Abstract
Oxidative base lesions, such as 8-oxoguanine (8-oxoG), accumulate in nuclear and mitochondrial DNAs under oxidative stress, resulting in cell death. However, it is not known which form of DNA is involved, whether nuclear or mitochondrial, nor is it known how the death order is executed. We established cells which selectively accumulate 8-oxoG in either type of DNA by expression of a nuclear or mitochondrial form of human 8-oxoG DNA glycosylase in OGG1-null mouse cells. The accumulation of 8-oxoG in nuclear DNA caused poly-ADP-ribose polymerase (PARP)-dependent nuclear translocation of apoptosis-inducing factor, whereas that in mitochondrial DNA caused mitochondrial dysfunction and Ca2+ release, thereby activating calpain. Both cell deaths were triggered by single-strand breaks (SSBs) that had accumulated in the respective DNAs, and were suppressed by knockdown of adenine DNA glycosylase encoded by MutY homolog, thus indicating that excision of adenine opposite 8-oxoG lead to the accumulation of SSBs in each type of DNA. SSBs in nuclear DNA activated PARP, whereas those in mitochondrial DNA caused their depletion, thereby initiating the two distinct pathways of cell death.
Keywords: 8-oxoguanine, mitochondria, MUTYH, nucleus, OGG1
Introduction
Reactive oxygen species (ROS), generated as byproducts of mitochondrial respiration or as a consequence of exposure to environmental agents, are known to oxidize DNA. Oxidative damage to cellular DNA often causes mutagenesis as well as programmed cell death; and the former might result in carcinogenesis, whereas the latter often causes degenerative disorders (Ames et al, 1993; Nakabeppu et al, 2006, 2007).
8-Oxoguanine (8-oxoG) is one of the major oxidative base lesions in DNA or nucleotides (Kasai and Nishimura, 1984), and is highly mutagenic as it can pair with adenine as well as cytosine (Maki, 2002). Studies of error-avoiding mechanisms directed against 8-oxoG revealed that organisms are equipped with elaborate means of minimizing the accumulation of such lesions in DNA, and that these strategies are conserved from bacteria to mammals. In mammals, MutT homolog-1 (MTH1) hydrolyzes oxidized purine nucleoside triphosphates to the monophosphate forms (Nakabeppu et al, 2006). 8-OxoG DNA glycosylase-1 (OGG1) excises 8-oxoG paired with cytosine in DNA (Boiteux and Radicella, 2000; Nakabeppu et al, 2006), whereas MutY homolog (MUTYH) removes adenine misincorporated opposite 8-oxoG in template DNA (Slupska et al, 1999; Nakabeppu et al, 2006). These three enzymes play major roles in suppressing spontaneous mutagenesis initiated by oxidation of nucleic acids. Mutant mice lacking one of these genes exhibit an increased spontaneous mutation rate and an increased susceptibility to carcinogenesis (Nakabeppu et al, 2006).
Several isoforms of human OGG1 (hOGG1) are known to be generated by alternative splicing, all of which share their N-terminal end with the common mitochondrial targeting signal. The two major isoforms are hOGG1-1a and hOGG1-2a; the former carries the nuclear localization signal at its C-terminal end and is localized in the nucleus, while the latter carries a hydrophobic/acidic region at its C-terminal end and is localized in mitochondria (Nishioka et al, 1999). 8-OxoG accumulated in either nuclear (nDNA) or mitochondrial DNA (mtDNA) is known to be efficiently repaired in mammals by OGG1-initiated base excision repair (BER) (Stuart et al, 2005).
8-OxoG accumulates in cellular DNA during aging and the level of accumulation is known to increase in patients with various neurodegenerative diseases such as Parkinson's disease (PD) or Alzheimer's disease (AD) (Shimura-Miura et al, 1999; Nunomura et al, 2001), suggesting that an accumulation of 8-oxoG in cellular DNA contributes to cell death. MTH1-null mice exhibited a strong accumulation of 8-oxoG in mtDNA of striatal nerve terminals of dopamine neurons, accompanied by the increased degeneration of these terminals after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration (Yamaguchi et al, 2006). Furthermore, in MTH1-null mouse embryonic fibroblasts (MEFs), accumulation of 8-oxoG in cellular DNA was induced by H2O2, resulting in mitochondrial dysfunction and finally cell death (Yoshimura et al, 2003). However, it is not known which form of DNA was responsible for such cell death, whether nDNA or mtDNA or both, nor is it known how the death order was executed.
In the present study, we found that OGG1-null MEFs exhibited an increased rate of cell death under oxidative stress, in comparison to the wild type. To elucidate the effects on cell death of 8-oxoG accumulated in either nDNA or mtDNA, we established and characterized MEF lines which selectively accumulate 8-oxoG in either type of DNA by expressing nuclear or mitochondrial forms of hOGG1.
Results
Ogg1−/− MEFs are highly susceptible to cell death under oxidative stress
Spontaneously immortalized MEF lines were established from wild-type and Ogg1−/− mouse embryos (Supplementary Figure S1). 8-OxoG DNA glycosylase activity was not detected in the extracts prepared from Ogg1−/− MEFs (Supplementary Figure S1). We then examined whether the deficiency in OGG1 altered susceptibility to cell death caused by 2-methyl-1.4-naphthoquinone (menadione), which produces ROS within cells (Supplementary Figure S2; Frei et al, 1986). The median lethal dose of menadione (LD50=18.5 μM) for Ogg1−/− MEFs was significantly lower than that for the wild type (32 μM) (Figure 1A).
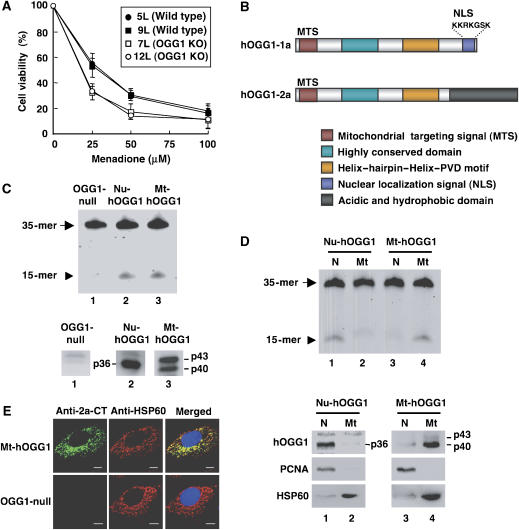
Figure 1.
Establishment of Ogg1−/− MEFs expressing either the nuclear or mitochondrial form of human OGG1. (A) Cell viability 24 h after exposure to menadione. Results from three independent experiments, each of which was run in triplicate, are shown (mean±s.d.). The Tukey–Kramer test revealed that cell viability with 25 and 50 μM menadione was significantly different between wild-type and Ogg1−/− MEFs, with a P-value <0.05. (B) Structural comparison of hOGG1 proteins. hOGG1-1a is a nuclear form with a C-terminal nuclear localization signal (NLS), while hOGG1-2a is a major mitochondrial form with a C-terminal acidic and hydrophobic domain. MTS, mitochondrial targeting signal. (C) 8-OxoG DNA glycosylase in whole-cell extracts was determined as an activity introducing a nick adjacent to 8-oxoG opposite cytosine in duplex oligonucleotides, in which 8-oxoG was placed at the sixteenth residue in a 5′-fluorescence-labeled oligonucleotide (upper panel). The arrow and arrowhead indicate the substrate (35-mer) and the cleaved product (15-mer), respectively. Western blotting analysis was performed using anti-HCD (lanes 1 and 2) or anti-2a-CT (lane 3), (lower panels). Whole-cell extracts (20 μg of protein) from OGG1-null (lane 1), Nu-hOGG1 (lane 2), and Mt-hOGG1 MEFs (lane 3) were analyzed. (D) 8-OxoG DNA glycosylase activity in nuclear or mitochondrial extracts (upper panel). Detection of hOGG1 in nuclear or mitochondrial extracts (lower panels). PCNA (35.5 kDa) or HSP60 (60 kDa) were detected as a nuclear or mitochondrial marker. Nuclear (N) and mitochondrial (Mt) extracts (5 μg of protein) from Nu-hOGG1 (lanes 1 and 2) and Mt-hOGG1 (lanes 3 and 4) were analyzed. (E) Intracellular distribution of hOGG1-2a in Mt-hOGG1 MEFs. Bar, 10 μm.
Establishment of Ogg1−/− MEF lines expressing either the nuclear or mitochondrial form of human OGG1 protein
To establish cell lines carrying a functional 8-oxoG DNA glycosylase in either nuclei alone or mitochondria alone, a cDNA expression vector for either the nuclear (hOGG1-1a) or mitochondrial (hOGG1-2a) form of hOGG1 (Figure 1B) was introduced into Ogg1−/− MEFs (7L). The 8-oxoG DNA glycosylase activity that was absent in OGG1-null MEFs was recovered only in the nuclear extract of Nu-hOGG1 MEFs or only in the mitochondrial extract of Mt-hOGG1 MEFs (Figure 1C and D, upper panels). A 36-kDa hOGG1-1a polypeptide was detected only in a nuclear extract prepared from Ogg1−/− MEFs carrying hOGG1-1a cDNA. A 40-kDa polypeptide and to a lesser extent, its precursor, a 43-kDa polypeptide, were detected only in a mitochondrial extract prepared from MEFs carrying hOGG1-2a cDNA (Figure 1C and D, lower panels), and these were mostly colocalized with a mitochondrial protein, HSP60 (Figure 1E). These cell lines were therefore designated Nu-hOGG1 MEF and Mt-hOGG1 MEF, respectively. Ogg1−/− MEFs into which an empty vector was introduced as a control, were designated the OGG1-null MEF line.
8-OxoG accumulated in nDNA or mtDNA independently induces cell death
Next, we examined the 8-oxoG content in nDNA and mtDNA of each MEF line after exposure to menadione. The content of 8-oxoG in nDNA was determined by HPLC-MS/MS (Figure 2A). In all types of MEFs, the nuclear content of 8-oxoG increased to more than twofold above the basal level during the 1 h exposure to menadione. The content in Nu-hOGG1 MEFs, as well as in wild-type MEFs, was significantly decreased within 1 h after the exposure, and returned to the basal level 24 h after exposure, suggesting that there are rapid and slow rates of repair of 8-oxoG in nDNA. In contrast, the levels remained high in OGG1-null and Mt-hOGG1 MEFs 24 h after exposure.
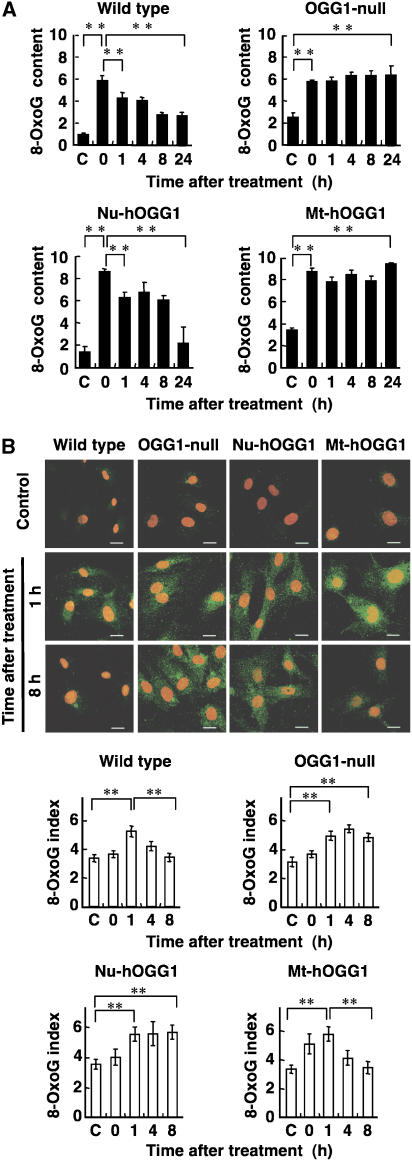
Figure 2.
OGG1 deficiency results in the accumulation of 8-oxoG in nDNA or mtDNA. (A) 8-OxoG accumulation in nDNA after exposure to menadione. The content of 8-oxoG in nDNA (8-oxoG residues per 106 residues of guanine) prepared from wild-type, OGG1-null, Nu-hOGG1, and Mt-hOGG1 MEFs was determined by HPLC-MS/MS analysis at certain time points (0–24 h) after a 60-min exposure to 50 μM menadione. Results from one of two independent experiments are presented (mean±s.d., n=3 per experiment). C, control with no exposure. Student's t-test, **P<0.01. (B) 8-OxoG accumulation in mtDNA after exposure to menadione. The top panels show 8-oxoG immunoreactivity for mtDNA of wild-type, OGG1-null, Nu-hOGG1, and Mt-hOGG1 MEFs at 1 and 8 h, after a 60-min exposure to 50 μM menadione. Nuclei were counterstained with PI. Control, no exposure. Bar, 20 μm. The bottom graphs show the 8-oxoG index representing mitochondrial 8-oxoG immunoreactivity in each cell (mean±s.e.m., 30 cells). Student's t-test, **P<0.01.
The content of 8-oxoG in mtDNA was monitored by immunofluorescence microscopy (Figure 2B). Cytoplasmic 8-oxoG immunoreactivities were colocalized with mitochondrial transcription factor A (TFAM), which is known to bind mtDNA (Alam et al, 2003), in Nu-hOGG1 MEFs 1 h after exposure to menadione (Supplementary Figure S3A). Since the 8-oxoG immunoreactivities were greatly reduced by pretreatment with either MutM, a bacterial 8-oxoG DNA glycosylase or DNase I, we concluded that the cytoplasmic 8-oxoG immunoreactivities represent the level of 8-oxoG in mtDNA (Supplementary Figure S3B). The 8-oxoG index indicating the level of 8-oxoG present in mtDNA increased to 1.5-fold or more above the basal level within 1 h after exposure to menadione in all types of MEFs. This may be because mitochondria damaged by menadione gradually undergo increased formation of ROS. In Mt-hOGG1 MEFs, as well as in wild-type MEFs, the 8-oxoG index was significantly decreased 8 h after exposure, whereas indices in OGG1-null and Nu-hOGG1 MEFs remained high. These results indicated that repair of 8-oxoG in mtDNA has faster kinetics than in nDNA, suggesting that the chromosomal structure of nDNA might be responsible for the slow repair of 8-oxoG in this latter material.
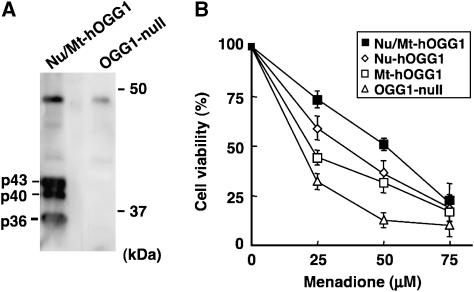
We further established Ogg1−/− MEFs expressing both the nuclear and mitochondrial forms of hOGG1, namely cells of the Nu/Mt-hOGG1 MEF line, and then compared their viabilities after exposure to menadione (Figure 3A and B). The LD50 were 18 μM for OGG1-null, 23 μM for Mt-hOGG1, 35 μM for Nu-hOGG1, and 51 μM for Nu/Mt-hOGG1 MEFs. The former three types of MEFs exhibited the same order of sensitivity to menadione in a colony-forming assay (data not shown). These results demonstrated that the nuclear and mitochondrial forms of hOGG1 independently initiated BER of the 8-oxoG accumulated in each type of DNA, and incrementally improved cell viability under oxidative stress. MEFs expressing both forms exhibited the strongest resistance, suggesting that a deficiency in the repair ability in either nDNA or mtDNA induces cell death.
Figure 3.
The accumulation of 8-oxoG in nDNA or mtDNA independently causes cell death. (A) Establishment of OGG1-null MEFs expressing both the nuclear and mitochondrial forms of hOGG1. Expression of both hOGG1 proteins was confirmed by western blotting analysis using anti-HCD. (B) Cell viability 24 h after exposure to menadione. Results from three independent experiments, each of which was run in triplicate, are presented (mean±s.d.). The Tukey–Kramer test revealed that the cell viability was significantly different between each type of MEF with 25 and 50 μM menadione (P-value<0.05) except between Nu-hOGG1 and Mt-hOGG1 MEFs with 50 μM menadione.
Twenty-four hours after exposure to 25 μM menadione, 42.3% of cells remaining in the culture of OGG1-null MEFs were dead cells that were stained with both Hoechst and PI, and their nuclei were condensed but not fragmented, indicating that they were pyknotic (data not shown). Cultures of Nu-hOGG1 and Mt-hOGG1 MEFs contained more cells than the culture of OGG1-null MEFs, and 26 and 30.9% of the remaining cells in the former cultures showed evidence of pyknosis, respectively. There were far fewer pyknotic cells (6.3%) in the Nu/Mt-hOGG1 MEF culture.
8-OxoG accumulated in nDNA activates poly-ADP-ribose polymerase accompanied by nuclear translocation of apoptosis-inducing factor, resulting in caspase-independent cell death
To elucidate whether caspase(s) is involved in menadione-induced cell death, MEF lines were exposed to menadione or staurosporine in the presence of a general caspase inhibitor, Z-VAD-fmk. Z-VAD-fmk efficiently suppressed staurosporine-induced cell death, which is dependent on caspases (Yoshimura et al, 2003), but did not suppress menadione-induced death (Supplementary Figure S4). We thus concluded that the menadione-induced cell death was independent of caspase activity.
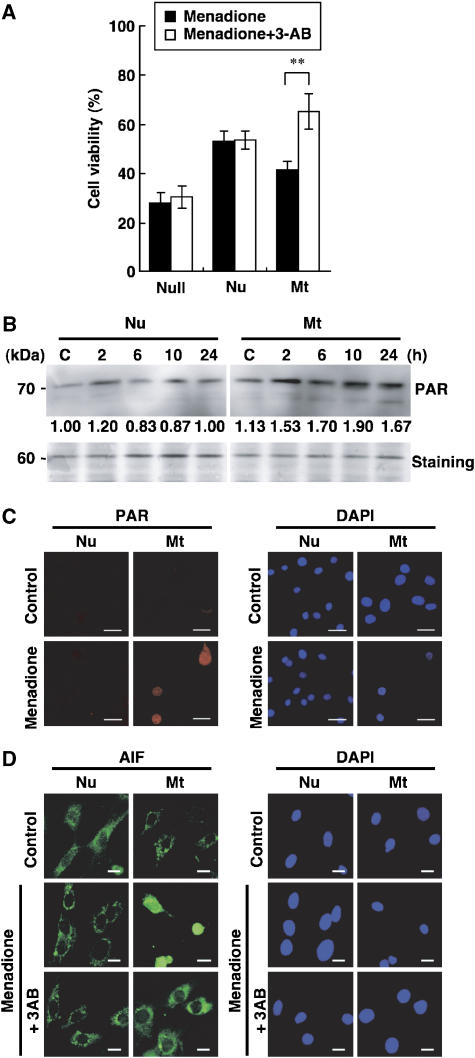
Next, MEFs were pre-cultured in the presence of 3-aminobenzamide (3-AB), an inhibitor of poly-ADP-ribose polymerase (PARP), and then exposed to menadione (Figure 4A). Only the Mt-hOGG1 MEFs exhibited a significantly increased viability (+25.6%), in comparison with that in the absence of 3-AB. In contrast, OGG1-null but not Nu-hOGG1 MEFs exhibited only a slightly increased viability. Western blotting and immunofluorescence microscopy revealed that the exposure to menadione increased poly-ADP ribosylation only in Mt-hOGG1 MEFs (Figure 4B and C).
Figure 4.
Menadione-induced death of Mt-hOGG1 MEFs is mediated by PARP activation with PARP-dependent nuclear translocation of AIF. (A) Suppression of menadione-induced death of Mt-hOGG1 MEFs by a PARP inhibitor. Each type of MEF was exposed to 50 μM menadione in the presence or absence of 10 mM 3-AB and was incubated in fresh medium with or without 3-AB for 24 h, and cell viability was determined. Results from three independent experiments, each of which was run in triplicate, are presented (mean±s.d.). Student's t-test, **P<0.01. (B) Increased poly ADP-ribosylation in Mt-hOGG1 MEFs after exposure to menadione. Nuclear extracts (5 μg of protein) prepared from each type of MEF at the indicated times after exposure to 50 μM menadione were subjected to western blotting with anti-PAR. PAR immunoreactivity of the 70-kDa band in each lane (upper panels) was normalized by the staining intensity of the 60-kDa band in each respective lane of a gel stained with GelCode Blue Stain (lower panel, staining), and the relative PAR immunoreactivity of each sample to that of the untreated Nu-hOGG1 sample (C) is shown under each lane in the upper panels. (C) Intracellular localization of PAR. Each type of MEF was exposed to 50 μM menadione for 60 min, and then incubated in fresh medium for 6 h. Nuclei were stained with DAPI. Control, no exposure. Bar, 20 μm. (D) Intracellular localization of AIF. Each type of MEF was exposed to 50 μM menadione in the presence or absence of 10 mM 3-AB for 60 min, and then incubated in fresh medium with or without 3-AB for 6 h. Nuclei were stained with DAPI. Control, no exposure. Bar, 20 μm.
It is known that PARP-dependent cell death is accompanied by nuclear translocation of apoptosis-inducing factor (AIF) (Yu et al, 2002). We thus examined the intracellular localization of AIF. As shown in Figure 4D, only Mt-hOGG1 MEFs exhibited an exclusive nuclear localization of AIF 6 h after exposure to menadione, which was completely abolished in the presence of 3-AB.
8-OxoG accumulated in mtDNA activates calpains accompanied by mitochondrial dysfunction, resulting in caspase-independent cell death
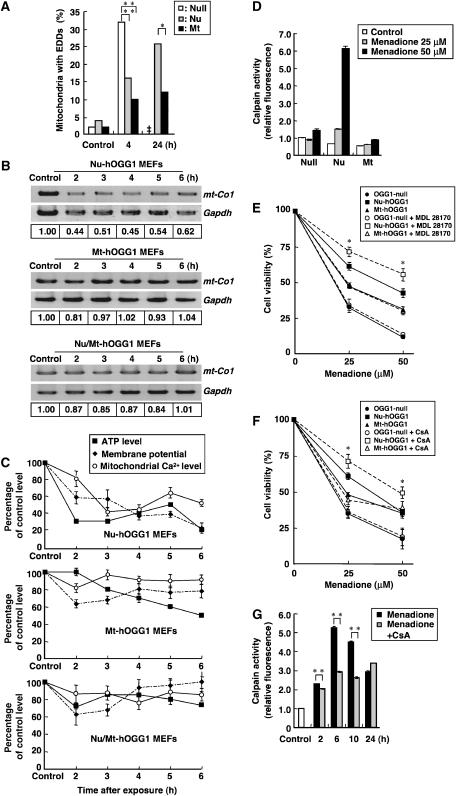
Menadione-induced ultrastructural alterations were examined by electron microscopy. No alteration in the nuclear structure was observed (data not shown), whereas mitochondrial cristae in OGG1-null and Nu-hOGG1 MEFs were highly degenerated and electron-dense deposits (EDDs), which are the hallmark of mitochondrial dysfunction under oxidative damage (Yoshimura et al, 2003), had accumulated in degenerating mitochondria from 4 to 24 h after exposure to menadione (Figure 5A). Swollen mitochondria were also observed in Nu-hOGG1 MEFs after the exposure (data not shown). In OGG1-null MEFs, mitochondria were hardly identified 24 h after exposure, indicating that most of their mitochondria were degraded. Since degenerating mitochondria were infrequently observed in Mt-hOGG1 MEFs, we concluded that 8-oxoG accumulated in mtDNA triggered this mitochondrial degeneration.
Figure 5.
Menadione-induced death of Nu-hOGG1 MEFs is accompanied by mitochondrial dysfunction and activation of the calpain pathway. (A) Suppression of mitochondrial degeneration by hOGG1-2a. At least 50 mitochondrial sections of MEFs were examined, and percentages of mitochondria with EDDs are shown. In OGG1-null MEFs, most mitochondria were degenerated and hardly identifiable 24 h after the exposure (‡). Fisher's exact probability test, *P<0.05, **P<0.01. (B) Depletion of mtDNA in each type of MEF after exposure to menadione. mt-Co1 DNA was amplified from total cellular DNA prepared from MEFs, which were harvested 2–6 h after a 60-min exposure to 50 μM menadione. The intensity of mt-Co1 DNA was normalized to that of nuclear Gapdh DNA, and the relative intensity of each sample to that of the control sample is shown at the bottom of each lane. (C) Time-dependent alterations in the intracellular ATP level, mitochondrial membrane potential, and mitochondrial Ca2+ level in each type of MEF after exposure to menadione. The ATP index in each type of MEF after a 60-min exposure to 50 μM menadione was determined, and levels are shown as a percentage of the untreated control (mean±s.e.m., n=6). The mitochondrial membrane potential was monitored by JC-1 and the mitochondrial Ca2+ level was monitored with rhod-2. Fluorescence intensities of JC-1 and rhod-2 of 30 and 50 cells, respectively, were examined, and are shown as a percentage of the untreated control (mean±s.e.m.). Top panel, Nu-hOGG1 MEFs; middle panel, Mt-hOGG1 MEFs; bottom panel, Nu/Mt-hOGG1 MEFs. (D) Activation of calpain after exposure to menadione. Calpain activity in a whole-cell extract prepared from OGG1-null (Null), Nu-hOGG1 (Nu), and Mt-hOGG1 MEFs (Mt) was determined 6 h after exposure to menadione (25, 50 μM). Control, no exposure. Relative calpain activities normalized to that of the untreated control for OGG1-null MEFs are shown (mean±s.d., n=3 per experiment). (E) Suppression of cell death by calpain inhibitor. OGG1-null, Nu-hOGG1, and Mt-hOGG1 MEFs were preincubated in the presence or absence of 20 μM MDL28170 (+MDL28170) for 60 min and were exposed to menadione for 60 min. Cell viability at 24 h after the exposure was determined by the trypan blue exclusion test. Results from three independent experiments, each of which was run in triplicate, are presented (mean±s.d.). Student's t-test with or without MDL28170, *P<0.05. (F) Suppression of cell death by CsA. OGG1-null, Nu-hOGG1, and Mt-hOGG1 MEFs were preincubated for 60 min in the presence or absence of 100 nM CsA, and then exposed to 50 μM menadione for 60 min. Cell viability at 24 h after the exposure was determined by the trypan blue exclusion test, and results from three independent experiments, each of which was run in triplicate, are presented (mean±s.d.). Student's t-test with or without CsA, *P<0.05. (G) Suppression of calpain activation by CsA. Nu-hOGG1 MEFs preincubated for 60 min in the presence or absence of 100 nM CsA were exposed to menadione (25, 50 μM) for 60 min, then calpain activity was determined periodically (mean±s.d., n=3 per experiment). Relative calpain activities normalized to that of an untreated control (open bar) are shown. Student's t-test, **P<0.01.
To delineate the process of mitochondrial degeneration and cell death triggered by 8-oxoG accumulation in mtDNA, we examined the levels of mtDNA, ATP, mitochondrial membrane potential, and Ca2+ in Nu-hOGG1, Mt-hOGG1, and Nu/Mt-hOGG1 MEFs. In Nu-hOGG1 MEFs but not the other two MEF lines, the level of mt-Co1 DNA encoding cytochrome c oxidase I was significantly decreased within 2 h after the exposure, compared with nuclear Gapdh DNA (Figure 5B). The intracellular ATP level was also rapidly decreased to 30% of the level of the control in Nu-hOGG1 MEFs within 2 h of the exposure, whereas the level in Mt-hOGG1 or Nu/Mt-hOGG1 MEFs gradually decreased to 60 or 80% that of the control within 6 h of the exposure (Figure 5C). Furthermore, in Nu-hOGG1 MEFs, the mitochondrial membrane potential and mitochondrial Ca2+ level started decreasing 2 h after the exposure (Figure 5C). These results indicate that 8-oxoG accumulation in mtDNA resulted in rapid depletion of mtDNA and intracellular ATP, followed by a decline in the mitochondrial membrane potential, namely, mitochondrial membrane permeability transition (MMPT), which further allowed Ca2+ to leave mitochondria prior to cell death.
The Ca2+ efflux from mitochondria might have resulted in activation of harmful calcium-dependent proteases in the cytoplasm such as calpains, which are known to mediate some types of cell death (Saito et al, 1993; Bizat et al, 2003). We thus examined calpain activation in each MEF line after exposure to menadione. Calpain activity in whole-cell extracts had remarkably increased 10-fold in Nu-hOGG1 MEFs 6 h after exposure to 50 μM menadione (Figure 5D). A calpain inhibitor, MDL28170, significantly suppressed the menadione-induced death of Nu-hOGG1 MEFs, but not that of the other two types of MEFs (Figure 5E). We also observed that the menadione-induced death of OGG1-null MEFs was partly suppressed in the presence of both 3-AB and MDL28170 (Supplementary Figure S5).
We next examined whether cyclosporin A (CsA), which suppresses MMPT by inhibiting cyclophilin D present in the MMPT pore (Baines et al, 2005), would increase the viability of each type of MEF after exposure to menadione. As a result, the viability of Nu-hOGG1 MEFs but not that of the other two types was significantly improved in the presence of CsA (Figure 5F). The increased calpain activity in Nu-hOGG1 MEFs after the exposure was also markedly suppressed in the presence of CsA (Figure 5G).
Furthermore, we observed that Nu-hOGG1 and Mt-hOGG1 MEFs exhibited a partial resistance to H2O2-induced cell death accompanying the activation of calpain in the former or the nuclear translocation of AIF in the latter, respectively (Supplementary Figure S6), thus confirming that the two distinct cell death pathways are also induced by H2O2 as well as by the intracellular ROS generated in cells after exposure to menadione.
Accumulation of 8-oxoG in either nuclear or mtDNA results in buildup of DNA single-strand breaks, which depends on the action of MUTYH
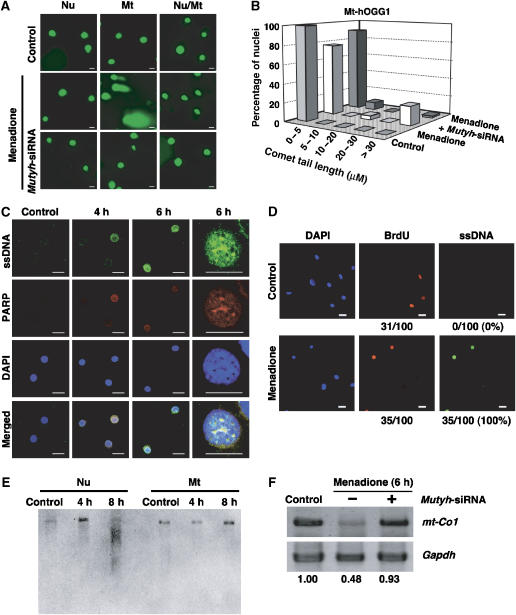
PARP was activated before cell death triggered by the accumulation of 8-oxoG in nDNA, suggesting that DNA single-strand breaks (SSBs), which are known to activate PARP (de Murcia and Menissier de Murcia, 1994), have accumulated in the nDNA. We therefore examined the extent of nDNA fragmentation after exposure to menadione using a comet assay employing alkaline denaturation of the nuclei, which converts abasic sites to SSBs and releases single-stranded DNA (ssDNA). As shown in Figure 6A, nuclei with a long comet tail were observed only in Mt-hOGG1 MEFs 6 h after exposure. Following the exposure, less than 5% of nuclei in Nu-hOGG1 MEFs exhibited a very short tail (<5 μm) (Supplementary Figure S7), whereas 20% of nuclei in Mt-hOGG1 MEFs had a significantly longer comet tail (>30 μm) (Figure 6B). Double-immunofluorescence microscopy with antibodies against PARP and ssDNA revealed that PARP and ssDNA were mostly colocalized in the nuclei of Mt-hOGG1 MEFs within 4–6 h after the exposure (Figure 6C). As shown in Figure 6D, about one-third of Mt-hOGG1 MEFs incorporated 5-bromodeoxyuridine (BrdU) within 6 h after exposure to menadione, and all of the BrdU-positive cells exhibited nuclear accumulation of ssDNA only after exposure to menadione, thereby indicating that SSBs in the nDNA were generated during S phase of the cell cycle.
Figure 6.
DNA SSBs are generated by the action of MUTYH in either nDNA or mtDNA with 8-oxoG accumulation. (A) Menadione-induced nDNA fragmentation. Nu-hOGG1 (Nu), Mt-hOGG1 (Mt), and Nu/Mt-hOGG1 MEFs (Nu/Mt) transfected with (bottom panels) or without (middle panels) Mutyh-siRNAs were cultured for 24 h, and then exposed to 50 μM menadione for 60 min. Six hours after the exposure, the extent of nDNA fragmentation was examined using a comet assay. Control, no exposure. Results from one of two independent experiments are presented. Bar, 20 μm. (B) Suppression of menadione-induced nDNA fragmentation in Mt-hOGG1 MEFs by Mutyh-siRNA. Comet tail lengths of 50 nuclei of Mt-hOGG1 MEFs (control, menadione, menadione+Mutyh siRNA) shown in panel A were measured, and percentages of nuclei with various tail lengths (0–5, 5–10, 10–20, 20–30, >30 μm) are shown in the bar graph. (C) Menadione-induced nuclear accumulation of ssDNA and PARP activation in Mt-hOGG1 MEFs. Mt-hOGG1 MEFs were exposed to 50 μM menadione for 60 min, and then subjected to immunofluorescence microscopy with anti-ssDNA and anti-PARP. nDNA was stained with DAPI and merged images are shown (merged). Control, no exposure. Bar, 20 μm. (D) Menadione-induced accumulation of SSBs in S-phase nuclei. Mt-hOGG1 MEFs were exposed to menadione (50 μM) for 60 min in the presence of BrdU (1 μg/ml), and were then cultured in fresh medium containing BrdU (1 μg/ml) for 5 h. Cells were subjected to immunofluorescent microscopy with anti-BrdU (red) and anti-ssDNA (green). Nuclei were counterstained with DAPI (blue). Control, no exposure; Bar, 20 μm. The number of BrdU-positive nuclei and that of ssDNA-positive nuclei per 100 nuclei are shown under each panel. Percentages of ssDNA-positive nuclei among BrdU-positive nuclei are shown in parentheses. (E) Menadione-induced mtDNA fragmentation. Total cellular DNA was prepared from each type of MEF 4 or 8 h after a 60-min exposure to 50 μM menadione, and PstI-digested DNA was subjected to Southern blotting for mtDNA. Control, no exposure. (F) Suppression of menadione-induced depletion of mtDNA by Mutyh-siRNA. Total cellular DNA was prepared from Nu-hOGG1 MEFs 6 h after a 60-min exposure to 50 μM menadione, and mt-Co1 and Gapdh DNA were amplified. Control, no exposure. The intensity of mt-Co1 DNA was normalized to that of nuclear Gapdh DNA, and the relative intensity of each sample to that of the control sample is shown at the bottom of each lane.
During replication, adenine can be inserted opposite 8-oxoG accumulated in DNA as a result of an OGG1 deficiency, thereby forming a considerable number of A:8-oxoG pairs. MUTYH excises adenine opposite 8-oxoG through its adenine DNA glycosylase activity, and forms abasic sites that are converted to SSBs by AP endonuclease. Since SSBs observed in nDNA or mtDNA after exposure to menadione are likely to be generated by the action of MUTYH, we examined the effects of MUTYH knockdown on either nDNA fragmentation or depletion of mtDNA using Mutyh-siRNA. The levels of Mutyh mRNA were found to be significantly decreased to 2.3% that of the control cells transfected with scrambled-siRNA (Supplementary Figure S8). In the presence of the Mutyh-siRNAs, the number of nuclei with a long comet tail and an accumulation of ssDNA in Mt-hOGG1 MEFs exposed to menadione were significantly decreased (Figure 6A and B; Supplementary Figure S9).
The extent of mtDNA fragmentation after exposure to menadione was evaluated by Southern blotting (Figure 6E). Fragmented mtDNA was detected in Nu-hOGG1 MEFs 8 h after the exposure, whereas such fragmentation was not observed in Mt-hOGG1 MEFs. This result was consistent with the depletion of mt-Co1 DNA in Nu-hOGG1 MEFs shown in Figure 5B. Moreover, menadione-induced depletion of mt-Co1 DNA in Nu-hOGG1 MEFs was also efficiently suppressed in the presence of Mutyh-siRNAs (Figure 6F).
These findings indicated that accumulation of 8-oxoG in either nDNA or mDNA results in the buildup of SSBs, which depends on the action of MUTYH, before cell death.
Cell death triggered by 8-oxoG accumulated in either nDNA or mtDNA is mediated by MUTYH
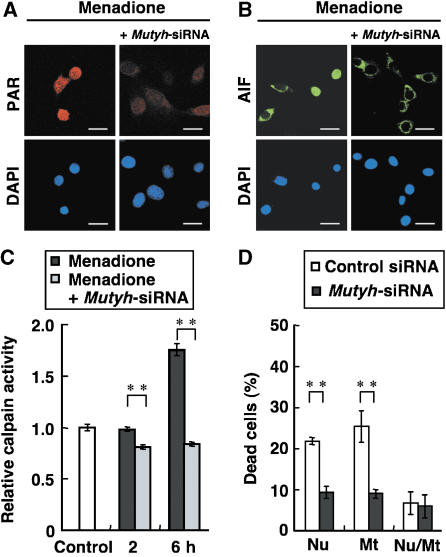
We next examined whether Mutyh knockdown would abrogate the activation of cell death executors, such as AIF or calpains, in either Mt-hOGG1 or Nu-hOGG1 MEFs after exposure to menadione. Nuclear translocation of AIF as well as accumulation of PAR in the nuclei of Mt-hOGG1 MEFs, which were induced as a result of PARP activation after the exposure, were efficiently suppressed in the presence of Mutyh-siRNAs (Figure 7A and B). Furthermore, activation of calpains in Nu-hOGG1 MEFs after the exposure was also suppressed to the basal level or lower (Figure 7C).
Figure 7.
Cell death triggered by 8-oxoG accumulated in cellular DNA is mediated by MUTYH. (A) Suppression of menadione-induced poly-ADP ribosylation in Mt-hOGG1 MEFs by Mutyh-siRNA. Mt-hOGG1 MEFs were exposed to 50 μM menadione for 60 min in the presence or absence of Mutyh-siRNA, and were subjected to immunofluorescence microscopy with anti-PAR. nDNA was stained with DAPI. Bar, 20 μm. (B) Suppression of mendione-induced nuclear translocation of AIF in Mt-hOGG1 MEFs by Mutyh-siRNA. Mt-hOGG1 MEFs were exposed to 50 μM menadione for 60 min in the presence or absence of Mutyh-siRNA and were subjected to immunofluorescence microscopy with anti-AIF. nDNA was stained with DAPI. Bar, 20 μm. (C) Suppression of menadione-induced activation of calpain in Nu-hOGG1 MEFs by Mutyh-siRNA. Calpain activity was determined in Nu-hOGG1 MEFs exposed to 50 μM menadione for 60 min in the presence or absence of Mutyh-siRNA. Control, no exposure. Relative calpain activities normalized to that of an untreated control are shown. Results from one of two independent experiments are presented (mean±s.d., n=3 per experiment). Student's t-test, **P<0.01. (D) Suppression of menadione-induced cell death by Mutyh-siRNAs. Each type of MEF was transfected with Mutyh-siRNAs or control-siRNA, cultured for 24 h, and then exposed to 25 μM menadione. Percentages of dead cells were determined 24 h after the exposure. Results from one of two independent experiments are presented (mean±s.d., n=3 per experiment). Student's t-test, **P<0.01.
We found that the percentage of dead cells in the Nu-hOGG1 (20.7%) and Mt-hOGG1 MEFs (24.3%) cultures after exposure to menadione had decreased to 9.3 and 9%, respectively, in the presence of only Mutyh-siRNAs but not control siRNA (Figure 7D). However, cultures of MEFs expressing both nuclear and mitochondrial hOGG1 (Nu/Mt-hOGG1) contained essentially the same percentage of dead cells in the presence of either Mutyh-siRNAs or control siRNA, and their levels were almost equivalent to those of Nu-hOGG1 and Mt-hOGG1 MEFs in the presence of the Mutyh-siRNAs.
Discussion
Our major conclusion in the present study is that accumulation of 8-oxoG in nDNA and mtDNA independently triggers two distinct cell death pathways; one depends on PARP and the nuclear translocation of AIF, and the other depends on the opening of the MMPT pore and calpains. Moreover, we demonstrated that both are caspase-independent and are initiated by the accumulation of SSBs in either nDNA or mtDNA, which is mediated by MUTYH.
Cell death triggered by 8-oxoG accumulated in nDNA
Mt-hOGG1 MEFs were deficient in BER of 8-oxoG in their nDNA but not in their mtDNA. Therefore, only the nDNA showed an accumulation of 8-oxoG after exposure to menadione, as well as a massive buildup of SSBs, followed by PARP activation with increased poly-ADP ribosylation of cellular proteins. Thus, these MEFs underwent caspase-independent cell death after nuclear translocation of AIF. In the presence of PARP inhibitor or Mutyh-siRNA, the nuclear translocation of AIF was efficiently diminished, resulting in the suppression of the cell death command. Furthermore, Mutyh-siRNA suppressed the massive buildup of SSBs or ssDNA in the nDNA.
PARP is a known molecular nick sensor that binds specifically to SSBs, and its specific activity catalyzing poly-ADP ribosylation of cellular proteins or of PARP itself can increase approximately 500-fold (de Murcia and Menissier de Murcia, 1994). Our data clearly indicate that MUTYH is responsible for the generation of SSBs in nDNA, with accumulation of 8-oxoG. Since MUTYH functions as an adenine DNA glycosylase, by excising adenine inserted opposite 8-oxoG in template DNA during replication, many abasic sites can be generated opposite 8-oxoG (Tominaga et al, 2004). During BER, abasic sites are concomitantly converted to SSBs by the action of AP endonuclease or AP lyases, thereby activating PARP.
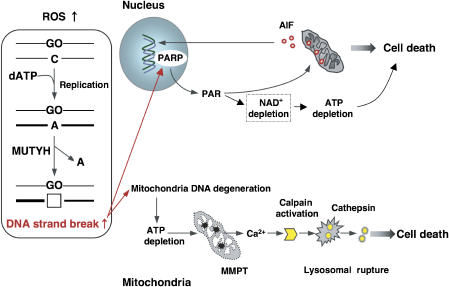
Recently, it has been reported that PAR polymer induces mitochondrial AIF release and translocation to the nucleus (Yu et al, 2006), thus executing the cell death command, thereby leading us to conclude that accumulation of 8-oxoG in nDNA causes massive buildup of SSBs through MUTYH-initiated BER, resulting in PARP-dependent cell death. PARP catalyzes poly-ADP ribosylation using nicotinamide adenine dinucleotide (de Murcia and Menissier de Murcia, 1994). Excessive activation of PARP leads to its auto-consumption, resulting in delayed depletion of ATP, as observed with 8-oxoG accumulation in nDNA (Figure 5F), and thus an energy crisis might also contribute to cell death (Figure 8).
Figure 8.
Model of cell death triggered by accumulation of 8-oxoG in nDNA and mtDNA.
Cell death triggered by 8-oxoG accumulated in mtDNA
Nu-hOGG1 MEFs were deficient in BER of 8-oxoG in mtDNA but not nDNA, thereby causing 8-oxoG to accumulate only in mtDNA after exposure to menadione. These cells exhibited depletion of mtDNA and ATP after the treatment, followed by MMPT and Ca2+ release from mitochondria, thus activating calpains to execute caspase-independent cell death.
Accumulation of 8-oxoG in mtDNA rapidly resulted in the depletion of mtDNA, which was efficiently suppressed in the presence of Mutyh-siRNA; therefore, we again conclude that MUTYH-initiated BER is responsible for the depletion of mtDNA. This depletion results in the simultaneous depletion of ATP, probably because of a decreased supply of mitochondrially encoded proteins, as well as tRNAs and rRNAs, which are all essential for mitochondrial respiratory function.
Depletion of ATP is known to open the MMPT pore, allowing Ca2+ to leave mitochondria (Simbula et al, 1997), indicating that mitochondrial dysfunction initiated by accumulation of 8-oxoG in mtDNA causes MMTP, thereby increasing the cytoplasmic concentration of Ca2+, which in turn activates calpains. Activated calpains are known to induce lysosomal rupture, causing release of cathepsin, which ultimately executes the cell death command (Yamashima, 2004).
MUTYH-initiated cell death and its implication in carcinogenesis and neurodegeneration
Several germline mutations in the human MUTYH gene have been found in patients with autosomal recessive familial adenomatous polyposis (Al-Tassan et al, 2002). We recently confirmed that MUTYH-deficient mice exhibit an increased occurrence of spontaneous or ROS-induced adenoma/adenocarcinoma in the small intestine and colon (Sakamoto et al, 2007). Based on our present findings, we propose that a loss in MUTYH function in stem or progenitor cells in the intestinal epithelium results in escape from cell death under oxidative stress; however, accumulated 8-oxoG results in mutations in proto-oncogenes or tumor-suppressor genes in these proliferative cells, thereby promoting tumorigenesis.
We and others have shown that an accumulation of 8-oxoG in mtDNA with increased expression of MTH1, OGG1 and MUTYH in the brains of PD and AD patients (Shimura-Miura et al, 1999; Iida et al, 2002; Fukae et al, 2005; Arai et al, 2006), and involvement of the calpain–cathepsin pathway, are implicated in various neurodegenerative disorders including PD and AD (Saito et al, 1993; Mouatt-Prigent et al, 1996). We have also demonstrated that 8-oxoG accumulation in mtDNA of striatal dopaminergic nerve terminals triggers their retrograde degeneration in a mouse model of PD (Yamaguchi et al, 2006). We therefore propose that an accumulation of 8-oxoG in mtDNA under oxidative stress causes mitochondrial dysfunction and activation of the calpain–cathepsin pathway, ending in neuronal loss (Figure 8).
We previously reported that alternative transcription initiation and splicing generating 10 different transcripts encoded by the human MUTYH gene yielded seven isoforms of MUTYH protein (Ohtsubo et al, 2000). We recently found that expression of an isoform lacking 106 residues of the N-terminus of a major form of MUTYH was significantly increased in mitochondria of surviving dopamine neurons in the substantia nigra of PD brains (Arai et al, 2006). This mitochondrial isoform is not likely to be fully active as an adenine DNA glycosylase. Therefore, it is very important to elucidate whether its expression in PD brains exacerbates neuronal loss or rather protects neurons in these patients. A study to address this question is now under way.
Materials and methods
Antibodies, western blotting and nicking assay
The rabbit polyclonal antibodies anti-2a-CT against the C-terminal region unique to hOGG1-2a and anti-HCD against the highly conserved domain of hOGG1 isoforms, have been described (Nishioka et al, 1999). Antibodies against PCNA (PC10; Abcam), HSP60 (LK-1; StressGen), AIF (E-1; Santa Cruz), poly-ADP-ribose polymers (PARs) (MC-100; Trevigen), ssDNA (IBL, Japan), and 8-oxoG (N45.1; Japan Institute for the Control of Aging) were also used. Western blot analysis (Tsuchimoto et al, 2001) and subcellular fractionation (Kang et al, 1995) were performed as described. Gels stained with GelCode Blue Stain (Pierce) were used to obtain loading controls. The nicking assay to detect 8-oxoG DNA glycosylase activity was also performed as described (Nishioka et al, 1999).
Immunostaining
hOGG1-2a protein was detected using anti-2a-CT in combination with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes). To detect ssDNA, the slides were incubated with 5 mg/ml RNase for 60 min at 37°C, and reacted with anti-ssDNA in combination with Alexa Fluor 488-conjugated goat anti-rabbit IgG. Nuclei were counterstained with 4′-diamino-2-phenylindole (DAPI, 50 ng/ml; Vector). HSP60 or PAR was detected using anti-HSP60 or anti-PAR in combination with Alexa Fluor 546-conjugated goat anti-mouse or anti-rabbit IgG. Digitized images were separately captured from identical fields using an LSM-510 Meta confocal microscopy system (Carl Zeiss). AIF was detected using anti-AIF in combination with Alexa Fluor 488-conjugated goat anti-mouse IgG, and the signal was observed using a Bio-Rad Radiance 2100 confocal microscopy system.
Quantification of 8-oxoG in nDNA and mtDNA
Purified nDNA was enzymatically hydrolyzed to nucleosides and subjected to HPLC-MS/MS analysis. The amount of 8-oxo-2′-deoxyguanosine was measured and the number of 8-oxoG residues per 106 guanine residues in nDNA was determined as described (Tsuruya et al, 2003). Confocal microscopy (Radiance 2100) with anti-8-oxoG and Alexa Fluor 488-conjugated goat anti-mouse IgG was used to quantify the 8-oxoG level in mtDNA, and the 8-oxoG index representing mitochondrial 8-oxoG immunoreactivity per cell was determined as described (Yoshimura et al, 2003).
Cell viability assays and morphological examination of cells
Cultured cells were incubated in medium containing various concentrations of menadione for 60 min, and then cultured in fresh medium for 24 h. Cell viability was determined by the trypan blue exclusion test. Numbers of unstained living cells were counted and cell viability in each experiment was determined as a percentage of the untreated control. To determine the percentage of dead cells in each culture, the number of dead cells stained with both Hoechst33342 and PI was divided by the total number of cells stained with Hoechst. MDL28170 was obtained from BIOMOL and CsA and 3-AB were from Sigma. Electron microscopy was performed as described (Yoshimura et al, 2003).
Intracellular ATP levels
Intracellular ATP level was measured using a CellTiter-GloR Assay kit (Promega), according to the manufacturer's instructions. MEFs were cultured in two sets of 96-well plates for each experiment, and exposed to medium containing 50 μM menadione for 60 min, and then cultured in fresh medium. One set of plates was subjected to the luciferase reaction to monitor the intracellular ATP level using a spectrofluorometer (Wallac 1420 ARVOsx multilabel counter), and the other set was subjected to staining with Hoechst33342 and PI. To determine the ATP index representing the relative amount of ATP per cell, the luminescence signal was divided by the number of living cells stained only with Hoechst33342.
Determination of mitochondrial membrane potential and levels of Ca2+ in mitochondria
The mitochondrial membrane potential was detected using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1; Molecular Probes). JC-1 forms red fluorescent aggregates in energized mitochondria with high membrane potential, whereas it dissociates to monomers with green fluorescence at low membrane potential. Levels of Ca2+ in mitochondria were monitored by rhod-2 AM (Molecular Probes) according to the procedure of Hajnoczky et al (1995). Rhod2-AM, a Ca2+-sensitive fluorescent indicator, has a net positive charge, and thus accumulates in the negatively charged mitochondrial matrix. MEFs were incubated in medium containing 50 μM menadione for 60 min, and then cultured in fresh medium for various periods. At certain time points, MEFs were incubated in the presence of 5 μg/ml JC-1 or 4.5 μM rhod-2 AM for 20 min at 37°C, and were observed under an Axio-Skop2-equipped Axio Cam after washing twice with PBS. Signal intensities of JC-1 (red-fluorescent) and rhod-2 were measured using Image Gauge V4.0 (Fuji Film).
Amplification of mt-Co1 DNA
Using a primer set for the mt-Co1 gene (mCOX1s: 5′-TGATTATTCTCAACCAATCAC-3′; mCOX1a: 5′-TGGTGGAGGGCAGCCATGAAG-3′), a 1438-bp fragment of mtDNA (residues 5393–6831) was amplified from total cellular DNA (500 ng) prepared using ISOGEN (Nippon Gene). The relative amount of mt-Co1 DNA normalized with nuclear Gapdh DNA was amplified using a set of primers (mGA5-1: 5′-CTGCCATTTGCAGTGGCAAAG-3′; mGA3-1:5′-TGGTATTCAAGAGAGTAGGGA-3′).
Calpain activity
Calpain activity was measured using a Calpain Activity Assay kit (BioVision). The calpain substrate (Ac-LLY-AFC) was incubated with cell lysates (10 μg protein) for 60 min at 37°C in the dark. Fluorescence of released free AFC was monitored using a Wallac 1420 ARVOsx multilabel counter (Perkin Elmer).
Southern blotting and the comet assay
Southern blotting of mtDNA was performed as described (Miyako et al, 2000). nDNA Fragmentation was detected using a Comet assay™ kit (Trevigen), according to the manufacturer's instructions. The slide was observed under an Axio-Skop2-equipped Axio Cam. Comet tail lengths were measured using Image Gauge V4.0. For each experiment, 50 cells were classified according to the five stages (0–30 μm) based on their tail length, and the percentage of cells in each stage was determined.
siRNA and transfection
siRNAs against mouse MUTYH (#86583, #86769) and a scrambled-siRNA (#4603G) as control siRNA were purchased from Ambion Inc. MEFs were transfected 24 h before exposure to menadione with both Mutyh-siRNAs, using an siPORT Amine, Silencer TM siRNA Transfection kit (Ambion).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure 9
Supplementary Information
Acknowledgments
We are grateful to Dr K Takeda for his generous gift of pPGK-PURO, and to Dr W Campbell for useful comments on this paper. We thank N Adachi for HPLC MS/MS analysis, A Matsuyama for animal care, and M Sasaki and M Ohtsu at the Laboratory for Technical Support of our institute for their expertise in performing the EM and DNA sequence analyses. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant numbers 16012248, 17014070, 18013038), and the Japan Society for the Promotion of Science (grant numbers 16390119, 18300124).
References
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP (2002) Inherited variants of MYH associated with somatic G:C → T:A mutations in colorectal tumors. Nat Genet 30: 227–232 [DOI] [PubMed] [Google Scholar]
- Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D (2003) Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res 31: 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Shigenaga MK, Hagen TM (1993) Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA 90: 7915–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Fukae J, Hatano T, Kubo S, Ohtsubo T, Nakabeppu Y, Mori H, Mizuno Y, Hattori N (2006) Up-regulation of hMUTYH, a DNA repair enzyme, in the mitochondria of substantia nigra in Parkinson's disease. Acta Neuropathol 112: 139–145 [DOI] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434: 658–662 [DOI] [PubMed] [Google Scholar]
- Bizat N, Hermel JM, Humbert S, Jacquard C, Creminon C, Escartin C, Saudou F, Krajewski S, Hantraye P, Brouillet E (2003) In vivo calpain/caspase cross-talk during 3-nitropropionic acid-induced striatal degeneration: implication of a calpain-mediated cleavage of active caspase-3. J Biol Chem 278: 43245–43253 [DOI] [PubMed] [Google Scholar]
- Boiteux S, Radicella JP (2000) The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys 377: 1–8 [DOI] [PubMed] [Google Scholar]
- de Murcia G, Menissier de Murcia J (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci 19: 172–176 [DOI] [PubMed] [Google Scholar]
- Frei B, Winterhalter KH, Richter C (1986) Menadione- (2-methyl-1,4-naphthoquinone-) dependent enzymatic redox cycling and calcium release by mitochondria. Biochemistry 25: 4438–4443 [DOI] [PubMed] [Google Scholar]
- Fukae J, Takanashi M, Kubo S, Nishioka K, Nakabeppu Y, Mori H, Mizuno Y, Hattori N (2005) Expression of 8-oxoguanine DNA glycosylase (OGG1) in Parkinson's disease and related neurodegenerative disorders. Acta Neuropathol 109: 256–262 [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP (1995) Decoding of cytosolic calcium oscillations in the mitochondria. Cell 82: 415–424 [DOI] [PubMed] [Google Scholar]
- Iida T, Furuta A, Nishioka K, Nakabeppu Y, Iwaki T (2002) Expression of 8-oxoguanine DNA glycosylase is reduced and associated with neurofibrillary tangles in Alzheimer's disease brain. Acta Neuropathol 103: 20–25 [DOI] [PubMed] [Google Scholar]
- Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K (1995) Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J Biol Chem 270: 14659–14665 [DOI] [PubMed] [Google Scholar]
- Kasai H, Nishimura S (1984) Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res 12: 2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H (2002) Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu Rev Genet 36: 279–303 [DOI] [PubMed] [Google Scholar]
- Miyako K, Takamatsu C, Umeda S, Tajiri T, Furuichi M, Nakabeppu Y, Sekiguchi M, Hamasaki N, Takeshige K, Kang D (2000) Accumulation of adenine DNA glycosylase-sensitive sites in human mitochondrial DNA. J Biol Chem 275: 12326–12330 [DOI] [PubMed] [Google Scholar]
- Mouatt-Prigent A, Karlsson JO, Agid Y, Hirsch EC (1996) Increased M-calpain expression in the mesencephalon of patients with Parkinson's disease but not in other neurodegenerative disorders involving the mesencephalon: a role in nerve cell death? Neuroscience 73: 979–987 [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Sakumi K, Sakamoto K, Tsuchimoto D, Tsuzuki T, Nakatsu Y (2006) Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol Chem 387: 373–379 [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K (2007) Oxidative damage in nucleic acids and Parkinson's disease. J Neurosci Res 85: 919–934 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y (1999) Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell 10: 1637–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, Jones PK, Ghanbari H, Wataya T, Shimohama S, Chiba S, Atwood CS, Petersen RB, Smith MA (2001) Oxidative damage is the earliest event in Alzheimer disease. J Neuropathol Exp Neurol 60: 759–767 [DOI] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y (2000) Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res 28: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Tominaga Y, Yamauchi K, Nakatsu Y, Sakumi K, Yoshiyama K, Egashira A, Kura S, Yao T, Tsuneyoshi M, Maki H, Nakabeppu Y, Tsuzuki T (2007) MUTYH-null mice are susceptible to spontaneous and oxidative-stress-induced intestinal tumorigenesis. Cancer Res 67: 6599–6604 [DOI] [PubMed] [Google Scholar]
- Saito K, Elce JS, Hamos JE, Nixon RA (1993) Widespread activation of calcium-activated neutral proteinase (calpain) in the brain in Alzheimer disease: a potential molecular basis for neuronal degeneration. Proc Natl Acad Sci USA 90: 2628–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura-Miura H, Hattori N, Kang D, Miyako K, Nakabeppu Y, Mizuno Y (1999) Increased 8-oxo-dGTPase in the mitochondria of substantia nigral neurons in Parkinson's disease. Ann Neurol 46: 920–924 [PubMed] [Google Scholar]
- Simbula G, Glascott PA Jr, Akita S, Hoek JB, Farber JL (1997) Two mechanisms by which ATP depletion potentiates induction of the mitochondrial permeability transition. Am J Physiol 273: 479–488 [DOI] [PubMed] [Google Scholar]
- Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH (1999) Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol 181: 6210–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA (2005) Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res 33: 3722–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga Y, Ushijima Y, Tsuchimoto D, Mishima M, Shirakawa M, Hirano S, Sakumi K, Nakabeppu Y (2004) MUTYH prevents OGG1 or APEX1 from inappropriately processing its substrate or reaction product with its C-terminal domain. Nucleic Acids Res 32: 3198–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y (2001) Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res 29: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruya K, Furuichi M, Tominaga Y, Shinozaki M, Tokumoto M, Yoshimitsu T, Fukuda K, Kanai H, Hirakata H, Iida M, Nakabeppu Y (2003) Accumulation of 8-oxoguanine in the cellular DNA and the alteration of the OGG1 expression during ischemia-reperfusion injury in the rat kidney. DNA Repair 2: 211–229 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Kajitani K, Dan Y, Furuichi M, Ohno M, Sakumi K, Kang D, Nakabeppu Y (2006) MTH1, an oxidized purine nucleoside triphosphatase, protects the dopamine neurons from oxidative damage in nucleic acids caused by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Cell Death Differ 13: 551–563 [DOI] [PubMed] [Google Scholar]
- Yamashima T (2004) Ca2+-dependent proteases in ischemic neuronal death: a conserved ‘calpain-cathepsin cascade' from nematodes to primates. Cell Calcium 36: 285–293 [DOI] [PubMed] [Google Scholar]
- Yoshimura D, Sakumi K, Ohno M, Sakai Y, Furuichi M, Iwai S, Nakabeppu Y (2003) An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress. J Biol Chem 278: 37965–37973 [DOI] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL (2006) Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA 103: 18314–18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297: 259–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure 9
Supplementary Information