Abstract
We have identified telomerase activity in extracts of three evolutionarily diverse kinetoplastid species: Trypanosoma brucei, Leishmania major, and Leishmania tarentolae. Telomerase activity was initially detected in extracts from insect form cells of all three kinetoplastid species by using a modification of the one-tube telomere repeat amplification protocol [Kim, N., et al. (1994) Science 266, 2011–2015], although better results were subsequently achieved with the two-tube telomere repeat amplification protocol [Autexier, C., Pruzan, R., Funk, W. & Greider, C. (1996) EMBO J. 15, 5928–5935]. The activity in T. brucei extracts was sufficiently robust to enable its detection in a direct assay of telomerase; enzyme processivity was found to be relatively low. The in vitro properties of telomerase suggest a possible templating domain sequence for the telomerase RNA of T. brucei. Telomerase activity is likely to contribute to telomere maintenance in these parasitic organisms and provides a new target for chemotherapeutic intervention.
Chromosomal termini, or telomeres, in most eukaryotes consist of DNA–protein complexes that are essential for genomic integrity and cell viability. Part of the telomeric DNA that is lost during each round of cell replication is replaced primarily by the action of a ribonucleoprotein enzyme complex telomerase (1). Telomerase activity, first described in the ciliated protozoan Tetrahymena thermophila (1), is widely distributed among phylogenetically diverse eukaryotes including yeasts, amphibians, mammals, and plants, and was recently reported in the pathogenic malaria parasite Plasmodium falciparum (2–8). Telomerase adds dNTPs to the 3′ end of the G-rich strand of chromosomal DNA by reverse transcription of a telomeric sequence template within the telomerase RNA subunit. Mutations in the template region of the telomerase RNA gene cause progressive shortening of the telomeres and cell senescence in T. thermophila (9). In mice, knock-out of telomerase RNA leads to increased telomere shortening, chromosomal aberrations, and sterility (10). The telomerase RNA subunit is generally difficult to identify because there is only limited sequence conservation at the level of secondary structure even between related species (ref. 11, and reviewed in ref. 12), necessitating more traditional biochemical approaches for its identification.
The cloning and characterization of the protein catalytic subunit (telomerase reverse transcriptase, TERT) of Euplotes aediculatus and Saccharomyces cerevisiae telomerases by Lingner et al. (13) made possible the identification of telomerase reverse transcriptase homologues in Schizosaccharomyces pombe and humans (14) and the ciliates T. thermophila and Oxytricha trifallax (15, 16). The amino acid sequences of these homologues indicate that the telomerase protein subunit is a specialized reverse transcriptase, evolutionarily most related to reverse transcriptases in the non-long terminal repeat retrotransposon family (ref. 14, and reviewed in ref. 17). In humans, the majority of somatic cells do not express telomerase enzyme activity, unlike germ line and some proliferating cells, including components of blood and epidermal tissue express telomerase (18–20). Enzyme activity also is frequently found in immortalized cell lines and in ≈85% of tumors (18). Stable ectopic expression of the human telomerase reverse transcriptase (hTERT) gene in normal primary somatic human cells results in some transfectants being able to continue to grow for many cell generations, bypassing the usual senescence (21). In various yeast and human cells, deleting or mutating the TERT gene causes cell growth arrest and telomere shortening, culminating in senescent or aberrant cellular phenotypes (13, 14, 22, 23). These results point to the importance of telomerase expression during proliferation of eukaryotic cell growth, and it has been suggested that suppression of telomerase activity, through the development of specific inhibitors of telomerase, may impair the proliferation not only of malignant cells but also of protozoan pathogens (7, 24–26).
Because the commonly prescribed antikinetoplastid drugs are extremely toxic to the mammalian host, we decided to study telomerase activity in these medically and agriculturally significant protozoa. As descendants of one of the most ancient eukaryotic lineages, they differ notably from other known eukaryotes in their genomic organization, processing of all nuclear pre-mRNAs by trans-splicing, and extensive editing of mitochondrial RNAs (reviewed in refs. 27 and 28). Their genomes are essentially diploid, although aneuploidy is commonly detected by electrophoretic karyotyping of clonal laboratory strains (reviewed in ref. 29; M.I.N.C. and F. da Silveira, unpublished data). In T. brucei and in Leishmania spp., the variability and dynamism of the telomeric regions and their characteristic telomere-associated sequences are thought to be responsible for chromosome rearrangements and consequent polymorphisms in chromosome lengths and gene expression (29–32).
Kinetoplastid telomeres, including those of human and animal pathogens, are composed of tandem, repeated copies of a 6-bp sequence 5′-TTAGGG-3′ (33, 34), which is also found at human telomeres (35). Although little is known about their telomere structure and function, parasitic protozoa in general have achieved a state of immortality similar to cancer cells, because parasite growth is limited primarily by host immune response or culture conditions.
Here we describe telomerase activity in semipurified extracts of three evolutionarily divergent kinetoplastid species: T. brucei, Leishmania major, and Leishmania tarentolae. We find that telomerase activity is detectable in extracts prepared from cultured insect forms of these parasites and that enzyme processivity is low compared with ciliate telomerase. A possible minimal template sequence for the telomerase RNA of T. brucei is extrapolated from the in vitro properties of telomerase activity. These findings suggest that telomerase contributes to telomere maintenance in kinetoplastid parasites.
MATERIALS AND METHODS
Parasite Species and Culture Conditions.
Promastigotes of L. tarentolae strain UC were cultivated in axenic medium with 3.7% Brain Heart Infusion (Difco) supplemented with 10 μg/ml hemin at 26°C for 48 h (doubling time of ≈6–9 h). Promastigotes of L. major strain LmV39 were cultivated in HOSMEM II medium (36) supplemented with 10% heat-inactivated FBS (Gemini Biological Products, Calabasa, CA) at 28°C for 52 h (doubling time of ≈8–10 h). Procyclics of T. brucei strain IsTat1.1 were cultivated in BSM medium (37) supplemented with 5% heat-inactivated FBS at 28°C for 60 h (doubling time of ≈12 h).
Telomerase Extraction.
Whole-cell extracts of L. tarentolae were prepared after harvesting the cells by centrifugation at 5,000 × g for 5 min at 4°C. Cells were washed in PBS supplemented with 2% glucose. Pellets of ≈5 × 1010 cells were immediately frozen in liquid nitrogen and slowly thawed at 2°C in 1× TME (10 mM Tris⋅HCl, pH 8.0/1.2 mM MgCl2/1 mM EGTA, pH 8.0) containing a mixture of protease inhibitors (0.7 μg/ml pepstatin, 10 μg/ml PMSF, 0.5 μg/ml leupeptin). Cells were lysed with 0.5% Nonidet P-40 at 2°C, and lysis was checked by reverse phase optical microscopy. TMG at a 2× concentration (1× TMG = 20 mM Tris⋅HCl pH 8.0/2.4 mM MgCl2/20% glycerol vol/vol) supplemented with 1 mM DTT, 1 mM EDTA, and 1 mM spermidine, was added to the lysed cell suspension. Extract was separated from cell debris by centrifugation at 18,000 × g for 20 min at 4°C. S100 was prepared by ultracentrifugation at 100,000 × g for 90 min at 4°C, aliquotted, and stored in liquid nitrogen. Protein concentration (Bradford) of the resulting S100 was ≈36.3 mg/ml. Extracts of ≈1 × 1010 L. major promastigotes or 2 × 1010 T. brucei procyclics suspended in buffer A (20 mM Tris⋅HCl, pH 7.5/1 mM EGTA, pH 8.0/1 mM EDTA, pH 8.0/1 mM spermidine/0.3 mM spermine/5 mM 2-mercaptoethanol/10 ng/ml leupeptin; ref. 38) were obtained by blending cells within a mortar in the presence of liquid nitrogen. S100 extracts were prepared as above. Protein concentrations of these S100 fractions were 18.4 mg/ml for L. major and 12.6 mg/ml for T. brucei. Each of the above-mentioned S100 supernatants were loaded onto a DEAE-agarose column (Biogel, Bio-Rad) equilibrated with 1× TMG. Columns were washed with 6 vol of 1× TMG containing 0.4 M sodium acetate (pH 8.0) for L. tarentolae and T. brucei and in 0.5 M sodium acetate (pH 8.0) for L. major. Activity was eluted in 1 column-volume each 0.5 M and 0.6 M sodium acetate (pH 8.0) in 1× TMG for L. tarentolae and T. brucei, respectively, and 0.7 M sodium acetate (pH 8.0) in 1× TMG for L. major. Eluates were then desalted on Microcon-30 columns (Amicon) to 100 mM final salt concentration.
Telomere Repeat Amplification Protocol (TRAP).
One-tube TRAP. Telomerase activity in DEAE-agarose fractions was assayed by using a modification of TRAP (18). The TS forward primer (5′-AATCCGTCGAGCAGAGTT-3′) and a modified reverse primer, CX-ext (5′-GTGCCCTTACCCTTACCCTTACCCTAA-3′) (39) were purchased from Operon Technologies (Alameda, CA) and gel purified before use. One-tube reactions (50 μl final volume) were set up as described (18). During standardization of TRAP conditions, we used different amounts of each extract and [α-32P]dGTP or [α-32P]dCTP. Best results were achieved by using 0.5–2 μl of each DEAE fraction (equivalent to 3.45 μg of protein or ≈3 × 106 L. tarentolae cell equivalents; 3.75 μg of protein or 5 × 106 L. major cell equivalents; and 2.0 μg of protein or 2.5 × 106 T. brucei cell equivalents, respectively) and 0.3 μCi (1 Ci = 37 GBq) per tube of [α-32P]dGTP or [α-32P]dCTP. The extracts were incubated at 28 °C for 1 h to allow extension of the TS primer by telomerase, followed by PCR amplification using the reverse CX-ext primer, as described (18). The products were fractionated in 10% sequencing gels and autoradiographed or analyzed by PhosphorImager (Molecular Dynamics). The TRAP controls were single-primer reactions (no TS and no CX-ext) and reactions in which the extract was omitted (nE). Activity in the extracts was tested for RNase A sensitivity by incubation with 100 ng of RNase A (Sigma) for 5 min at 37°C before or after the telomerase reaction step and with or without 1 unit of RNase inhibitor RNasin (Promega) before addition of RNase A.
Two-tube TRAP.
We adapted a two-tube procedure (40) with the following modifications. Telomerase reaction mix (40 μl) contained 1× modified TRAP buffer (50 mM Tris⋅HCl, pH 8.3/1 mM DTT/1 mM spermidine/1 mM MgCl2), 2 mM dATP, 2 mM dTTP, 10 μM dGTP, 40 pmol of TS primer, and 12 μl of a 1:1 dilution of the relevant DEAE fraction. Mixtures were incubated for 1 h at 28°C. Ten microliters of the telomerase reaction was added to a 50-μl final volume PCR mix containing 1× modified TRAP buffer, 50 μM each dNTP, 20 pmol of TS primer, 20 pmol of CX-ext primer, 0.1 μCi/μl [α-32P]dGTP or [α-32P]dCTP and 1 unit of Taq polymerase (Boehringer Mannheim). PCR conditions and nuclease controls were the same as for the one-tube TRAP described above (18). To analyze the telomerase assay step separately from the PCR step, we also performed template-directed termination reactions in which the DEAE fractions were assayed under conditions in which the dNTPs were replaced by equimolar concentration of corresponding ddNTPs.
Conventional Telomerase Assay for T. brucei DEAE Fractions.
To directly assay telomerase activity in extracts, a modification of non-PCR-based assay protocols was used (23, 41). Ten microliters of a 1:1 dilution of a DEAE fraction (corresponding to ≈5 × 106 cell equivalents) was assayed in a total volume of 20 μl containing 1× telomerase buffer (25 mM Tris⋅HCl, pH 8.3/1 mM MgCl2/1 mM EGTA), 0.8 mM dATP, 0.8 mM dTTP, 6.5 μM dGTP, 1.5 μM [α-32P]dGTP (800 Ci/mmol) and 1.0 μM telomeric primer. Sequences of the telomeric primers permuted at the 3′ end and a T. thermophila telomeric primer are shown in Table 1. All primers were obtained from Operon Technologies (Alameda, CA) and gel-purified before use. The reactions were incubated for 1–3 h at 28°C and stopped with 50 mM EDTA. Telomerase products were phenol-extracted once followed by extraction with phenol/chloroform. Products were ethanol-precipitated in the presence of 10 μg of tRNA or glycogen as carrier, washed with 70% ethanol, and resolved in 10% sequencing gels. RNase A sensitivity, chain termination, and control reactions were as described above.
Table 1.
Primers used as substrates in telomerase assays
| Primer | Telomeric primer sequence |
|---|---|
| tel 1 | 5′-GTTAGGGTTAGGGTTAGG-3′ |
| tel 2 | 5′-GGTTAGGGTTAGGGTTAG-3′ |
| tel 3 | 5′-GGGTTAGGGTTAGGGTTA-3′ |
| tel 4 | 5′-AGGGTTAGGGTTAGGGTT-3′ |
| tel 5 | 5′-TAGGGTTAGGGTTAGGGT-3′ |
| tel 6 | 5′-TTAGGGTTAGGGTTAGGG-3′ |
| Tet | 5′-GGGGTTGGGGTTGGGGTT-3′ |
Size Standards.
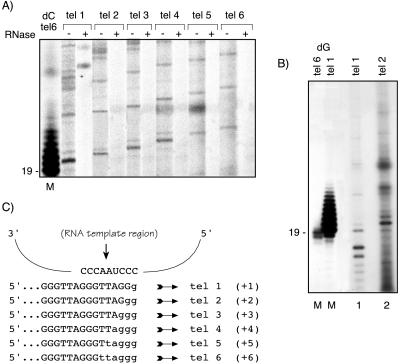
Telomeric primers (tel 6 and tel 1, 20 pmol of each) were elongated for 5 min at 25°C with 10 units of terminal deoxynucleotidyltransferase in the buffer supplied by the manufacturer (Promega) and 10 μCi of [α-32P]dCTP (3,000 Ci/mmol) or 0.4 μCi of [α-32P]dGTP (800 Ci/mmol). Note that in Figs. 2 and 3A, where the marker was tel 6-labeled by addition of dC residues, the smallest strong telomerase product from the tel 2 primer comigrates with a 21-nt band. In Fig. 3B, where the markers tel 6 and tel 1 were labeled with dG residues, the corresponding smallest band migrates with a 20-nt marker fragment. This and other data showed that the migration profile of the labeled standard markers depended on whether dG or dC residues were added by terminal transferase. This observation was crucial for the accurate identification of the +1 and +2 products resulting from elongation of tel 1 and tel 2 by T. brucei telomerase (Fig. 3B, lanes 1 and 2).
Figure 2.
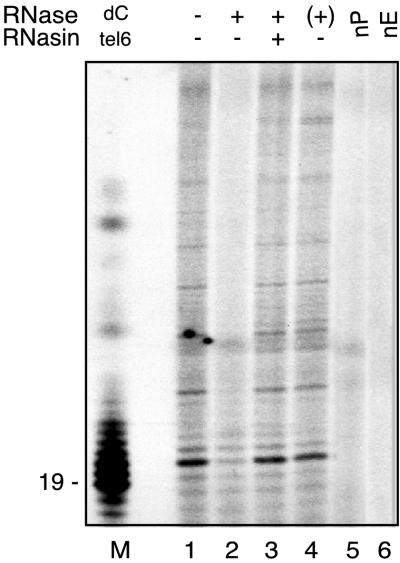
T. brucei activity monitored directly by telomerase primer-extension assay. Reactions were performed with DEAE fraction and primer tel 2. Lane 1, standard reaction; lane 2, extract pretreated with 100 ng of RNase A; lane 3, extract incubated with RNasin before addition of RNase A; lane 4, RNase A treatment after telomerase reaction (+); lane 5, nP, no input primer, lane 6, nE, extract substituted by reaction buffer; lane M, terminal deoxynucleotidyltransferase used to label tel 6 with [α-32P]dCTP (19 indicates the position of the primer plus 1-nt molecular weight marker).
Figure 3.
Determination of T. brucei telomerase template by direct synthesis. (A) Semipurified DEAE fraction was used as telomerase source to elongate permuted telomeric primers (see Table 1 for primer sequences and names). The most intense bands correspond to the putative template translocation/pause position. Duplicated reactions were performed in which extracts were or were not RNase A-pretreated as indicated (+ or −). Lane M is oligonucleotide tel 6 labeled with terminal transferase and [α-32P]dCTP. (B) Two different oligonucleotide (tel 6, lane 1 and tel 1, lane 2) were 3′ end labeled with terminal transferase and [α-32P]dGTP to demonstrate that primers labeled with dGTP more accurately indicate the primer +1 nucleotide position (19 on the left side of the gel). Lanes 3 and 5 show, respectively, standard telomerase reactions using oligonucleotides tel 1 and tel 2 as enzyme substrates. (C) Hypothetical alignment of the permuted primers in the putative T. brucei telomerase RNA template region. Capital letters represent the sequence of the input primers and small letters indicate nucleotides added by a single round of telomerase polymerization, according to the results obtained in A and B. The number of residues added to the 3′ end of each primer to reach the translocation/pause position is indicated in parentheses according to the results obtained in A and B.
RESULTS
Identification and Initial Characterization of Telomerase Activity in Kinetoplastid Extracts by a Modified TRAP.
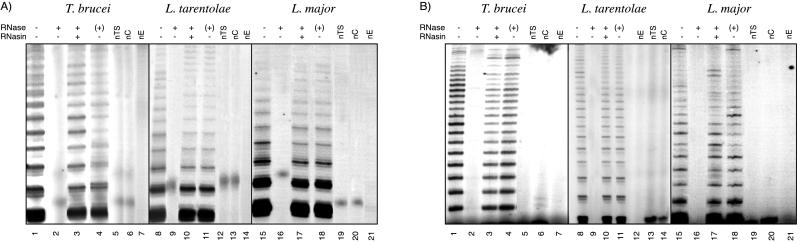
In initial attempts to detect telomerase activity in parasite extracts, the one-tube PCR-based TRAP (18) was used. Briefly, after elongation of TS (forward) primer by telomerase, the products were amplified by using the reverse primer (CX) that was separated from the first reaction mixture by a wax barrier. Use of CX as the reverse primer repeatedly gave false-positive products, presumably because of staggered annealing (primer-dimers) between TS and CX primers. Another reverse primer, CX-ext (39), eliminated the PCR artifacts and made the one-tube version of the TRAP more reliable for detection of telomerase activity. Fig. 1A shows the results of TRAP performed by using the CX-ext reverse primer and the relevant DEAE fractions of L. tarentolae, L. major, and T. brucei. The average sizes and intensities of the products fractionated in 10% denaturing gels were variable and differed with each parasite species, with T. brucei and L. tarentolae extracts showing a higher proportion of longer products than did L. major extracts (in Fig. 1A, compare lanes 1 and 8 with lane 15). With extracts from all three organisms, enzyme activity was abolished by pretreatment with 100 ng of RNase A (Fig. 1A, lanes 2, 9, and 16), whereas the addition of RNase A after the telomerase reaction (Fig. 1A, lanes 4, 11, and 18) or the incubation of extracts with the RNasin only (data not shown) did not affect the profile of TRAP products. Furthermore, RNasin (1 unit per reaction) protected the activity of each species from RNase A action (Fig. 1A, lanes 3, 10, and 17). Control reactions that omitted either one of the primers or the extract indicated that the activity was not the result of an artifact (Fig. 1A, lanes 5–7, 12–14, and 19–21). Semiquantitative assays showed that extracts of T. brucei and L. tarentolae, diluted 1:100 and 1:10, respectively, still yielded detectable TRAP products (data not shown).
Figure 1.
Telomerase activity in kinetoplastid parasites. Lanes 1–6, T. brucei DEAE eluate; lanes 8–13, L. tarentolae DEAE eluate; lanes 15–20, L. major DEAE eluate. Telomerase products were fractionated in 10% sequencing gels to reveal the periodicity of banding pattern. In lanes 2, 9, and 16, extracts were pretreated with RNase A; lanes 3, 10, and 17, RNasin incubated with extract before addition of RNase; lanes 4, 11, and 18, RNase A was added after telomerase step incubation (+). nTS, reaction performed without the forward primer; nC, reaction performed in the absence of CX-ext; nE, reaction in which the extracts were omitted. (A) One-tube TRAP using primer CX-ext as reverse primer and semipurified extracts. (B) Two-tube modified TRAP using CX-ext reverse primer. The assays were performed with half the amount of DEAE fractions used in A.
We also used another variant of the TRAP (a two-tube TRAP) in which the telomerase-replication step was carried out in a separate reaction tube from the subsequent PCR reaction (40). The results shown in Fig. 1B demonstrate that, although we used only half the amount of each parasite extract as in Fig. 1A, telomerase products appeared as discrete robust bands. Also, the higher sensitivity of the assay permitted detection of much longer products than those shown in Fig. 1A (Fig. 1B, lanes 1, 8, and 15). This higher sensitivity was apparent in serial titrations, in which activity was detectable in L. tarentolae and T. brucei extracts down to 1:10 and 1:100 dilutions, respectively, and in L. major extracts down to a 1:5 dilution (data not shown). The elongation activity was also sensitive to pretreatment with low levels of RNase A (Fig. 1B, lanes 2, 9, and 16), and the products were again unaffected by the addition of RNase A after the reaction (Fig. 1B, lanes 4, 11, and 18), or when RNasin was incubated with extracts prior to RNase A addition (Fig. 1B, lanes 3, 10, and 17). Omission of either TTP or dATP or replacement of one of the dNTPs by its ddNTP analog during the telomerase step abolished the synthesis of elongation products (data not shown). Similarly, reactions in which either one of the primers or the extract was omitted failed to generate any detectable PCR products (Fig. 1B, lanes, 5–7, 12–14, and 19–21). The results obtained with the two-tube TRAP confirmed that the polymerization activities detected by using parasite extracts fulfilled the essential criteria for telomerase activity.
Moderate Processivity of T. brucei Telomerase Activity Revealed by Using the Conventional Primer Extension Assay.
Several different primer-extension assay protocols were used to identify and characterize T. brucei, L. tarentolae, and L. major telomerase activity (1, 2, 42). We chose T. brucei activity as representative because it was the most readily detectable. By using DEAE-fractionated extracts of T. brucei and modifications of conventional assay protocols for human telomerase (23, 26, 41), we optimized reaction conditions and demonstrated additional features of the polymerization reaction, providing further evidence that the activity was attributable to telomerase. As shown in Fig. 2 (lanes 1, 3 and 4) telomerase products appeared as a series of radiolabeled bands longer than the input primer, forming ladders with a 6-nt periodicity. Polymerization primed by the tel 2 oligodeoxynucleotide was sensitive to pretreatment with a low concentration of RNase A (100 ng) and was protected from such pretreatment by the addition of RNasin before RNase A treatment (Fig. 2, lanes 2 and 3). The appearance of elongation products was unaffected when RNase A was added after substrate elongation (Fig. 2, lane 4). Only about six telomeric repeats were added to the 3′ end of the input primer. Thus, enzyme processivity may be moderately low, although it depended on the dGTP concentration in the reaction (data not shown); highest processivity was seen with over 8.0 μM dGTP.
Analysis of T. brucei Telomerase Activity and Templating Properties.
In previous reports, telomerases from the ciliate T. thermophila and human cells were shown to polymerize by transcribing up to the 5′-terminal nucleotide of the telomerase RNA templating domain (1, 5, 42, 43). In these species, after reaching this position, a strong band in the product profile marked the translocation and/or dissociation of telomerase from the elongated product. Consequently, when given substrates of identical length but consisting of different permutations of the telomeric repeat, telomerases produce ladders of products shifted according to the alignment of the 3′ end of the substrate relative to the telomerase template translocation or pause position. Because such results can be diagnostic of telomerase copying its RNA template region (refs. 1, 2, 5, 42, and 44, and reviewed in ref. 11), we performed similar primer-extension assays by using a series of primers, each bearing one of the six possible permutations of the telomeric repeat (5′-TTAGGG-3′) at its 3′ end (Table 1). The shifts in banding patterns obtained in each reaction were consistent with templated synthesis of TTAGGG telomeric repeats. By analogy to other previously characterized telomerases, the strongest band is likely to correspond to the last nucleotide (the 5′ end) of the internal template region of the telomerase RNA (Fig. 3A, − RNase A lanes and Fig. 3B, lanes 1 and 2). On the basis of these results with the set of permuted primers, we propose the 5′ residue of the template and a minimal 9-nt templating domain for T. brucei telomerase RNA to be 3′-CCCAAUCCC-5′, as shown in Fig. 3C. Until we obtain the sequence of the T. brucei telomerase RNA for direct comparison with our proposed template region, we will not be certain that we are not observing alternative pausing, as seen with yeast telomerases (45, 46). A Tetrahymena telomeric primer (see Table 1) and the G-rich nontelomeric TS primer (see Materials and Methods) also were used as substrates for T. brucei telomerase elongation but in both cases resulted in the addition of only one detectable repeat to each primer 3′ end (data not shown).
In some reactions, labeled products shorter than the input primer appeared sporadically (Fig. 3B, strong bands −2 and −3 in lane 1), and their origin is unknown. As their formation was not RNase A-sensitive (data not shown), they are probably not telomerase products shortened by a telomerase-associated DNA endonuclease, as has been demonstrated for Euplotes and Tetrahymena telomerase activities (47–49), but instead they must have arisen by some other DNA-labeling and -shortening mechanisms.
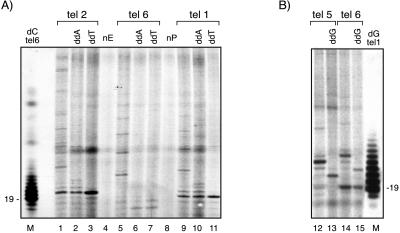
The telomerase-mediated DNA synthesis reaction also was analyzed by substituting one dNTP with its ddNTP analog (Fig. 4A and B). Consistent with the predicted T. brucei RNA template sequence shown in Fig. 3C, when telomerase assays were primed with tel 1 (ending in -AGG) and tel 2 (ending in -TAG) (Fig. 4A, lanes 9 and 1), reactions containing ddATP terminated after addition of 4 and 5 nt, respectively (Fig. 4A, lanes 2 and 10). Reactions containing ddTTP, however, terminated synthesis after addition of only 1 nt and 2 nt, respectively (Fig. 4A, lanes 11 and 3) suggesting that ddTTP is not efficiently incorporated by T. brucei telomerase activity. Both ddATP and ddTTP apparently blocked extension primed with tel 6 before it reached the position at which the first [α-32P]dG residue was incorporated, because no labeled product was visualized (Fig. 4A, compare lane 5 with lanes 6 and 7). These results are consistent with incorporation of radiolabeled dG by telomerase and polymerization up to the proposed 5′ end of the RNA template followed by translocation and initiation of a new round of synthesis until the addition of a chain-terminating ddNTP. Control reactions in which the input primer or the extract was omitted did not produce any radiolabeled reaction products (Fig. 4A, lanes 4 and 8). By using [α-32P]TTP as label, the elongation of tel 5 was blocked by ddGTP after addition of 3 nt to the primer, resulting in a chain termination product 2 nt shorter than the putative template translocation position (Fig. 4B, lanes 12 and 13). The results of telomerase extension of tel 6 in the presence of [α-32P]TTP (see lanes 14 and 15 of Fig. 4B) are consistent with a limiting level of the labeling dNTP in the reaction causing the telomerase extension reaction to pause or to stall after addition of a single residue. However, the addition of ddGTP as chain terminator in these reactions again yields a product of the predicted size (22 nt), corresponding to the expected termination 2 nt before the putative template translocation position (Fig. 4B, lanes 14 and 15).
Figure 4.
T. brucei telomerase-mediated template termination synthesis with ddNTPs. Telomerase reactions containing either [α-32P]dGTP (A) or [α-32P]TTP (B) and the telomerase substrate oligonucleotides indicated were tested for the effects of ddNTP chain terminators. Control reactions lacking chain terminators (lanes 1, 5, 9, 12, and 14) were compared with reactions in which dATP was substituted by ddATP (lanes 2, 6, and 10), TTP was substituted by ddTTP (lanes 3, 7, and 11), or dGTP was substituted by ddGTP (lanes 13 and 15). Size markers (lanes M) are terminal deoxynucleotidyltransferase-labeled oligonucleotides tel 6 with [α-32P]dCTP (A) and tel 1 labeled with [α-32P]dGTP (B). Lanes nE and nP are mock reactions lacking extract or primer, respectively.
In conclusion, a specific telomerase detection assay by using semipurified T. brucei extracts was successfully developed. This assay has given us useful information about enzyme activity and processivity. Furthermore, it has made possible the prediction of a minimum model for the RNA template region used by T. brucei telomerase.
DISCUSSION
We have identified telomerase activity in semipurified whole cell extracts of L. tarentolae, L. major, and T. brucei. The telomerase activities from all three parasite species generated products with a 6-nt periodicity consistent with synthesis of the expected TTAGGG telomeric repeats. Telomerase activity in DEAE-fractionated extracts was assayed by using modifications of the one- and two-tube TRAPs (18, 40), with the two-tube TRAP being the more sensitive. A conventional assay for telomerase using T. brucei extracts produced direct evidence of telomerase products without an intervening PCR amplification step, allowing us to verify and characterize telomerase activity and to propose a likely template region for the telomerase RNA of this species. This information should help identify a candidate telomerase RNA gene for this medically important human pathogen. Under our reaction conditions, we could not detect telomerase products by using crude S100 extracts, probably because of the presence of high concentrations of nucleases including RNase H (J.D. and N.A., unpublished observations). We were unable to detect telomerase activity in Leishmania extracts by using the direct assay. Perhaps this is a reflection of the greater number of linear chromosomes, and especially minichromosomes, in T. brucei.
The primer recognition and elongation properties of kinetoplastid telomerases appear to follow the general features of ciliate, human, and yeast telomerases (1, 2, 5, 42). Like their higher eukaryotic counterparts, the trypanosomatid activities generated a pattern of pausing after addition of each consecutive telomeric repeat to the substrate (Figs. 1, 2, and 3). This pattern most likely reflects either translocation or dissociation and reassociation with the substrate oligonucleotide between rounds of elongation. The preferential use of the permuted trypanosome telomeric sequence primers over other sequences (for example, Tetrahymena telomeric oligonucleotide and TS oligonucleotide) by the T. brucei activity (data not shown) also is consistent with similar preferences of telomerases of other organisms for templates that match the sequence of the endogenous telomeres. For example, telomerase derived from vegetatively growing Euplotes crassus adds only a single dG residue to the 3′ end of nontelomeric oligos (50). These observations combined with the results of the chain termination reactions (Fig. 4) are consistent with the addition of the expected TTAGGG repeats found at telomeres in T. brucei (33, 34).
In addition to their presumed role in the maintenance of chromosome integrity, telomeres and subtelomeric regions in the kinetoplastid protozoa may have further significance for parasite survival. In T. brucei and in Leishmania spp., genes whose expression is regulated in proximity to telomeres have been characterized. For example, in bloodstream-form trypanosomes, the prinicipal expression sites for the variable surface glycoprotein (VSG) antigens are located at telomeres. Whereas silent or basic copy VSG genes are located at internal chromosomal positions or at inactive telomeres, a necessary but not sufficient condition for their expression is their translocation to a telomeric site known as the expression site (ES) (51, 52). The expressed VSG gene is located 1–3 kb from the start of the repetitive hexameric telomeric sequence, organized in a long polycistronic operon (51). Another example in which the chromosomal environment is exploited to control gene expression is the Leishmania circular and/or linear LD1 (Leishmania DNA 1) elements or minichromosomes. These are a multicopy family of amplicons of unknown function that arise spontaneously from a subtelomeric region of the 1.9-Mb chromosome (reviewed in ref. 32). LD1 elements were found containing one or more different genes that are frequently duplicated and overexpressed after the translocation of LD1 to another chromosome locus (32, 53). Thus, there is substantial evidence that gene expression in kinetoplastid parasites can be controlled by dynamic genomic reconfigurations that remove or insert critical genes at subtelomeric regions, where the telomeric environment can be exploited to silence or, as in African trypanosomes, selectively activate transcription.
Efforts should be directed at testing whether perturbing telomere replication by targeting telomerase will affect telomeric gene expression and subsequent parasite growth. Although it is known that some organisms can use other nontelomerase-mediated mechanisms for telomere maintenance (reviewed in refs. 51 and 54), telomerase could be an effective target for the development of new therapeutic strategies for the treatment of parasitic disease. The kinetoplastid parasites, with their relevant animal models and ability to tolerate genetic manipulation, should provide powerful experimental systems for reaching these goals.
Acknowledgments
We thank members of the Blackburn and Agabian laboratories for the helpful discussions. M.I.C. is a postdoctoral fellow of the Pew for Latin American Fellows Program and FAPESP 96/94122–9 (Fundaçao Amparo a Pesquisa do Estado de Sao Paulo, Brazil). This research was funded in part by National Institutes of Health Grants AI21975 (to N.A.) and GM26259 (to E.H.B.).
ABBREVIATION
- TRAP
telomere repeat amplification protocol
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Greider C, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 2.Cohn M, Blackburn E H. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 3.Prowse K R, Greider C. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantell L, Greider C. EMBO J. 1994;13:3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morin G B. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald M, Mcknight T, Shippen D. Proc Natl Acad Sci USA. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botius E, Bakhsis N, Sherf A. Mol Cell Biol. 1998;18:919–925. doi: 10.1128/mcb.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shippen-Lentz D, Blackburn E H. Science. 1990;274:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 9.Yu G L, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 10.Blasco M A, Lee H-W, Hande P, Samper E, Landsorp P, DePinho R, Greider C. Cell. 1997;91:25–43. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Romero D P, Blackburn E H. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 12.Nugent C, Lundblad V. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 13.Lingner J, Hughes T, Shevchenko A, Mann M, Lundblad V, Cech T. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T, Morin G, Chapman K, Weinrich S, Andrews W, Lingner J, Harley C, Cech T. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 15.Collins K, Gandhi L. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan T, Sperger J, Chapman K, Cech T. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura T, Cech T. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 18.Kim N, Piatyszek M, Prowse K, Harley C, West M, Ho P, Coviello G, Wright W, Weinrich S, Shay J. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 19.Broccoli D, Young J, de Lange T. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor R S, Ramirez R D, Ogoshi M, Chaffins M, Piatyszek M A, Shay J W. J Invest Dermatol. 1996;106:759–765. doi: 10.1111/1523-1747.ep12345811. [DOI] [PubMed] [Google Scholar]
- 21.Bodnar A, Oullette M, Frolkis M, Holt S, Chiu C, Morin G, Harley C, Shay J, Lichtsteiner S, Wright W. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 22.Counter C M, Meyerson M, Eaton E N, Weinberg R A. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinrich S, Pruzan R, Oullete M, Tesmer V, Holt S, Bodnar A, Lichtsteiner S, Kim N, Trager J. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 24.Harley C B, Futcher A B, Greider C. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 25.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 26.Strahl C, Blackburn E H. Mol Cell Biol. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 28.Stuart K, Allen T E, Heidmann S, Seiwert S D. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanzer M, Fischer K, Le Blancq S. Mol Biochem Parasitol. 1995;70:1–8. doi: 10.1016/0166-6851(95)00021-r. [DOI] [PubMed] [Google Scholar]
- 30.Crozatier M, Van der Ploeg L, Gommers-Ampt J, Borst P. Mol Biochem Parasitol. 1990;42:1–12. doi: 10.1016/0166-6851(90)90107-w. [DOI] [PubMed] [Google Scholar]
- 31.Henderson E. In: Telomeres. Blackburn E, Greider C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 11–34. [Google Scholar]
- 32.Segovia M, Ortiz G. Parasitol Today. 1997;13:342–348. doi: 10.1016/s0169-4758(97)01111-3. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn E H, Challoner P B. Cell. 1984;36:447–457. doi: 10.1016/0092-8674(84)90238-1. [DOI] [PubMed] [Google Scholar]
- 34.van der Ploeg L H T, Lin A Y C, Borst P. Cell. 1984;36:459–468. doi: 10.1016/0092-8674(84)90239-3. [DOI] [PubMed] [Google Scholar]
- 35.Meyne J, Ratliff R L, Moyzis R K. Proc Natl Acad Sci USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berens R, Man J. J Parasitol. 1978;64:160–164. [PubMed] [Google Scholar]
- 37.Bienen E J, Hammadi E, Hill G C. Exp Parasitol. 1981;51:408–417. doi: 10.1016/0014-4894(81)90128-4. [DOI] [PubMed] [Google Scholar]
- 38.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krüpp G, Köhne K, Tamm S, Klapper W, Heidorn K, Rott A, Parwaresch R. Nucleic Acids Res. 1997;25:919–921. doi: 10.1093/nar/25.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Autexier C, Pruzan R, Funk W, Greider C. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 41.Counter C M, Avillion A, Lefeuvre C, Stewart N, Greider C, Harley C, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilley D, Lee M S, Blackburn E H. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 43.Feng J, Funk W, Wang S, Weinrich S, Avilion A, Chiu C, Adams R, Chang E, Allsopp R, Yu J, et al. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 44.Shippen-Lentz D, Blackburn E H. Mol Cell Biol. 1989;9:2761–2764. doi: 10.1128/mcb.9.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prescott J, Blackburn E H. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 46.Fulton T B, Blackburn E H. Mol Cell Biol. 1998;18:4961–4970. doi: 10.1128/mcb.18.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melek M, Greene E, Shippen D. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greene E, Bednenko J, Shippen D. Mol Cell Biol. 1998;18:1544–1552. doi: 10.1128/mcb.18.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharyya A, Blackburn E. Proc Natl Acad Sci USA. 1997;94:2823–2827. doi: 10.1073/pnas.94.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bednenko J, Melek M, Greene E, Shippen D. EMBO J. 1997;16:2507–2518. doi: 10.1093/emboj/16.9.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Ploeg L, Gottesdiener K, Tse D, Chung H, Weiden M. Mechanisms of Eukaryotic DNA Recombination. London: Academic; 1992. pp. 179–187. [Google Scholar]
- 52.Borst P, Greaves D. Science. 1987;235:658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- 53.Tripp C A, Myler P J, Stuart K. Mol Biochem Parasitol. 1991;47:151–156. doi: 10.1016/0166-6851(91)90174-5. [DOI] [PubMed] [Google Scholar]
- 54.Pryde F, Gorham H, Louis E. Genom Evol. 1995;34:822–828. [Google Scholar]