doi: 10.1002/gene.1066. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijl J, Thompson A, Ramirez-Solis R, Krosl J, Grier DG, Lawrence HJ, Sauvageau G. Analysis of HSC activity and compensatory Hox gene expression profile in Hoxb cluster mutant fetal liver cells. Blood. 2006;108:116–122. doi: 10.1182/blood-2005-06-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi A, Cimino G, Pieters R, Pui CH. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96:24–33. [PubMed] [Google Scholar]
- Brun AC, Bjornsson JM, Magnusson M, Larsson N, Leveen P, Ehinger M, Nilsson E, Karlsson S. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood. 2004;103:4126–4133. doi: 10.1182/blood-2003-10-3557. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daser A, Rabbitts TH. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin Cancer Biol. 2005;15:175–188. doi: 10.1016/j.semcancer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Djabali M, Selleri L, Parry P, Bower M, Young B, Evans GA. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Genet. 1993;4:431. doi: 10.1038/ng0893-431. [DOI] [PubMed] [Google Scholar]
- Domer PH, Fakharzadeh SS, Chen CS, Jockel J, Johansen L, Silverman GA, Kersey JH, Korsmeyer SJ. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci U S A. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Drynan LF, Pannell R, Forster A, Chan NM, Cano F, Daser A, Rabbitts TH. Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis. Embo J. 2005;24:3136–3146. doi: 10.1038/sj.emboj.7600760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004a;6:437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004b;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- FitzGerald KT, Diaz MO. MLL2: A new mammalian member of the trx/MLL family of genes. Genomics. 1999;59:187–192. doi: 10.1006/geno.1999.5860. [DOI] [PubMed] [Google Scholar]
- Forster I, Rajewsky K. The bulk of the peripheral B-cell pool in mice is stable and not rapidly renewed from the bone marrow. Proc Natl Acad Sci U S A. 1990;87:4781–4784. doi: 10.1073/pnas.87.12.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton SJ, Grier DG, McGonigle GJ, Thompson A, Morrow M, De Silva I, Moulding DA, Kioussis D, Lappin TR, Brady HJ, Williams O. Continuous MLL-ENL expression is necessary to establish a “Hox Code” and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;65:9245–9252. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, Korsmeyer SJ. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–1104. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, Rallapalli R, Yano T, Alder H, Croce CM, Huebner K, et al. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997;15:549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br J Haematol. 2000;109:13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- Ruault M, Brun ME, Ventura M, Roizes G, De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284:73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Xia ZB, Popovic R, Chen J, Theisler C, Stuart T, Santillan DA, Erfurth F, Diaz MO, Zeleznik-Le NJ. The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc Natl Acad Sci U S A. 2005;102:14028–14033. doi: 10.1073/pnas.0506464102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92:108–117. [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.