Abstract
Nod1 and Nod2 are intracellular proteins that are involved in host recognition of specific bacterial molecules and are genetically associated with several inflammatory diseases. Nod1 and Nod2 stimulation activates NF-κB through RICK, a caspase-recruitment domain-containing kinase. However, the mechanism by which RICK activates NF-κB in response to Nod1 and Nod2 stimulation is unknown. Here we show that RICK is conjugated with lysine-63-linked polyubiquitin chains at lysine 209 (K209) located in its kinase domain upon Nod1 or Nod2 stimulation and by induced oligomerization of RICK. Polyubiquitination of RICK at K209 was essential for RICK-mediated IKK activation and cytokine/chemokine secretion. However, RICK polyubiquitination did not require the kinase activity of RICK or alter the interaction of RICK with NEMO, a regulatory subunit of IκB kinase (IKK). Instead, polyubiquitination of RICK was found to mediate the recruitment of TAK1, a kinase that was found to be essential for Nod1-induced signaling. Thus, RICK polyubiquitination links TAK1 to IKK complexes, a critical step in Nod1/Nod2-mediated NF-κB activation.
Keywords: NLR, Nod1, Nod2, RICK, TAK1
Introduction
Members of the nucleotide-binding domain-like receptor (NLR) protein family are found in both mammals and plants (Inohara et al, 2005; Fritz et al, 2006). Several mammalian NLRs, like plant NLRs, are involved in host resistance against microbial pathogens and genetically associated with human immune diseases (Inohara et al, 2005; Fritz et al, 2006). Two NLR family members, Nod1 and Nod2, play an essential role in the recognition of specific bacterial peptidoglycan (PGN)-related molecules (Chamaillard et al, 2003; Inohara et al, 2003; Girardin et al, 2003a, 2003b). Nod1 and Nod2 mediate transcriptional activation of innate immune genes including antimicrobial peptides, proinflammatory cytokines and chemokines, that recruit immune cells to the sites of microbial stimulation (Chamaillard et al, 2003; Kobayashi et al, 2005; Boughan et al, 2006; Marks et al, 2006; Masumoto et al, 2006; Voss et al, 2006). Although the precise structure of the bacterial molecules that stimulate NLR signaling remains largely unknown, the core recognition moieties of Nod1 and Nod2 have been identified as γ-glutamyl-meso-diaminopimelic acid (iE-DAP), and muramyl dipeptide (MDP), respectively (Chamaillard et al, 2003; Girardin et al, 2003a, 2003b; Inohara et al, 2003). Previous studies using synthetic and natural PGN-related molecules containing MDP and/or iE-DAP revealed that these molecules induce/enhance innate and adaptive immune responses (Goto and Aoki, 1987; Fritz et al, 2007).
The induction of immune response genes through Nod1 and Nod2 signaling largely depends on NF-κB, a proinflammatory transcriptional factor (Inohara et al, 1999, 2000; Ogura et al, 2001; Masumoto et al, 2006). Transcriptional activation of proinflammatory genes is initiated by nuclear translocation of NF-κB following its release from the IκB proteins (Karin and Ben-Neriah, 2000; Hoffmann and Baltimore, 2006). Previous studies demonstrated that RICK (RIP2/CARDIAK/RIPK2) is a critical downstream mediator of Nod1 and Nod2 signaling (Inohara et al, 1999; Ogura et al, 2001; Chin et al, 2002; Kobayashi et al, 2002; Park et al, 2007). RICK is composed of N-terminal kinase and C-terminal caspase-recruitment domains (CARD) linked via an intermediate (IM) region (Inohara et al, 1998; McCarthy et al, 1998; Thome et al, 1998). RICK physically associates with Nod1 and Nod2 through CARD–CARD interaction and activates IκB kinase (IKK), leading to phosphorylation and degradation of IκBs (Inohara et al, 1999, 2000; Ogura et al, 2001). Functional studies revealed that close proximity of RICK molecules by self-oligomerization of Nod1 and Nod2 is important for NF-κB activation (Inohara et al, 2000). RICK is known to interact with NEMO (IKKγ/IKBKG) and this interaction is essential for NF-κB activation in RICK-mediated signaling (Inohara et al, 2000). Furthermore, Nod2 stimulation with MDP induces NEMO polyubiquitination and this event is required for optimal NF-κB activation (Abbott et al, 2004). However, the molecular mechanism by which the IKK complex is activated via RICK to induce NF-κB activation in response to NLR stimulation is unknown. Here we show that RICK is conjugated with K63-linked polyubiquitin chains upon Nod1 and Nod2 stimulation. RICK polyubiquitination was essential for NF-κB activation and chemokine/cytokine secretion in response to Nod1 stimulation. Furthermore, we provide evidence for a bridging function of RICK ubiquitination by linking TAK1 to the IKK complex that is critical for NF-κB activation during Nod signaling.
Results
Nod1 and Nod2 stimulation induce lysine 63-linked polyubiquitination of RICK
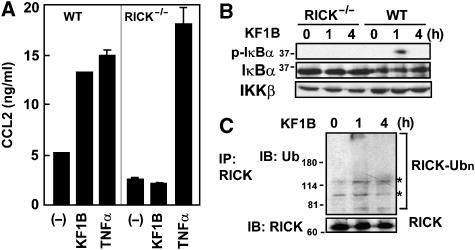
In order to determine the mechanism by which Nod1 stimulation induces NF-κB activation, we used mouse embryonic fibroblasts (MEFs) because we previously found that non-immune cells are much more sensitive to Nod1-stimulatory molecules than hematopoietic cells (Hasegawa et al, 2006; Masumoto et al, 2006). Stimulation of MEFs with KF1B, a synthetic specific Nod1-stimulatory compound, induced secretion of the chemokine CCL2 (Figure 1A) and phosphorylation of IκBα (Figure 1B). Such responses were dependent on RICK in that the induction of CCL2 production induced by KF1B, but not TNFα, was abolished in RICK-deficient MEFs (Figure 1A).
Figure 1.
Nod1 stimulation induces ubiquitination of RICK. (A) MEFs from WT and RICK-deficient mice were stimulated with 5 μg/ml KF1B (synthetic Nod1-stimulatory compound), 10 ng/ml TNFα or left alone. Twenty-four hours post-stimulation, the levels of CCL2 in culture supernatant were determined by ELISA. The results shown are given as mean±s.d. of triplicate cultures and are representative of three experiments. (B) WT or RICK-deficient MEFs were stimulated with 5 μg/ml KF1B for the indicated times. Post-stimulation, cells were lysed and immunoblotted with anti-phospho-IκBα, anti-IκBα or IKKβ Ab. (C) MEFs were stimulated with or without KF1B for the indicated times. Post-stimulation, cells were lysed and RICK was immunoprecipitated with rabbit anti-RICK Ab. Ubiquitinated (upper panel) and total (lower panel). RICK proteins in the immunoprecipitates were immunodetected by mouse monoclonal anti-RICK and anti-Ub Abs, respectively. Nonspecific signals detected with the Ab are indicated by asterisks.
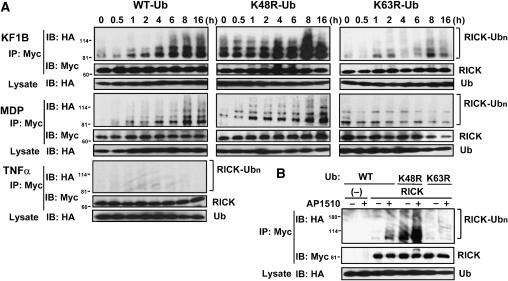
Recently, ubiquitination of RIP, a mediator of TNFα signaling, was found to be important for IKK activation (Ea et al, 2006; Li et al, 2006). Because our previous studies suggest functional and structural similarities between RIP and RICK, we first hypothesized that Nod1 stimulation might result in ubiquitination of RICK. As shown in Figure 1, RICK from MEFs stimulated with KF1B co-immunoprecipitated with high-molecular-weight ubiquitinated proteins (Figure 1C, upper panel). To further assess a role of ubiquitination in Nod1 and Nod2 signaling, we tested if RICK is ubiquitinated upon Nod1 and Nod2 stimulation, using human embryonic kidney (HEK) 293 cells constitutively expressing Nod1 and Nod2. Nod1- and Nod2-expressing HEK293 cells were transfected with plasmids producing Myc-RICK and HA-Ub and stimulated with specific synthetic Nod1 and Nod2 agonists, KF1B or MDP, respectively. Immunoblotting analysis revealed that RICK was polyubiquitinated after KF1B and MDP stimulation (Figure 2A). RICK polyubiquitination was detected 1 h after stimulation with Nod1 and Nod2 agonists. The polyubiquitination of RICK by Nod1 and Nod2 stimulation was specific in that it was not detected in parental HEK293 cells stimulated with TNFα (Figure 2A). The ubiquitination of RICK is not due to overexpression of proteins because we also found significant ubiquitination of expressed RICK whose levels were similar to those of endogenous RICK in MEFs (see below, Figures 5D and 7D). To determine the linkage type of polyubiquitin chains on RICK, Nod1- and Nod2-expressing cells were transfected with RICK-Myc and HA-tagged wild-type (WT) or lysine-mutant Ub proteins containing single lysine mutations at position 48 (K48R) or 63 (K63R). Upon Nod1 and Nod2 stimulation with KF1B and MDP, respectively, RICK was strongly polyubiquitinated with K48R Ub, but much less with K63R Ub (Figure 2A). Thus, the polyubiquitin chains on RICK were linked primarily through lysine 63 of ubiquitin.
Figure 2.
Nod1 and Nod2 stimulation induce K63-linked polyubiquitination of RICK. (A) HEK293 cells constitutively expressing Nod1 (for KF1B stimulation), Nod2 (for MDP stimulation) and parental HEK293 cells (for TNFα stimulation) were transfected with expression plasmid of Myc-RICK in the presence of expression plasmids of HA-tagged WT, K48R, K63R mutant Ub proteins. Cells were stimulated with 100 ng/ml KF1B, 100 ng/ml MDP (synthetic Nod2-stimulatory compound) or 10 ng/ml TNFα, or left alone (0 h control) for the indicated times. Myc-RICK proteins in transfected cells were immunoprecipitated with rabbit anti-Myc Ab and ubiquitinated RICK proteins were then detected by immunoblotting with anti-HA Ab (upper panels labeled as RICK-Ubn). As a control, RICK proteins in the same samples and Ub proteins in total lysate were detected by immunoblotting with the indicated Abs. (B) Oligomerization-dependent RICK ubiquitination. HEK293T cells were transfected with control pcDNA3-RICK-Fpk3-Myc (−) or pcDNA3-RICK-Fpk3-Myc (RICK) in the presence of HA-Ub expression plasmids. Eighteen hours post-transfection, cells were treated with or without 200 nM AP1510. Twenty-four hours post-transfection, proteins were immunoprecipitated and ubiquitinated RICK was analyzed as described in panel A.
Previous studies showed that enforced oligomerization of RICK by Nod1 and Nod2 induces NF-κB activation (Inohara et al, 2000). Similarly, oligomerization of RICK-ΔCARD-Fpk3, a RICK fusion protein in which the CARD is replaced with three tandem FKBP-related dimerization domains, results in NF-κB activation that is dependent on the FKBP-specific dimerizer AP1510 (Inohara et al, 2000; Figure 2B). Using this approach, we tested if oligomerization of RICK-ΔCARD-Fpk3 results in RICK polyubiquitination in HEK293T cells that coexpress HA-Ub. Immunoblotting analysis revealed that RICK-ΔCARD-Fpk3 was polyubiquitinated in an AP1510-dependent manner whereas the control Fpk3 protein was not (Figure 2B). These results suggest that induced proximity of RICK (1–435) lacking the CARD domain mimics the interaction between RICK and Nod proteins and induces RICK signaling.
The kinase domain of RICK is essential for RICK ubiquitination
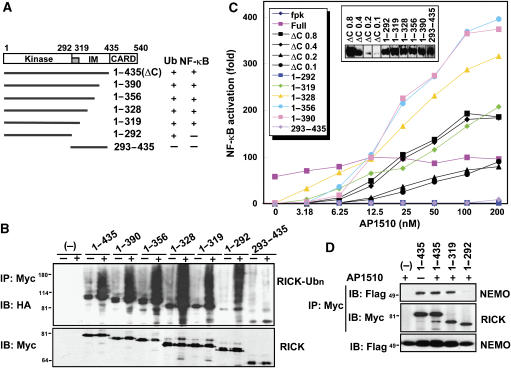
To determine which region of RICK is required for polyubiquitination, we constructed RICK-ΔCARD-Fpk3-mutant proteins lacking various regions of RICK and tested their ability to be ubiquitinated in the absence and presence of the dimerizer AP1510 (Figure 3A). Immunoblotting analysis showed that all RICK-ΔCARD-Fpk3 proteins containing residues 1–292 of RICK were ubiquitinated in an AP1510-dependent manner, whereas that encompassing residues 293–435 was not (Figure 3B). These results indicate that the region composed of amino-acid residues 1–292 located within the kinase region of RICK contains the polyubiquitinated site(s). To determine the role of RICK polyubiquitination in NF-κB activation, the ability of RICK fusion proteins to activate NF-κB was tested using a reporter assay. Transfection of HEK293T cells with the different constructs revealed that all RICK proteins carrying the region between amino acid 1 and 319 induced NF-κB activation in an AP1510-dependent manner (Figure 3C). In contrast, enforced oligomerization of the fusion proteins carrying the region between 1 and 292 and that between 293 and 435 did not result in NF-κB activation (Figure 3C). These results indicate that both the kinase domain (residues 1–292), which contains the polyubiquitinated site(s), and the IM region (residues 293–319) are critical for NF-κB activation.
Figure 3.
The kinase domain of RICK is essential for RICK ubiquitination, but is not sufficient to activate NF-κB. (A) Schematic diagram of the structural domains of RICK and different deletion mutants. The results of ubiquitination and NF-κB activation experiments are summarized on the right. The region, which requires NF-κB activation in the IM domain is indicated by a gray box. (B) HEK293T cells were transfected with pcDNA3-RICK-(1–435)-Fpk3-Myc (ΔC), RICK-(1–292)-Fpk3-Myc, RICK-(1–319)-Fpk3-Myc, RICK-(1–328)-Fpk3-Myc, RICK-(1–356)-Fpk3-Myc, RICK-(1–390)-Fpk3-Myc RICK-(293–435)-Fpk3-Myc and control vector in the presence of HA-Ub expression plasmid and treated with AP1510 as in Figure 2B. Twenty-four hours post-transfection, proteins were immunoprecipitated and ubiquitinated RICK were analyzed as described in Figure 2. (C) HEK293T cells were transfected with pcDNA3-RICK-(1–435)-Fpk3-Myc (ΔC), RICK-(1–292)-Fpk3-Myc, RICK-(1–319)-Fpk3-Myc, RICK-(1–328)-Fpk3-Myc, RICK-(1–356)-Fpk3-Myc, RICK-(1–390)-Fpk3-Myc RICK-(293–435)-Fpk3-Myc and control vector in the presence of reporter plasmids. Eight hours post-transfection, cells were treated with medium containing the indicated amounts of AP1510. Twenty-four hours post-transfection, ligand-dependent NF-κB activation was determined with reporter assay. The level of NF-κB-dependent transcription activity in cells transfected with control vector is given as 1. The expression levels of Myc-RICK proteins are shown in an inset. (D) HEK293T cells were transfected with control vector (−), pcDNA3-RICK-(1–435)-Fpk3-Myc, RICK-(1–319)-Fpk3-Myc and RICK-(1–292)-Fpk3-Myc in the presence of reporter plasmids, and treated with AP1510 as in Figure 2B. Twenty-four hours post-transfection, RICK proteins were immunoprecipitated with rabbit polyclonal anti-Myc Ab, and then Flag-NEMO (upper panel) and Myc-RICK (middle panel) were immunodetected with mouse monoclonal anti-Flag and anti-Myc Abs, respectively. As control, Flag-NEMO in total lysate was immunodetected with anti-Flag Ab and shown in the lower panel.
Previous studies showed that NF-κB activation mediated by RICK requires NEMO (Inohara et al, 2000). Therefore, we hypothesized that at least one of the regions required for NF-κB activation interacts with NEMO. To explore this hypothesis, we tested the ability of RICK-Fpk3-Myc constructs carrying different regions of RICK to interact with NEMO by co-immunoprecipitation assay. Consistent with a previous study (Inohara et al, 2000), a RICK truncation mutant expressing a region between amino-acid residues 1–319 interacted with NEMO (Figure 3D). Significantly, a RICK mutant composed of residues 1–292 did not associate with NEMO (Figure 3D). These results indicate that the interaction between RICK and NEMO requires the IM region-spanning residues 293–319 of RICK.
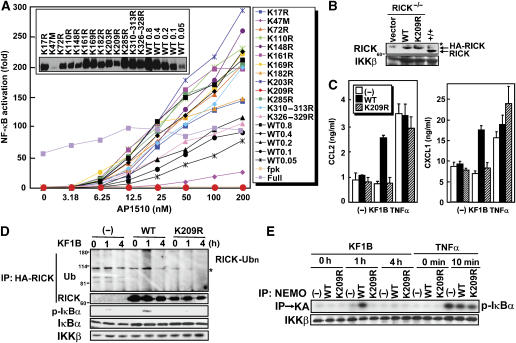
Lysine 209 in the kinase domain of RICK is polyubiquitinated in a signal-dependent manner
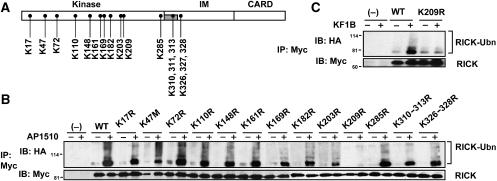
To map the polyubiquitinated lysine residue(s) in the kinase domain of RICK, we first performed close inspection of the amino-acid sequence of RICK. Alignment of amino-acid sequences from the kinase domain and IM region of RICK revealed 10 lysine residues that are evolutionarily conserved in 11 animal species (Supplementary Figure 1). Therefore, we introduced site-directed mutations in the 10 absolutely conserved and seven additional highly conserved lysine residues and tested if the mutations affect the ability of RICK-ΔCARD-Fpk3 to be ubiquitinated in an AP1510-dependent manner (Figure 4A). As shown in Figure 4B, the point mutant K209R, but not the remaining 16 lysine mutations, abolished the polyubiquitination of RICK. Notably, the K47M mutation, which replaces the essential lysine residue in the conserved ATP-binding site and results in loss of RICK kinase activity (Inohara et al, 1998), did not impair RICK polyubiquitination (Figure 4B). Thus, the conserved K209, but not the kinase activity of RICK, is essential for polyubiquitination. To determine if K209 is responsible for polyubiquitination of RICK during Nod1 signaling, we introduced the K209R mutation into a full-length RICK construct and tested the ability of WT and K209R-mutant RICK proteins to be ubiquitinated in Nod1-expressing HEK293 cells upon Nod1 stimulation with KF1B. Consistent with the results presented in Figure 4B, the K209R mutation abolished KF1B-induced polyubiquitination of RICK (Figure 4C). These results indicate K209 is essential for RICK polyubiquitination in Nod1 signaling.
Figure 4.
Lysine 209 is ubiquitinated in the kinase domain of RICK. (A) Location of lysine residues, which were replaced by arginines in RICK site-directed mutants. (B) HEK293T cells were transfected with control vector (−), pcDNA3-RICK-ΔCARD-Fpk3-Myc (WT) and various lysine-to-arginine mutants in the presence HA-Ub expression plasmid and treated with AP1510 as in Figure 2B. Twenty-four hours post-transfection, proteins were immunoprecipitated and ubiquitinated RICK were immunodetected as described in Figure 1. (C) HEK293 cells constitutively expressing Nod1 were transfected with the expression plasmids of WT and K209R-mutant Myc-RICK in the presence of HA-Ub expression plasmid. Eighteen hours post-transfection, cells were stimulated with or without 100 ng/ml KF1B. Twenty-four hours post-transfection, proteins were immunoprecipitated and ubiquitinated RICK were immunodetected as described in Figure 1.
Polyubiquitination of RICK is essential for Nod1 and Nod2-mediated NF-κB activation
To test if polyubiquitination at K209 of RICK is required for NF-κB activation, we assessed the ability of lysine-mutant RICK-ΔCARD-Fpk3 proteins to activate NF-κB after stimulation with different doses of AP1510. All the point mutant RICK proteins were expressed at levels comparable with that of WT protein, except the kinase-dead K47M mutant that was expressed at significantly lower levels (Figure 5A, inset). Consistent with its lower expression, the K47M mutant showed reduced levels of NF-κB activation when compared with WT protein (Figure 5A). However, the K47 mutant did show a dose-dependent increase in NF-κB and since K47 is essential for the kinase activity of RICK (Inohara et al, 1998), we conclude that the kinase activity of RICK is required for neither NF-κB activation nor polyubiquitination. Notably, the K209R mutation, but not those of the remaining conserved lysines, abolished AP1510-dependent NF-κB activation (Figure 5A). These results indicate that polyubiquitination of RICK at K209 is essential for NF-κB activation.
Figure 5.
Polyubiqutination at K209 is essential for RICK-mediated signaling. (A) HEK293T cells were transfected with pcDNA3-RICK-ΔCARD-Fpk3-Myc (WT) or the mutants in the presence of reporter plasmids. Eight hours post-transfection, cells were treated with medium containing the indicated amount of AP1510. Twenty-four hours post-transfection, ligand-dependent NF-κB activation was determined with reporter assay. The level of NF-κB-dependent transcription activity in cells transfected with control vector is given as 1. Myc-tagged RICK proteins detected with anti-Myc Ab are shown in the inset. (B) RICK-deficient MEFs were infected with retrovirus vectors expressing WT or K209R HA-RICK or control vector and selected by hygromycine. RICK proteins detected with anti-RICK Ab. A nonspecific signal detected with the Ab is indicated by an asterisk. (C) RICK-deficient MEFs were infected with control (−) or retrovirus vectors expressing WT or K209R HA-RICK. Infected MEFs were selected by hygromycin and stimulated with 5 μg/ml KF1B, 10 ng/ml TNFα or left alone. Twelve hours post-stimulation, secretion levels of CCL2 and CXCL1 were determined by ELISA. The results shown are given as mean±s.d. of triplicate cultures and are representative of three experiments. (D) MEFs stably expressing RICK or RICK K209R were stimulated with 5 μg/ml KF1B for the indicated time. Post-stimulation, cells were lysed, RICK was immunoprecipitated and ubiquitinated RICK was immunodetected by anti-Ub Ab (upper panel). Cell lysate was also immunoblotted with anti-phospho-IκBα, anti-IκBα or IKKβ Ab (bottom three panels). A nonspecific signal detected with the Ab is indicated by an asterisk. (E) MEFs stably expressing RICK or RICK K209R were stimulated with 5 μg/ml KF1B or TNFα for indicated times, and then anti-NEMO Ab was used to immunoprecipitate the IKK complex. The IKK assay was carried out by using GST-IκBα and γ-32P-ATP as substrates (upper). The amount of IKKβ in the immunoprecipitates was detected by immunoblotting.
To test if K209 polyubiquitination on RICK is important for Nod1 signaling, we assessed the ability of WT and K209R-mutant RICK proteins to complement RICK function in embryonic fibroblasts deficient in RICK. In these experiments, MEFs from RICK-deficient mice were infected with a retroviral vector producing WT or K209R-mutant RICK proteins. The expression levels of both WT and K209R RICK proteins in RICK−/− MEFs were similar to that of endogenous RICK in WT MEFs (Figure 5B). Notably, expression of WT RICK conferred responsiveness of MEFs deficient in RICK to KF1B, and resulted in secretion of chemokines CCL2 and CXCL1 (Figure 5C). In contrast, the K209R RICK mutant failed to rescue responsiveness to the Nod1 agonist (Figure 5C). Immunoblotting showed that WT but not K209R-mutant RICK was polyubiquitinated upon Nod1 stimulation and resulted in IKK activation (Figure 5D and E). Because control TNFα induced similar levels of CCL2 and CXCL1 secretion from cells infected with WT and K209R-mutant RICK constructs, these findings indicate that polyubiquitination of RICK at K209 is essential for Nod1 signaling but dispensable for that induced by TNFα stimulation.
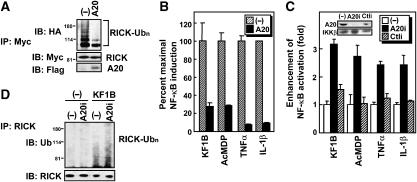
A20 inhibits Nod1- and Nod2-mediated NF-κB activation and polyubiquitination of RICK
A20 is a deubiquitinase that removes K63-linked polyubiquitin chains and functions as a negative feedback regulator of inflammatory responses (Lee et al, 2000; Boone et al, 2004; Wertz et al, 2004). Previous studies showed that A20 expression is induced by NF-κB activation (Krikos et al, 1992) and Nod1 stimulation (Masumoto et al, 2006). To test if A20 negatively regulates Nod signaling through deubiquitination of RICK, we tested the ability of A20 to affect RICK polyubiquitination and NF-κB activation in response to Nod1 or Nod2 stimulation. To assess the latter, A20 was coexpressed with Myc-RICK and HA-ubiquitin in HEK293T cells and the level of polyubiquitinated RICK was determined by immunoblotting. We found that expression of A20 decreased RICK polyubiquitination (Figure 6A). HEK293T cells express low, but significant, levels of endogenous Nod1 and Nod2 (Viala et al, 2004). Consistent with the latter, treatment of HEK293T cells with high amount of lipophilic Nod1- and Nod2-stimulatory molecules KF1B (5 μg/ml) and AcMDP (acetyl-(6-O-stearoyl)-muramyl-Ala-D-Glu-NH2) (5 μg/ml) induced NF-κB activation (Figure 6B). Using this system, we tested the ability of A20 to affect the NF-κB activation induced by these Nod1- and Nod2-stimulatory molecules in HEK293T cells, and found that NF-κB activation induced by KF1B and AcMDP, as well as that induced by TNFα and IL-1β, was reduced by A20 overexpression (Figure 6B). We next determined whether endogenous A20 mediates negative feedback of NF-κB activation after Nod1 stimulation. To test this, we used A20 small-RNA interference (RNAi) construct, which suppresses specifically the expression of A20 (Saitoh et al, 2005; also see inset in Figure 6C). Like that induced by TNFα and IL-1β, NF-κB activation induced by KF1B and AcMDP were enhanced by transfection with the A20 RNAi plasmid but not with control plasmid (Figure 6C). In addition, the ubiquitination level of endogenous RICK induced by Nod1 stimulation was upregulated by expression of A20 RNAi (Figure 6D). These findings suggest that A20 is a negative feedback regulator of Nod1 and Nod2 signaling.
Figure 6.
A20 functions as a negative regulator of Nod1 and Nod2 signaling via deubiquitination of RICK. (A) HEK293T cells were transfected with Flag-tagged A20 expression plasmids or control vector (−) in the presence of pcDNA3-RICK-ΔCARD-Fpk3-Myc (WT) and HA-Ub expression plasmid. Eighteen hours post-transfection, cells were treated with 200 nM AP1510 or left alone. Twenty-four hours post-transfection, proteins immunoprecipitated with rabbit anti-Myc Ab were subjected to SDS–PAGE and analyzed by western blot analysis using anti-HA, Myc and Flag Abs to detect ubiquitinated RICK proteins, RICK proteins, and A20 protein, respectively. (B) HEK293T cells were transfected with expression plasmids of A20 or control vector in the presence of reporter plasmids. Eight post-transfection, cells were treated with medium containing 1 μg/ml KF1B, 1 μg/ml AcMDP, 10 ng/ml TNF or 10 ng/ml IL-1β. Twenty-four hours post-transfection, ligand-dependent NF-κB activation was determined with reporter assay. The level of NF-κB-dependent transcription activity in the absence of A20 is given as 100%. The results shown are given as mean±s.d. of triplicate cultures and are representative of three experiments. (C) HEK293T cells were transfected with pcDNA3 (−), A20 interference RNA (RNAi) plasmid or control RNAi plasmid (Ctli) in the presence of reporter plasmids. Thirty-six hours after transfection, cells were treated with medium containing 1 μg/ml KF1B, 1 μg/ml AcMDP, 10 ng/ml TNFα or IL-1β. Forty-eight hours post-transfection, ligand-dependent NF-κB activation was determined with reporter assay. The level of NF-κB-dependent transcription activity in cells transfected with pcDNA3 is given as 1. Expression of A20 and control IKKβ proteins is shown in inset. The results shown are given as mean±s.d. of triplicate cultures and are representative of three experiments. (D) HEK293 cells constitutively expressing Nod1 were transfected with pcDNA3 (−) or A20 RNAi plasmid. Forty-eight hours after transfection, cells were treated with 100 ng/ml KF1B for 1 h. The cells were lysed and endogenous RICK was immunoprecipitated. Ubiquitinated and total RICK proteins were immunodetected with anti-Ub and anti-RICK Abs, respectively.
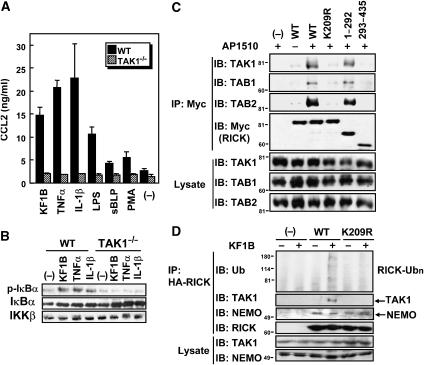
Polyubiquitination of RICK is required for the recruitment of TAK1
Recent studies showed that a TAK1 complex containing TAK1, TAB1, TAB2 and/or TAB3 is a general mediator of NF-κB activation induced by various inflammatory stimuli (Ishitani et al, 2003; Sato et al, 2005; Shim et al, 2005). Because TAK1 has been suggested to play a role in NF-κB activation induced by Nod2 signaling (Chen et al, 2004; Windheim et al, 2007), we hypothesized that TAK1 mediates Nod1-induced innate immune responses. To test this, we examined if KF1B induces the secretion of CCL2 from TAK1-deficient MEFs. We found that WT MEFs secreted CCL2 in response to KF1B stimulation but TAK1-deficient MEFs did not (Figure 7A). The impaired response in TAK1−/− MEFs was not due to the lack of expression of the general NF-κB regulators, IKKβ and IκBα, but was associated with impaired phosphorylation of IκBα (Figure 7B). These results indicate that TAK1 plays an essential role in Nod1 signaling. Because K63-linked polyubiquitination of RIP recruits TAK1 complex to the TNFR1 complex leading to NF-κB activation (Kanayama et al, 2004), we tested whether RICK polyubiquitination at K209 mediates the recruitment of TAK1 complex to RICK. To determine this, Myc-RICK-ΔCARD-Fpk3 proteins were coexpressed with TAK1, TAB1 and TAB2. We found that WT Myc-RICK-ΔCARD-Fpk3 protein co-immunoprecipitated with TAK1, TAB1 and TAB2 in the presence but not in the absence of the dimerizer AP1510 (Figure 7C). These results suggest that polyubiquitinated RICK is associated with the TAK1 complex. To test if polyubiquitination is required for recruitment of TAK1 complex to RICK, we tested the ability of K209R-mutant construct that is deficient in signal-dependent polyubiquitination, to interact with the TAK1 complex. The analysis revealed that the K209R mutant of RICK did not associate with TAK1 complex when compared with WT RICK (Figure 7C). These results suggest that polyubiquitination of RICK at K209 is essential for recruitment of the TAK1 complex to RICK. To test if the interaction of TAK1 with RICK is sufficient to induce NF-κB activation, we assessed if TAK1 is recruited to the RICK (1–292)-mutant construct that is polyubiquitinated upon stimulation but is unable to activate NF-κB and interact with NEMO (see Figure 3). Notably, RICK (1–292) mutant, but not RICK (293–435), associated with the TAK1 complex after addition of AP1510 (Figure 7C). To verify that this finding is not due to overexpression of recombinant proteins, we further investigated the interactions of TAK1 and NEMO with WT and K209R RICK proteins, which are expressed at levels comparable to that of endogenous RICK in RICK−/− MEFs. As expected, WT but not K209R-mutant RICK was polyubiquitinated and interacted with TAK1 upon Nod1 stimulation, whereas the interaction between RICK and NEMO was constitutive and was not affected by the mutation at K209 (Figure 7D). Together with the findings described in previous figures, this finding suggests that TAK1 recruitment to K209-polyubiquitinated RICK is essential for IKK activation upon Nod1 stimulation.
Figure 7.
RICK polyubiquitination at K209 is required for the recruitment of TAK1/TAB1/TAB2 complex. (A) WT or TAK1-deficient MEFs were stimulated with 5 μg/ml KF1B, 10 ng/ml TNFα, 10 ng/ml IL-1β, 100 ng/ml LPS, 1 μg/ml sBLP, 50 ng/ml PMA plus 0.7 μg/ml Ca2+ ionophore A23187 or left alone. Twenty-four hours post-stimulation, secretion levels of CCL2 were determined by ELISA. The results shown are given as mean±s.d. of triplicate cultures and are representative of three experiments. (B) WT or TAK1-deficient MEFs were stimulated with 5 μg/ml KF1B, 10 ng/ml TNFα or 10 ng/ml IL-1β for 1 h. Post-stimulation, cells were lysed and lysates immunoblotted with anti-phospho-IκBα, anti-IκBα or IKKβ Ab. (C) HEK293T cells were transfected with expression plasmids of RICK-ΔCARD-Fpk3-Myc (WT), RICK-K209R-ΔCARD-Fpk3-Myc (K209R), RICK-(1–292)-Fpk3-Myc, RICK-(293–435)-Fpk3-Myc and control vector (−) in the presence of expression plasmids of Ub, HA-TAK1, Flag-TAB1 and T7-TAB2 and treated with AP1510 as in Figure 2B. Twenty-four hours post-transfection, proteins immunoprecipitated with rabbit anti-Myc Ab were subjected to SDS–PAGE and analyzed by immunoblotting analysis using anti-HA or, anti-Flag, anti-T7 and anti-Ub Abs. (D) MEFs stably expressing RICK or RICK K209R were stimulated with 5 μg/ml KF1B for 1 h. The cells were lysed, RICK was immunoprecipitated and endogenous Ub, TAK1, NEMO and RICK were immunodetected with anti-Ub, anti-TAK1 Ab, anti-NEMO or anti-RICK antibodies.
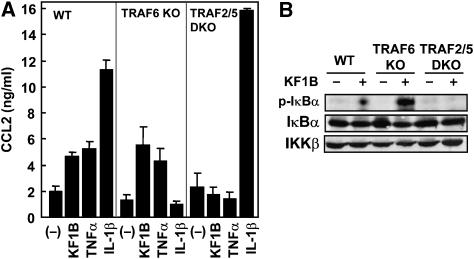
TRAF2/TRAF5 but not TRAF6 are required for Nod1-mediated NF-κB activation and CCL2 secretion
Recent findings suggest that TRAFs are ubiquitin ligases (E3s), which regulate several immune responses including inflammatory responses mediated by NF-κB. Although TRAF6 is suggested to be important for Nod2-mediated signaling, TRAF6-depeleted cells still respond to MDP, a Nod2-stimulatory molecule (Abbott et al, 2007). We also found that TRAF6−/− MEFs respond to KF1B, resulting in IκBα phosphorylation (Figure 8B) and induced secretion of CCL2 (Figure 8A). Thus, TRAF6 appears to be a non-essential E3 for both Nod1 and Nod2 signaling. Notably, Nod1 stimulation did not induce NF-κB activation and IκBα phosphorylation in MEFs lacking TRAF2 and TRAF5, which are essential for TNFα-induced NF-κB activation and are able to complement one another (Tada et al, 2001; Figure 8). These results suggest that TNFα- and Nod1-mediated signaling share common E3s to activate NF-κB.
Figure 8.
TRAF2/TRAF5, but not TRAF6, are required for Nod1-mediated NF-κB activation and CCL2 secretion. (A) WT, TRAF6−/− (TRAF6 KO) or TRAF2−/−TRAF5−/− (TRAF2/5 DKO) MEFs were stimulated with 5 μg/ml KF1B, 10 ng/ml TNFα, 10 ng/ml IL-1β or left alone. After 24 h incubation, the secretion levels of CCL2 were determined by ELISA. The results shown are given as mean±s.d. of triplicate cultures and are representative of three experiments. (B) WT and mutant MEFs were stimulated with 5 μg/ml KF1B for 1 h or left alone. The phosphorylated (upper panel) and whole (middle panel) populations of IκBα and IKKβ (lower panel) were immunodetected with specific Abs.
Discussion
RICK is an essential and specific mediator of Nod1 and Nod2 signaling (Chin et al, 2002; Kobayashi et al, 2002; Park et al, 2007). However, the mechanism by which RICK mediates Nod1- and Nod2-mediated immune responses has remained poorly defined. Here we found an essential role for RICK polyubiquitination in Nod1- and Nod2-mediated NF-κB activation. The K63-linked polyubiquitin chain at K209 in RICK was found to be important for the recruitment of the TAK1 complex to RICK and IKKs. Similarly, RIP (RIPK1), a RICK homologue, was shown to play a critical role in TNFα-induced NF-κB activation through its polyubiquitination at K377 located in the IM region between the N-terminal kinase and C-terminal death domains (Ea et al, 2006; Li et al, 2006). Like that of RICK, polyubiquitinations of RIP are both critical for recruitment of TAK1 complex. Therefore, our results indicate that Nod1/Nod2 and TNFα signaling is mediated through similar molecular events.
Previous studies suggested an important role for the IM domain located between the kinase domain and the CARD of RICK in Nod1-mediated NF-κB activation (Inohara et al, 2000). Here we further characterized the structural requirement of IM and found that the region between 293 and 319, near to the N-terminal kinase domain, is important for NF-κB activation. This is also comparable to the observation that the IM region of RIP is required for TNF signaling (Inohara et al, 2000). However, oligomerization of RICK-Fpk3 fusion protein carrying the IM region alone did not result in NF-κB activation (Figure 3C), whereas that of the equivalent region of RIP was sufficient to induce oligomerization-dependent NF-κB activation (Inohara et al, 2000). These results suggest that the kinase domain of RICK, but not that of RIP, is essential for NF-κB activation. Previous studies showed that the K47M kinase-inactive mutant of RICK still activates NF-κB (Inohara et al, 2000), suggesting that the kinase domain of RICK mediates a critical function, other than phosphorylation, which is required for the induction of NF-κB. Indeed, we found here that K63-linked polyubiquitination at K209 within the kinase domain of RICK is essential for RICK-mediated signaling. In contrast, the critical polyubiquitination site of RIP at K377 is located the IM region (Ea et al, 2006; Li et al, 2006). These results indicate that RICK and RIP differ in the domain localization of the critical K63-linked polyubiquitination site, providing a mechanistic explanation for the observed differences between the two related kinases in the induction of NF-κB. Although the localization of the K63-linked polyubiquitinated site is different in RICK and RIP, polyubiquitination of both kinases is regulated by upstream stimuli and required for recruitment of the critical TAK1 complex to these kinases. While polyubiquitination of RICK is essential for Nod1-mediated NF-κB activation, the E3s that mediate polyubiquitination of RICK remain unknown. Some possible candidates include the TRAFs, which are known to associate with RICK (McCarthy et al, 1998; Thome et al, 1998). Here we demonstrated that TRAF2−/−TRAF5−/− MEFs lack NF-κB activation upon Nod1 stimulation. However, we do not have convincing evidence that TRAF2 and TRAF5 directly ubiquitinate RICK. We could not find consensus sequences of TRAF2- and TRAF5-binding sites in regions near K209 in RICK. Although we still do not know how the E3(s) specifically recognize RICK as a substrate, K209 is highly conserved in RICK but not in other kinases and is located in the putative flexible loop region (Supplementary Figure 1). Further studies are required to determine the mechanism by which TRAFs are involved in Nod/RICK-mediated signaling, and conclusively identify the E3(s), which ubiquitinates K209 in RICK.
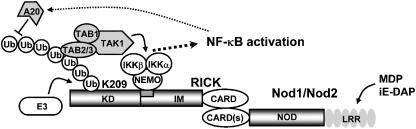
In summary, we have dissected two molecular signaling events that are critical for Nod signaling, namely recruitment of TAK1 to polyubiquitinated K209 located in the kinase domain, and recruitment of NEMO, which is mediated by the IM region of RICK. NEMO interacted with RICK-ΔCARD-Fpk3 in the absence of AP1510, which is required for polyubiquitination of K209 and concomitant NF-κB activation (Figure 3D). As a result, the interaction between RICK and NEMO through the IM region appears to be independent of the interaction between TAK1 and RICK and is not sufficient to activate NF-κB. Therefore, the association of RICK with both TAK1 and IKK complexes are required for signal transduction resulting in NF-κB activation. This conclusion leads us to propose the following model (Figure 9): the molecular events during Nod1 and Nod2 signaling are (1) the recruitment of RICK to Nod1 and Nod2 upon sensing of their specific microbial stimuli, (2) the close proximity of RICK induced by the self-oligomerization of Nod1 and Nod2, (3) polyubiquitination at K209 in RICK by unknown E3(s), (4) recruitment of the TAK1 complex to RICK, (5) further recruitment of IKK complex through the interaction between the IM region of RICK with NEMO, and finally (6) activation of IKKs by TAK1. In this model, Nod1 and Nod2 play an important role as switches to turn on the downstream cascade reaction and RICK functions as a bridging protein to link TAK1 and IKK complexes. Similarly, RIP may function as a bridging protein between TAK1 and IKK complexes in TNFα signaling. However, the K63-linked polyubiquitination site K377 is located in the IM region and, therefore, previous studies could not separate the two molecular events mediated by RIP, namely the recruitment of TAK1 and that of the IKKs. Further studies are required to explore the differences and similarities between Nod and TNF signaling pathways.
Figure 9.
Model for the role of RICK polyubiquitination in Nod signaling. For detail, see text.
The kinase domain of RICK possesses protein kinase activity (Inohara et al, 1998; McCarthy et al, 1998; Thome et al, 1998) and residues critical for catalytic activity are evolutionary conserved. However, previous and current studies showed that the residue critical for kinase activity is not essential for NF-κB activation, polyubiquitination of RICK and interaction of RICK with NEMO (Inohara et al, 2000). Therefore, the kinase activity of RICK might regulate other molecules including ERKs, p38 kinases and caspase-1 that have been linked to RICK-mediated signaling (Thome et al, 1998; Navas et al, 1999; Chin et al, 2002; Kobayashi et al, 2002). Previous studies suggested that the activation of these intracellular molecules is important for secretion of certain cytokines upon Nod1 and Nod2 stimulation (Inohara et al, 2001; Girardin et al, 2003a, 2003b; Viala et al, 2004). Alternatively, the kinase domain of RICK may regulate the turnover rate of the RICK protein. Consistent with the latter, we found lower but significant residual polyubiquitination of RICK independent from K63-Ub upon Nod1 and Nod2 stimulation (Figure 2), suggesting that RICK is partially polyubiquitinated in a K63-Ub-independent manner. Furthermore, K48-linked polyubiquitination of RIP during TNFα signaling has been suggested to regulate proteasomal degradation of RIP (Wertz et al, 2004). Therefore, our finding suggests that K48-linked polyubiquitination of RICK might regulate RICK protein levels during Nod signaling. The functional relevance of K63-Ub-independent polyubiquitination of RICK and the role of the kinase activity in RICK protein degradation warrant further investigation.
Materials and methods
Ligand compounds, plasmids and culture cells
Synthetic compounds, iE-DAP and KF1B have been described (Masumoto et al, 2006). MDP, AcMDP, sBLP (Pam3Cys-OH) TNFα, IL-1β, PMA and A23187 were obtained from commercial sources. AP1510 is a gift from Ariad (Cambridge, MA). pcDNA3-Fpk3-Myc, pcDNA3-RICK-Fpk3-Myc, RICK-(1–292)-Fpk3-Myc, pcDNA3-RICK-(K47M)-Fpk3-Myc, pcDNA3-Myc-RICK, pCGN-HA-Ub, pBxIV-luc, pcDNA3-β-gal and pMX2-puro-HA-Nod2 have been described (Inohara et al, 2000, 2003; Nishito et al, 2006). pCMV-SPORT6-UBB encoding human ubiquitin B (IMAGE 4943285) was obtained from Open Biosystems (Huntsville, AL). RICK-(1–319)-Fpk3-Myc, RICK-(1–328)-Fpk3-Myc, RICK-(1–356)-Fpk3-Myc, RICK-(1–390)-Fpk3-Myc and RICK-(293–435)-Fpk3-Myc were generated by subcloning of RICK amplified by PCR using specific primers and subcloned into the KpnI and XhoI sites of pcDNA-Fpk3-Myc. pMSCV-hygro-HA-RICK and pMSCV-puro-Nod1-Flag are generated by subcloning of the HA-RICK and Nod1-Flag genes into pMSCV-hygro and pMSCV-puro, respectively (Clontech). Point mutations of RICK and Ub, in which individual lysine residues were mutated to arginines, were constructed using the QuikChange™ site-directed mutagenesis kit (Stratagene). The fidelity of all constructs was confirmed by sequencing. pcDNA3-HA-TAK1, pcDNA3-T7-TAB1 and pcDNA3-Flag-TAB2 are gifts from Dr T Koseki (Tohoku University, Sendai, Japan). pcDNA3-A20 (Vincenz and Dixit, 1996) is a gift from Dr C Vincenz (University of Michigan, MI). pU6-A20i and pU6-Ctli, A20-specific and control siRNA plasmids (Saitoh et al, 2005) are gifts from Dr S Yamaoka (Tokyo Medical and Dental University, Tokyo, Japan).
HEK293T, parental 293 cells and MEFs were cultured as described (Inohara et al, 2000). RICK-, TAK1-, TRAF6- and TRAF2/TRAF5-deficient and control WT MEFs are described previously (Naito et al, 1999; Tada et al, 2001; Kobayashi et al, 2002; Shim et al, 2005). TAK1-deficient MEFs were provided by Dr S Ghosh (Yale University, New Heaven, CT) and TRAF6-deficient MEFs by Dr J Inoue (Tokyo University, Tokyo, Japan). HEK293 cell lines constitutively expressing Nod1-Flag and HA-Nod2 with NF-κB-dependent GFP reporter were generated by transfection of pMSCV-puro-Nod1-Flag and pMX2-puro-HA-Nod2, respectively, and NFκB-eGFP (Wang et al, 2002), followed by antibiotic selection.
Luciferase assay
NF-κB activation was determined using 0.5 × 105 HEK293T cells transfected with expression plasmids in the presence of reporter plasmids, NF-κB-dependent pBxIV-luc and control pEF1BOS-β-gal, as described (Inohara et al, 2000; Kobayashi et al, 2002).
Cytokine/chemokine secretion
WT and RICK-deficient MEFs were infected with retroviral vectors pMSCV-hygro-HA-RICK and pMSCV-hygro-HA-RICK K209R packaged in HEK293T cells cotransfected with EcoPack plasmid (a gift from Dr G Nolan, UCSF). Infection was carried out in the presence of 5 μg/ml polybrene for 3 h and incubation continued for 48 h with fresh medium. Hygromycin-resistant (hygror) MEFs were selected with 0.3 mg/ml hygromycin (Roche) for 5 days. A total of 1 × 104 hygror MEFs were cultured in 0.25 ml medium in 48-well plates and stimulated with inflammatory stimuli indicated in figure legends. Twenty-four hours post-stimulation, the secretion levels of CCL2 and CXCL1 in the medium were determined by sandwich ELISA kits (BD, San Diego, CA). Similarly, WT and TAK1-deficient MEFs were stimulated with the indicated inflammatory stimuli and CCL2 and CXCL1 secretion levels were determined 24 h post-stimulation.
Immunoprecipitation analyses and immunodetection
HEK293 cells were transfected with expression plasmids or siRNA plasmids and lysed with NP-40 buffer as described (Inohara et al, 1999) The proteins were immunoprecipitated with anti-Myc rabbit polyclonal (A-14; Santa Cruz) or rabbit anti-RICK (H-300; Santa Cruz) antibody (Ab). Myc-, HA- or Flag-tagged proteins were immunodetected with horseradish peroxidase-conjugated Abs specific for each tag (A-14 and Y-11 from Santa Cruz for Myc and HA tags, respectively, M2 from Sigma-Aldrich for Flag tag).
A total of 1 × 107 MEFs were stimulated with 5 μg/ml KF1B for the indicated time. MEFs were lysed with and RICK proteins were immunoprecipitated by rabbit anti-RICK or rabbit anti-HA Ab (Y-11; Santa Cruz). Ubiquitinated and total RICK proteins, TAK1 or NEMO in the immunoprecipitates were immunodetected by mouse monoclonal anti-Ub (Abcam, Cambridge, MA), anti-RICK (Alexis, San Diego, CA), anti-TAK1 (C-9; Santa Cruz) and anti-NEMO Abs (B-3; Santa Cruz), respectively. Cell lysates were immunoblotted with anti-phospho-IκBα, anti-IκBα (Cell Signaling) or IKKβ Ab (H-470; Santa Cruz).
In vitro kinase assay
A total of 2 × 106 MEFs stably expressing RICK or RICK K209R were stimulated with 5 μg/ml KF1B or 10 ng/ml TNFα for the indicated time, and then cells were lysed in Triton lysis buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM β-glycerophosphate, 10 mM NaF, 300 μM Na3VO4 and 1 mM DTT). The cell lysates were immunoprecipitated with mouse anti-NEMO Ab (BD Pharmingen). Resulting immunoprecipitates were washed three times with lysis buffer and once with kinase buffer (20 mM HEPES pH 7.6, 2 mM MgCl2, 2 mM MnCl2, 10 μM ATP, 10 mM β-glycerophosphate, 10 mM NaF, 300 μM Na3VO4 and 1 mM DTT). Immunoprecipitated IKK complexes were incubated with GST-IκBα as substrates in kinase buffer with γ-32P-ATP at 30°C for 30 min. The samples were fractionated on SDS–PAGE followed by autoradiography. The amounts of IKKβ in the immunoprecipitates were detected by immunoblotting.
Supplementary Material
Supplementary Figure 1
Supplementary Figure Legend
Acknowledgments
This work was supported by NIH grants R01 GM60421 (to N Inohara) and R01 DK067628 (to G Nuñez). We are grateful to L McAllister-Lucas (University of Michigan) for stimulating discussions; S Qiu, P Kuffer, Y Nishito and S Chen (University of Michigan) for technical assistance and S Yamaoka (Tokyo Medical and Dental University, Tokyo), T Koseki (Tohoku University, Japan), J Inoue (Tokyo University, Japan), C Vincenz (University Michigan), S Ghosh (Yale University) and Ariad Pharmaceuticals for materials. The authors have no conflicting financial interests.
References
- Abbott DW, Wilkins A, Asara JM, Cantley LC (2004) The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol 14: 2217–2227 [DOI] [PubMed] [Google Scholar]
- Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC (2007) Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol 27: 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A (2004) The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5: 1052–1060 [DOI] [PubMed] [Google Scholar]
- Boughan PK, Argent RH, Body-Malapel M, Park JH, Ewings KE, Bowie AG, Ong SJ, Cook SJ, Sorensen OE, Manzo BA, Inohara N, Klein NJ, Nunez G, Atherton JC, Bajaj-Elliott M (2006) Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor: critical regulators of beta-defensins during Helicobacter pylori infection. J Biol Chem 281: 11637–11648 [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N (2003) An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 4: 702–707 [DOI] [PubMed] [Google Scholar]
- Chen CM, Gong Y, Zhang M, Chen JJ (2004) Reciprocal cross-talk between Nod2 and TAK1 signaling pathways. J Biol Chem 279: 25876–25882 [DOI] [PubMed] [Google Scholar]
- Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, Cheng G (2002) Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416: 190–194 [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ (2006) Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE (2006) Nod-like proteins in immunity, inflammation and disease. Nat Immunol 7: 1250–1257 [DOI] [PubMed] [Google Scholar]
- Fritz JH, Le Bourhis L, Sellge G, Magalhaes JG, Fsihi H, Kufer TA, Collins C, Viala J, Ferrero RL, Girardin SE, Philpott DJ (2007) Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity 26: 445–459 [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zaehringer U, Coyl AJ, Di Stefano PS, Bertin J, Sansonetti PJ, Philpott DJ (2003a) Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300: 1584–1587 [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ (2003b) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278: 8869–8872 [DOI] [PubMed] [Google Scholar]
- Goto T, Aoki H (1987) The immunomodulatory activities of acylpeptides. In Immunostimulants: Now and Tomorrow, Azuma I, Jollès G (eds), pp 99–108. Tokyo, Japan: Japan Science Society Press [Google Scholar]
- Hasegawa M, Yang K, Hashimoto M, Park JH, Fujimoto Y, Nunez G, Fukase K, Inohara N (2006) Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem 281: 29054–29063 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D (2006) Circuitry of nuclear factor kappaB signaling. Immunol Rev 210: 171–186 [DOI] [PubMed] [Google Scholar]
- Inohara N, Chamaillard M, McDonald C, Nuñez G (2005) NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem 74: 355–383 [DOI] [PubMed] [Google Scholar]
- Inohara N, del Peso L, Koseki T, Chen S, Nunez G (1998) RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem 273: 12296–12300 [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino LD, Ni J, Nunez G (1999) Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J Biol Chem 274: 14560–14568 [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, Chen FF, Ogura Y, Nunez G (2000) An induced proximity model for NF-kappaB activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem 275: 27823–27831 [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Chen FF, Muto A, Nunez G (2001) Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem 276: 2551–2554 [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nuñez G (2003) Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem 278: 5509–5512 [DOI] [PubMed] [Google Scholar]
- Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K (2003) Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J 22: 6277–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ (2004) TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y (2000) Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol 18: 621–663 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA (2002) RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416: 194–199 [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA (2005) Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307: 731–734 [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM (1992) Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem 267: 17971–17976 [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A (2000) Failure to regulate TNF-induced NF-kappa-B and cell death responses in A20-deficient mice. Science 289: 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kobayashi M, Blonska M, You Y, Lin X (2006) Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. J Biol Chem 281: 13636–13643 [DOI] [PubMed] [Google Scholar]
- McCarthy JV, Ni J, Dixit VM (1998) RIP2 is a novel NF-kappa-B-activating and cell death-inducing kinase. J Biol Chem 273: 16968–16975 [DOI] [PubMed] [Google Scholar]
- Marks DJ, Harbord MW, MacAllister R, Rahman FZ, Young J, Al-Lazikani B, Lees W, Novelli M, Bloom S, Segal AW (2006) Defective acute inflammation in Crohn's disease: a clinical investigation. Lancet 367: 668–678 [DOI] [PubMed] [Google Scholar]
- Masumoto J, Yang K, Varambally S, Hasegawa M, Tomlins SA, Qiu S, Fujimoto Y, Kawasaki A, Foster SJ, Horie Y, Mak TW, Nunez G, Chinnaiyan AM, Fukase K, Inohara N (2006) Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med 203: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, Inoue J (1999) Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells 4: 353–362 [DOI] [PubMed] [Google Scholar]
- Navas TA, Baldwin DT, Stewart TA (1999) RIP2 is a Raf1-activated mitogen-activated protein kinase kinase. J Biol Chem 274: 33684–33690 [DOI] [PubMed] [Google Scholar]
- Nishito Y, Hasegawa M, Inohara N, Nunez G (2006) MEX is a testis-specific E3 ubiquitin ligase that promotes death receptor-induced apoptosis. Biochem J 396: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G (2001) Nod2, a Nod1/Apaf-1 family member that is restincted to monocytes and activates NF-κB. J Biol Chem 276: 4812–4818 [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Nunez G (2007) RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol 178: 2380–2386 [DOI] [PubMed] [Google Scholar]
- Saitoh T, Yamamoto M, Miyagishi M, Taira K, Nakanishi M, Fujita T, Akira S, Yamamoto N, Yamaoka S (2005) A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol 174: 1507–1512 [DOI] [PubMed] [Google Scholar]
- Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, Akira S (2005) Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol 6: 1087–1095 [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S (2005) TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev 19: 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, Hashimoto H, Mak TW, Yagita H, Okumura K, Yeh WC, Nakano H (2001) Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J Biol Chem 276: 36530–36534 [DOI] [PubMed] [Google Scholar]
- Thome M, Hofmann K, Burns K, Martinon F, Bodmer JL, Mattmann C, Tschopp J (1998) Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol 8: 885–888 [DOI] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL (2004) Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5: 1166–1174 [DOI] [PubMed] [Google Scholar]
- Vincenz C, Dixit VM (1996) 14-3-3 Proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J Biol Chem 271: 20029–20034 [DOI] [PubMed] [Google Scholar]
- Voss E, Wehkamp J, Wehkamp K, Stange EF, Schroder JM, Harder J (2006) NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem 281: 2005–2011 [DOI] [PubMed] [Google Scholar]
- Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X (2002) A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat Immunol 3: 830–835 [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430: 694–699 [DOI] [PubMed] [Google Scholar]
- Windheim M, Lang C, Peggie M, Cummings L, Cohen P (2007) Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J 404: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure Legend