Abstract
CD8+ cytotoxic and CD4+ helper/inducer T cells develop from common thymocyte precursors that express both CD4 and CD8 molecules. Upon T cell receptor signaling, these cells initiate a differentiation program that includes complex changes in CD4 and CD8 expression, allowing identification of transitional intermediates in this developmental pathway. Little is known about regulation of these early transitions or their specific importance to CD4 and CD8 T cell development. Here, we show a severe block at the CD4loCD8lo transitional stage of positive selection caused by loss of the nuclear HMG box protein TOX. As a result, CD4 lineage T cells, including regulatory T and CD1d-dependent natural killer T cells, fail to develop. In contrast, functional CD8+ T cells develop in TOX-deficient mice. Our data suggest that TOX-dependent transition to the CD4+CD8lo stage is required for continued development of class II major histocompatibility complex–specific T cells, regardless of ultimate lineage fate.
Changes in expression of two surface proteins, CD4 and CD8, which act as coreceptors in conjunction with the TCR, are commonly used markers for stages of T cell development. CD4+CD8+ double positive (DP) thymocytes that express a mature TCR of appropriate specificity undergo a positive selection process, up-regulating survival factors and differentiating into either CD4+CD8− or CD4−CD8+ single positive (SP; CD4SP or CD8SP, respectively) thymocytes. However, positive selection can be further subdivided into stages based on coreceptor expression, including partial down-regulation of both coreceptors to yield a CD4loCD8lo double dull (DD) phenotype, followed by reexpression of CD4 to produce a CD4+CD8lo transitional phenotype (1–3). Maintenance of CD4 expression coupled with complete loss of CD8 or reexpression of CD8 and silencing of CD4 in these transitional cells results in the mature CD4SP or CD8SP phenotype, respectively (4).
DD thymocytes express cell surface and molecular markers that indicate that they are the product of positive selection (1, 2). Indeed, production of the DD phenotype has been used as an assay for identification of self-peptides that can mediate positive selection (3). That there is a direct precursor–product relationship between DD and CD4+8lo cells is supported by the fact that development of DD thymocytes precedes that of CD4+8lo thymocytes during recovery from irradiation and, most directly, that DD thymocytes give rise to CD4+8lo cells in culture (1, 5). Based solely on coreceptor expression, however, this is also a heterogeneous cell population, likely containing both dying cells (6) and transitional intermediates that are products rather than precursors of CD4+8lo cells (2, 7). CD4+8lo cells themselves contain precursors for both CD4 and CD8 T cells (4, 8, 9), consistent with the induction of Zbtb7b (10, 11) and Runx3 (5), critical factors for CD4 and CD8 lineage development, respectively, in this cell subpopulation. Moreover, it has been shown that class II MHC–specific TCR-transgenic (Tg) DP thymocytes transition to CD4SP phenotype cells through DD and CD4+8lo stages when cultured with thymic epithelial cells expressing the cognate MHC specificity (5). However, in a two-step reaggregation system, these same DD thymocytes become CD4+8lo and then CD8SP when cognate MHC interactions are removed (5). Thus, a linear pathway of DD to CD4+8lo to SP appears to be the normal developmental progression during positive selection of both CD4 and CD8 lineage T cells. Thymocytes bearing some specificities, however, may directly transit from a DD to CD8SP phenotype (12).
Interestingly, the DD phenotype is a reflection of reduced Cd4 and Cd8 gene expression (reference 13 and unpublished data). Evidence from marker Tg mice has suggested that the Cd4 gene silencer may be transiently active in DD thymocytes (13). In addition, there is data to suggest that regulation of the Cd4 enhancer may differ between immature and mature T cells (13–16). Similarly, changes in Cd8 gene regulation occur as a result of positive selection (17–19). Thus, the DD phenotype may reflect a “gear shift” in coreceptor gene regulation. Whether the phenotype of these cells also has functional significance, however, remains unclear. Asymmetrical coreceptor expression at the later CD4+CD8lo stage, however, has been proposed to allow discrimination between class I and class II MHC–specific TCR due to differential dependence on CD8 or CD4 coreceptors for signaling (9, 20).
Several nuclear factors have been identified that play key roles in coreceptor gene regulation and development to the CD4 or CD8 lineages (for review see reference 21). Nevertheless, much remains unknown about how the specific phenotypic stages during positive selection are regulated, their linkage to TCR-mediated signaling, and their importance for T cell lineage diversification.
We have shown previously that the thymocyte selection–associated HMG box protein (TOX) is transiently up-regulated by calcineurin-mediated TCR signaling in DP thymocytes as they develop into SP thymocytes (22, 23). In this study, we describe TOX as a key regulator of the DD to CD4+8lo transition during positive selection. The absence of this nuclear protein prevents CD4 lineage T cell development, including CD1d-dependent natural killer T (NKT) and T regulatory (T reg) CD4 T cell sublineages, but has only modest effects on CD8 T cell development. The failure of the majority of developing polyclonal CD4 lineage T cells to switch to the CD8 lineage in the absence of TOX further indicates the critical importance of the CD4+8lo stage to developmental progression of class II MHC–restricted T cells.
RESULTS
Germline deletion of Tox
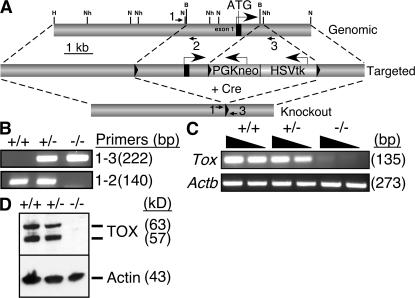
Using standard gene-targeting methodology, embryonic stem (ES) cells were produced in which loxP sites flanked a genomic region that included exon 1 and 1.7 kb upstream of the Tox gene. After transfection of Cre recombinase, ES cell clones were selected that deleted this genomic fragment as well as that encoding the co-integrated selectable markers (Fig. 1 A). Chimeric mice produced from these targeted ES cells were screened by Southern blotting (not depicted) and bred to produce heterozygous (Tox+/−) and homozygous (Tox−/−) mutant mice, as screened by genomic PCR (Fig. 1 B). Tox−/− mice did not express detectable Tox mRNA or TOX protein, the latter assessed using an antibody directed against a C-terminal peptide of TOX (22, 23), when compared with Tox+/+ or Tox+/− mice (Fig. 1, C and D).
Figure 1.
Generation of Tox−/− mice. (A) Schematic of gene-targeting strategy to delete genomic region surrounding exon 1 of the Tox gene. The location of loxP sites (triangles), exon 1 (black box), transcription start sites (ATG; large arrows), and genomic PCR primers (small arrows) is shown. Location and direction of selectable marker gene cassettes encoding thymidine kinase (HSVTk) and neomycin resistance (PGKneo) are also depicted. Restriction enzymes sites in Tox are indicated. H, HindIII; Nh, NheI; N, NcoI; B, BglII. (B) Genomic PCR for Tox in total thymocytes from Tox+/+ (+/+), Tox+/− (+/−), and Tox−/− (−/−) mice using combinations of primers shown in A demonstrates expected genomic structure of the targeted locus. (C and D) Loss of Tox mRNA demonstrated by RT-PCR (C) and TOX protein analyzed by immunoblotting (D) in total thymocytes from gene-targeted mice. Expression of the Actb gene and β-actin protein was used for controls. In this and all subsequent figures, protein masses are based on relative mobility in SDS-PAGE.
Loss of TOX does not inhibit B cell development or thymic β selection
Tox mRNA is detected in several tissues in addition to the thymus, including the brain (22). However, Tox−/− mice were viable and fertile, and had no obvious abnormalities in appearance or behavior. We have not detected expression of TOX in developing B cells (unpublished data). Consistent with this, loss of TOX had no discernable effect on stages of B cell development in the BM or appearance of mature B cells in the spleen (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071944/DC1).
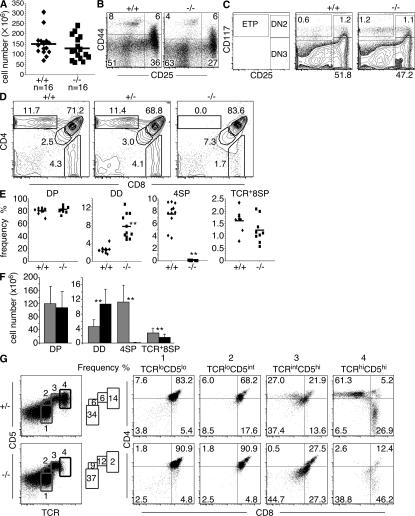
TOX is expressed in early DN subsets and then transiently during β selection, the process that initiates the DN to DP thymocyte progression (22). There was no significant reduction in thymic cellularity in Tox−/− mice (Fig. 2 A), nor loss of DN subsets as defined by expression of CD44 and CD25, or CD117+ early T lineage precursor (ETP) cells (for review see reference 24) (Fig. 2, B and C). Consistent with these results, the frequency and number of DP thymocytes in Tox−/− mice are comparable to that of wild-type animals (Fig. 2, D–F). Thus, TOX is dispensable for T cell commitment in the thymus as well as differentiation and cell expansion associated with β selection.
Figure 2.
TOX is required for CD4SP thymocyte development but not initiation of positive selection. (A) No statistically significant difference (P = 0.28) in total thymic cellularity of individual +/+ and −/− mice was observed. Mice ranging in age from 2.5 to 9 wk were analyzed as age-matched control and experimental pairs. Population means are shown as horizontal bars. (B) Maintenance of CD44- and CD25-defined DN subsets in lineage− (CD4−, CD8α−, B220−, DX-5−, and γδTCR−) thymocytes from Tox−/− mice. Numbers indicate frequency of DN1 (CD44+CD25), DN2 (CD44+CD25+), DN3 (CD44−CD25+), and DN4 (CD44+CD25−) subsets. (C) Maintenance of CD117- and CD25-defined DN subsets and ETP in lineage− (CD8α−, CD8β−, CD3ε−, TCR-β−, CD11b−, CD11c−, TER119−, CD19−, B220−, NK1.1−, and γδTCR−) thymocytes from Tox−/− mice. ETP (CD117+CD25−), DN2 (CD117+CD25+), DN3 (CD117−CD25+), and DN4 (CD117+CD25−) subsets are depicted. (D) Representative CD4/CD8 staining patterns of thymocytes derived from +/+, +/−, and −/− mice are shown. Numbers indicate frequency of indicated thymocyte subsets, expressed as a percentage in this figure and all subsequent figures unless otherwise indicated. (E) Compilation of the frequency of CD4- and CD8-defined thymocyte subsets in +/+ and −/− mice. CD8SP thymocytes were also gated on TCR+ cells to eliminate immature CD8SP thymocytes from the analysis. Statistically significant differences between the mean frequency of DD (P = 4.10−11) and 4SP (P = 1.10−12) +/+ and −/− thymocytes are indicated (**). (F) Absolute numbers of thymocyte subsets in +/+ (gray bars) and −/− (black bars) mice. Error bars refer to standard deviations (n = 16). Statistically significant differences between the mean absolute number of DD (P = 5.7 × 10−6), 4SP (P = 1.9 × 10−10), and 8SP (P = 0.0047) +/+ and −/− thymocytes are indicated (**) (G) Tox−/− thymocytes initiate positive selection based on marker up-regulation. Two-parameter analysis for expression of TCR-β and CD5 allows identification of developmental stages (labeled 1–4) that were then assessed for expression of CD4 and CD8.
Thymocyte-intrinsic TOX is required for completion of positive selection of the CD4 lineage
Analysis of later stages of T cell development revealed that Tox−/− mice had a near complete loss of CD4+CD8lo transitional and CD4SP thymocytes (Fig. 2, D–F). In contrast, the DD thymocyte subpopulation was expanded threefold on average (Fig. 2, D–F). Postpositive selection TCR+ CD8SP thymocytes were present in Tox−/− mice at modestly reduced frequency on average that failed to reach statistical significance (P = 0.17) (Fig. 2 E), although absolute cell numbers were significantly reduced (P = 0.005) (Fig. 2 F). No T cell developmental defects were observed in Tox+/− mice (Fig. 2 D). Staining for CD4 and CD8β gave similar results (not depicted).
Analysis of TCR and CD5 expression is useful to delineate stages of positive selection (25). In wild-type mice, TCRloCD5lo cells consisted of preselection DP thymocytes, TCRloCD5int cells were DP cells initiating positive selection, TCRintCD5hi cells were DD and CD4+8lo thymocytes in the process of positive selection, and TCRhiCD5hi cells were primarily postselection CD4SP and CD8SP thymocytes (stages 1–4, respectively; Fig. 2 G). In Tox−/− mice, preselection and early selection DP stage 1 and 2 cells, respectively, were present normally, stage 3 cells undergoing positive selection were increased in frequency but lacked CD4+8lo cells, and stage 4 CD4SP thymocytes were absent (Fig. 2 G). Some stage 4 cells were present in Tox−/− mice, although with somewhat reduced TCR expression, and these were primarily CD8SP thymocytes (Fig. 2 G). These data indicate that Tox−/− thymocytes initiated positive selection (stages 1–3) but failed to complete development to subsequent CD4+8lo and CD4SP stages.
These results were confirmed with an additional marker of positive selection, CD69 (26). In this instance, stages 1–4 were defined as TCR−/loCD69−, TCRintCD69lo/hi, TCRhiCD69hi, and TCRhiCD69lo/− cells, respectively, with a progression of coreceptor changes similar to the subpopulations defined above by expression of TCR and CD5 (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20071944/DC1). In Tox−/− mice, preselection DP stage 1 cells were present normally, stage 2 cells undergoing positive selection were increased in frequency but lacked CD4+8lo cells, and stage 3 and 4 CD4SP thymocytes were absent (Fig. S2). The stage 4 cells that were present in Tox−/− mice were primarily CD8SP thymocytes (Fig. S2).
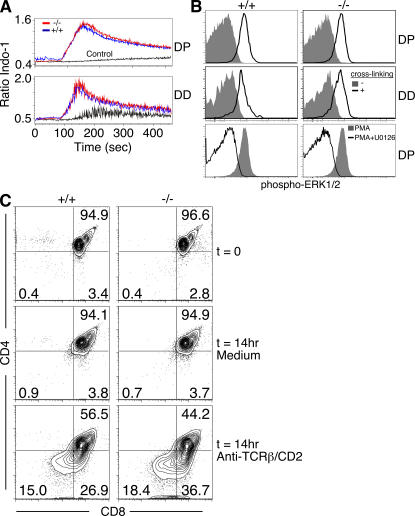
The normal initiation of positive selection in Tox−/− mice (Fig. 2 G and Fig. S2) suggested that there was no defect in TCR signaling in the mutant thymocytes. To address this more directly, we analyzed calcium flux and phosphorylation of extracellular signal–regulated kinases 1 and 2 (ERK1 and 2) in response to CD3 cross-linking, aspects of two signaling pathways known to be required for positive selection (21). Although TOX is poorly expressed by DP thymocytes (22), perturbations in β selection can impact the signaling properties of later-stage DP thymocytes (27). Because TOX is also up-regulated during β selection (22), we sought to determine if TOX deficiency altered TCR signaling in DP cells, as well as in DD thymocytes that normally up-regulate TOX. In these assays, wild-type and Tox−/− DP and DD thymocytes were indistinguishable (Fig. 3, A and B). To assess the outcome of TCR engagement on these cells, we analyzed the response of DP thymocytes to an anti–TCR-β/CD2 mAb combination that has been shown to deliver a differentiation signal leading to progression to a DD phenotype (28). DP thymocytes from both wild-type and Tox−/− mice developed into DD thymocytes in culture in response to this stimulus, further demonstrating that TCR signaling necessary for transition to a DD phenotype is intact in the absence of TOX (Fig. 3 C).
Figure 3.
TCR signaling is intact in the absence of TOX. (A) Analysis of anti-CD3ε mAb induced calcium flux in wild-type (blue line) or Tox−/− (red line) gated populations of DP or DD thymocytes, as indicated. Control (black line) refers to Tox−/− thymocytes that were treated with anti-CD3ε mAb but did not receive the cross-linking secondary antibody. (B) Induction of phospho-ERK1/2 in DP and DD thymocytes upon TCR cross-linking as in A. To validate the specificity of staining, PMA stimulation of DP thymocytes with or without the MAPK or ERK kinase (MEK) inhibitor U0126 is also shown. (C) Purified DP thymocytes were analyzed directly (t = 0) or induced to differentiate by overnight culture without (medium) or with coimmobilized anti-CD2 and anti–TCR-β antibodies as indicated.
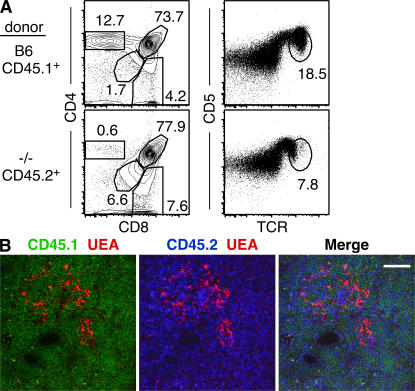
To ensure that the defects observed in T cell development were intrinsic to thymocytes, we reconstituted lethally irradiated wild-type mice with a 1:1 mix of Tox−/− (CD45.2+) and control Tox+/+ (CD45.1+) BM cells. In these mixed chimeric mice, developing Tox−/− cells recapitulated the phenotype of intact Tox−/− mice, including loss of CD4 T cell development, increase in DD thymocytes, and reduction of TCRhiCD5hi thymocytes (Fig. 4 A). Despite these developmental defects, cells that had initiated positive selection (corresponding to stages 2 and 3 in Fig. 2 G) were not diminished (Fig. 4 A). In contrast, Tox+/+ cells in the same animal developed normally (Fig. 4 A). Even under these competitive conditions, loss of TOX had no discernible effect on cell expansion associated with β selection, and did not prevent CD8SP thymocyte production (Fig. 4 A and not depicted).
Figure 4.
The Tox−/− phenotype is T cell intrinsic. (A) Thymocytes from irradiated wild-type (B6, CD90.1+) mice reconstituted with a 1:1 mixture of CD90.2+CD45.2+ mutant (−/−) and CD90.2+CD45.1+ wild-type (B6) BM-derived cells were analyzed as shown. (B) Tox−/− thymocytes appear in the thymic cortex and medulla in mixed chimeric mice. Medullary regions are defined by staining of medullary epithelium with Ulex europaeus agglutinin I (UEA; red), wild-type (B6) cells are detected with anti-CD45.1 mAb (green), and mutant cells are detected with anti-CD45.2 mAb (blue). Bar, 100 μm.
Histological analysis of the thymus of a mixed BM chimera revealed normal thymic architecture and distribution of Tox+/+ (CD45.1) and Tox−/− (CD45.2) thymocytes throughout the cortex and medulla (Fig. 4 B). Because Tox−/− cells fail to develop into CD4 T cells (Fig. 4 A), the CD45.2+ medullary thymocytes presumably correspond to CD8SP thymocytes. These results confirm that developmental defects observed in Tox−/− mice are cell intrinsic and, moreover, that CD4SP thymocytes do not develop in the absence of TOX even in the context of a normal thymic microenvironment.
Molecular signature of Tox−/− DD thymocytes places them after positive selection and pre-lineage commitment
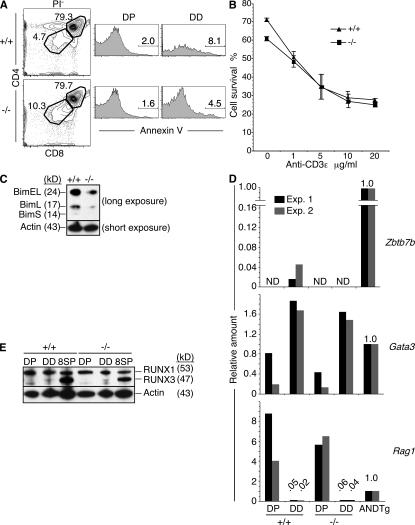
Dying thymocytes can take on a DD-like phenotype (6). However, there was no increase (and some decrease) in the frequency of annexin V+ cells, an early marker of apoptosis, among freshly isolated Tox−/− DP or DD thymocytes compared with their wild-type counterparts (Fig. 5 A). Mutant and wild-type thymocytes also showed similar extents of spontaneous cell death and anti-CD3ε/CD28 mAb–induced cell death (Fig. 5 B). Expression of all three isoforms of the proapoptotic factor Bim, which play a role in negative selection (29), were detected in wild-type DD thymocytes. A similar pattern of isoform expression has been previously reported in thymocytes undergoing TCR-mediated apoptosis (30), suggesting that the DD population may include some thymocytes undergoing negative selection (Fig. 5 C). However, expression of Bim was reduced in mutant DD thymocytes (Fig. 5 C), arguing against the possibility that the Tox−/− DD thymocytes result from enhanced negative selection. The reduction in apoptotic cells and expression of Bim in the Tox−/− DD cell population could be due to dilution with postpositive selection DD cells and/or the lack of CD4 lineage thymocytes that would normally be subjected to negative selection.
Figure 5.
Tox−/− DD thymocytes are after positive selection but do not express the CD4 lineage commitment factor. (A) Total viable thymocytes, as determined by PI exclusion (PI−), were analyzed for binding of annexin V to gated populations of DP and DD thymocytes. (B) Purified DP thymocytes were treated with various concentrations of plate-bound anti-CD3ε mAb in the presence of anti-CD28 mAb. Cell viability was determined by PI and annexin V staining after overnight culture. Plotted frequencies represent mean ± SD of duplicate cultures. (C) The proapoptotic factor Bim is poorly expressed by the Tox−/− DD thymocyte population compared with wild-type. Total cell lysates from purified DD thymocytes were analyzed for Bim by Western blotting. BimEL is the prominent isoform present, although BimL and BimS were also detectable with longer exposure. Expression of β-actin was used as a loading control. (D) Quantitative real-time RT-PCR for Zbtb7b, Gata3, and Rag1 mRNA in purified populations of DP or DD thymocytes. Results are compared with mRNA derived from AND TCR-Tg total thymocytes, which was arbitrarily set to a value of 1.0. Where bars are not graphically visible, values or not detectable (ND) (Ct > 33 cycles) is indicated. (E) Runx3 protein is up-regulated in Tox−/− CD8SP thymocytes. Total cell lysates from sorted thymocyte populations were analyzed for expression of Runx1 and Runx3 by immunoblotting using a pan anti-Runx antibody. Expression of β-actin is shown for comparison.
Because Tox−/− thymocytes initiated positive selection but specifically failed to progress in their differentiation program to the CD4 lineage, we investigated whether this failure was associated with an inability to up-regulate T cell lineage–specific factors. The Zbtb7b zinc finger transcription factor is specifically expressed in developing thymocytes of the CD4 lineage, and both gain-of-function and loss-of-function approaches have demonstrated that this nuclear protein is both necessary and sufficient for commitment to the CD4 T cell lineage (10, 11). DP thymocytes from either mutant or wild-type mice did not express Zbtb7b mRNA (Fig. 5 D). Zbtb7b mRNA, however, was present in developing AND TCR-Tg thymocytes (∼60% CD4SP) and at ∼25-fold lower amounts in wild-type DD thymocytes (Fig. 5 D). In contrast, DD thymocytes isolated from Tox−/− mice did not express detectable amounts of Zbtb7b mRNA, consistent with the inability of these postselection thymocytes to develop into CD4 T cells (Fig. 5 D).
Expression of Gata3, up-regulated by TCR activation during positive selection and required for CD4 lineage development or survival (31, 32), was up-regulated in both wild-type and mutant DD thymocytes when compared with their respective DP thymocyte populations (Fig. 5 D). Expression of Rag1 is extinguished early during positive selection, preventing additional TCR gene rearrangements once the selection process has initiated (33). Compared with DP thymocytes, DD thymocytes from both wild-type and Tox−/− mice down-regulated Rag1 (Fig. 5 D). These data indicate that Tox−/− DD thymocytes are postpositive selection but blocked at a stage before CD4 lineage commitment.
Runx3 protein is expressed in the CD8 but not CD4 T cell lineage in the thymus (5). Moreover, development of the CD8SP phenotype is dependent on this nuclear factor, both to initiate Cd4 gene silencing and to activate a Cd8 gene enhancer (5, 34, 35). As expected, Runx3 was undetectable in wild-type DP thymocytes but was highly expressed by CD8SP thymocytes (Fig. 5 E). Similarly, Runx3 protein was expressed by Tox−/− CD8SP but not DP thymocytes, suggesting that the CD8SP phenotype in Tox−/− mice was a reflection of CD4 silencing (Fig. 5 E). Both wild-type and Tox−/− DD cell populations expressed low levels of Runx3 (Fig. 5 E).
Functional CD8 T cells develop in the absence of TOX and CD4+8lo transitional thymocytes
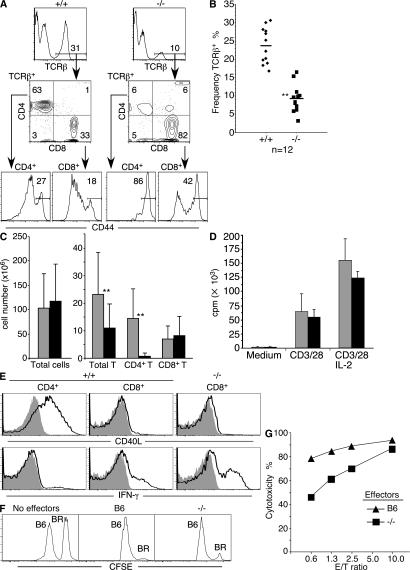
The severe inhibition of thymic CD4SP development in Tox−/− mice resulted in significant T lymphopenia in the spleen, which could be attributed specifically to loss of CD4 T cells (Fig. 6, A–C). Compared with wild-type mice, Tox−/− mice had, on average, 24- and 16-fold reductions in the frequency and absolute number of splenic CD4 T cells, respectively. In addition, the CD4 T cells present in Tox−/− mice were almost all CD44hi (Fig. 6 A). Because T cells undergoing homeostatic expansion in lymphopenic animals also up-regulate CD44 (36), it is possible that rare CD4 T cells that develop in the Tox−/− thymus undergo considerable expansion in the spleen. In contrast, splenic CD8 T cell numbers were not reduced in Tox−/− compared with wild-type animals (Fig. 6 C). However, the proportion of CD8 T cells that were CD44hi also increased somewhat in Tox−/− mice (Fig. 6 A). Nevertheless, isolated Tox−/− splenic CD8 T cells showed normal proliferative responses to anti-CD3/CD28 mAb stimulation in the absence or presence of IL-2 (Fig. 6 D).
Figure 6.
CD4 T lymphopenia but functional CD8 T cells in Tox−/− mice. (A) Profound loss of CD4 T cells and increase in frequency of CD44hi T cells in the spleens of Tox−/− mice as shown by four-color flow cytometry analysis. (B) Frequencies of TCR-β+ spleen cells are plotted for individual mice. Population means are shown as bars. A statistically significant reduction in the frequency of TCR-β+ spleen cells (P = 5.3 × 10−8) in −/− compared with +/+ mice is indicated (**). (C) Absolute numbers of total splenocytes, splenic T cells, and CD4 and CD8 T cell subsets in +/+ (gray bars) and −/− (black bars) mice. Error bars refer to standard deviations (n = 11). Statistically significant reductions in the number of TCR-β+ spleen cells (P = 0.026) and CD4 T cells (P = 3.10−4) in −/− compared with +/+ mice are indicated (**). (D) Tox−/− CD8 T cells are responsive to TCR activation. Proliferative response (mean [3H]-thymidine incorporation ± SD of triplicate cultures) of wild-type (gray bars) or Tox−/− (black bars) splenic CD8 T cells to anti-CD3ε and anti-CD28 mAb stimulation in the presence or absence of IL-2. (E) Mutant CD8 splenic T cells belong to the CD8 lineage. Gated populations of +/+ or −/− CD4 and CD8 splenic T cells were analyzed for induction of CD40L and IFN-γ production after culture in the absence (shaded) or presence (black line) of PMA and ionomycin. (F and G) Tox−/− CD8 T cells exhibit cytotoxic effector function. In vitro B10.BR (H-2k)-stimulated CD8 T cells from C57BL/6 or H-2b TOX−/− mice were tested for cytotoxicity against a mix of allogeneic CFSEhi B10.BR (BR) and syngeneic CFSElo C57BL/6 (B6) target cells. Cytolytic activity was determined by specific loss of CFSEhi-viable cells by flow cytometry using an E/T ratio of 10 in F, or as indicated in G, and calculated as in Materials and methods.
To determine whether the phenotypically defined T cells present in Tox−/− mice were true CD8 lineage T cells, we analyzed up-regulation of CD40L and IFN-γ. CD40L was induced on activated CD4 but not CD8 T cells derived from wild-type or mutant mice (Fig. 6 E). Conversely, IFN-γ production was more pronounced from activated wild-type or mutant CD8 T cells than wild-type CD4 T cells. In addition, Tox−/− CD8 T cells developed into cytolytic effector cells upon stimulation with allogeneic spleen cells (Fig. 6 F), although some reduction in cytolytic activity was noted when compared with wild-type effector cells (Fig. 6 G). By these criteria, CD8 T cells that develop in Tox−/− mice show an appropriate match of coreceptor expression with associated T lineage effector functions.
Lineage commitment in the absence of TOX
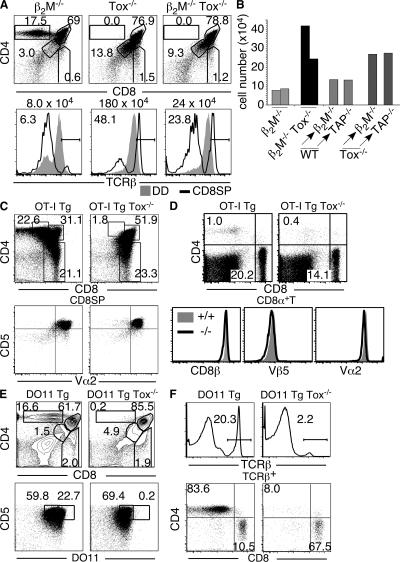
The loss of CD4SP thymocytes and lack of a compensatory increase in CD8SP thymocytes in Tox−/− mice suggested that class II MHC–restricted T cells were unlikely to be misdirected to the CD8 lineage in these animals. To address this directly, we generated β2-microglobulin (β2M)-deficient (β2M−/−) Tox−/− mice. Up-regulation of TCR-β from the DD to the CD8SP stages of development was evident in all strains, indicative of positive selection (Fig. 7 A). However, numbers of TCR+ CD8SP cells were greatly reduced in β2M−/− Tox−/− mice when compared with Tox−/− mice (Figs. 2 F and 7, A and B). Similar results were obtained in the spleen, where CD8 T cells in β2M−/− Tox−/− mice were reduced approximately fivefold when compared with Tox−/− mice (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20071944/DC1). These results demonstrate that development of the majority of CD8 lineage T cells in Tox−/− mice was class I MHC dependent.
Figure 7.
Preferential development of class I but not class II MHC–specific T cells in Tox−/− mice. (A) Reduced development of TCR-β+ CD8SP in the absence of class I MHC. TCR-β expression on CD8SP (black line) is compared with DD thymocytes (shaded). Gate frequencies refer to the CD8SP subsets. The absolute number of TCR-β+ CD8SP cells in each animal is indicated above the TCR-β profiles. (B) Absolute numbers of TCR+ CD8SP thymocytes in β2M−/−, β2M−/− Tox−/− mice, and lethally irradiated β2M−/− or TAP-1−/− (TAP−/−) mice reconstituted with either wild-type (WT) or Tox−/− BM. (C) CD8SP thymocytes develop in OT-I TCR-Tg Tox−/− mice. Expression of CD4 and CD8 on total thymocytes or CD5 and Vα2 on gated CD8SP thymocytes is shown. (D) CD8 splenic T cells develop in OT-I TCR-Tg Tox−/− mice. Expression of CD8β, Vβ5, and Vα2 on a gated subpopulation of CD8α+ splenic T cells from +/+ (shaded) and −/− (black line) mice. (E) Total thymocytes were analyzed for CD4, CD8, CD5, and DO11 TCR expression. (F) Splenocytes were stained for CD4, CD8, and TCR-β. Shown is the expression of CD4 and CD8 on TCR-β+ cells.
Nevertheless, numbers of TCR+ CD8SP cells were greater in β2M−/− Tox−/− mice than in β2M−/− mice (Fig. 7, A and B). The splenic CD8 T cell population in β2M−/− Tox−/− mice was also more pronounced than that seen in the β2M−/− strain (Fig. S3). In addition, class I MHC–deficient mice (β2M−/− or transporter associated with antigen processing [TAP]-1−/−) reconstituted with Tox−/− BM revealed an increase in numbers of TCR+ CD8SP cells compared with those reconstituted with wild-type BM, suggesting a thymocyte-intrinsic effect (Fig. 7 B). These data indicate class I MHC–independent development of a minor population of CD8 T cells in the absence of TOX.
To determine whether TOX deficiency affects the development of CD8 T cells of known specificity, we analyzed class I MHC–specific OT-I TCR-Tg mice. Development of CD5hiVα2hi CD8SP thymocytes was unaffected by the absence of TOX in OT-I TCR-Tg mice, despite the loss of CD4+8lo transitional cells in the mutant animals (Fig. 7 C). CD8αβ+ T cells that expressed high levels of transgene-encoded TCR (Vβ5 and Vα2) were also predominant in the spleens of both wild-type and Tox−/− OT-I mice (Fig. 7, C and D). In contrast, SP thymocytes bearing the class II MHC–restricted DO11 TCR failed to develop in the absence of TOX, despite the fact that the majority of thymocytes in DO11 TCR-Tg Tox−/− mice had received positive selection signals as assessed by CD5 up-regulation (Fig. 7 E). As a result of this developmental block, the spleens of DO11 TCR-Tg Tox−/− mice exhibited severe T lymphopenia (Fig. 7 F). The minor population of splenic T cells present in these animals was predominantly CD8+ (Fig. 7 F).
Expression of transgene-encoded TOX but not a DNA-binding domain mutant rescues CD4 T cell development in Tox−/− mice
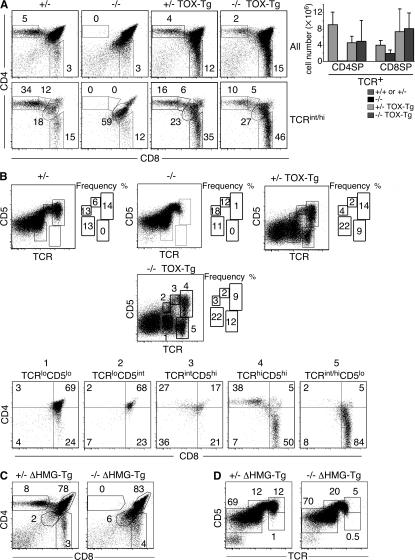
To definitively demonstrate that loss of TOX is responsible for the observed block in T cell development in mutant mice, we bred Tox−/− mice to TOX-Tg mice (22). As expected, expression of the TOX transgene reconstituted CD4 T cell development in Tox−/− mice, including the production of TCRint/hi CD4+8lo transitional and CD4SP thymocytes (Fig. 8 A). This was accompanied by a reduction in the DD population in Tox−/− TOX-Tg mice, relative to Tox−/− mice (Fig. 8 A). In addition, the TCRhiCD5hi thymocyte subpopulation that was absent in Tox−/− mice was restored by expression of transgene-encoded TOX (stage 4 in Figs. 2 D and 8 B). Previously, we reported that CD8SP thymocyte development was enhanced in TOX-Tg mice in the absence of positive selection (Fig. 8 A) (23). This increase in CD8SP thymocytes was also evident in Tox−/− TOX-Tg mice (Fig. 8 A). In addition, Tox−/− TOX-Tg mice had a population of TCRint/hiCD5lo/− (stage 5) CD8SP thymocytes (Fig. 8 B) not present in wild-type, Tox+/−, or Tox−/− mice, but evident in Tox+/− TOX-Tg and Tox−/− TOX-Tg mice (Figs. 2 D and 8 B). A similar population of cells was observed previously in TOX-Tg mice on a β2M−/− background, suggesting that these cells are formed by ectopic expression of TOX in the absence of positive selection (22). We also produced Tg mice that express a mutant of TOX that lacks the HMG box domain (ΔHMG-Tg) but maintained nuclear localization signals and was expressed at the level of protein (21). Expression of TOXΔHMG was unable to restore CD4 T cell development in Tox−/− mice (Fig. 8, C and D). Thus, the DNA-binding domain of TOX is critical for its function.
Figure 8.
Expression of transgene-encoded TOX rescues CD4 T cell development in Tox−/− mice. (A) Frequency of cell populations in Tox+/− (+/−), Tox−/− (−/−), Tox+/− TOX-Tg (+/− TOX-Tg), and Tox−/− TOX-Tg (−/− TOX-Tg) mice in total thymocytes (top panels) or in TCRint/hi-gated thymocytes (bottom panels). Mean cell numbers of mature TCR+ CD4SP and CD8SP thymocytes are shown, and error bars indicate standard deviation (n = 3). (B) Five subpopulations (labeled 1–5) of Tox+/− (+/−), Tox−/− (−/−), Tox+/− TOX-Tg (+/− TOX-Tg), and Tox−/− TOX-Tg (−/− TOX-Tg) thymocytes based on expression of TCR and CD5 were analyzed. Expression of CD4 and CD8 on Tox−/− TOX-Tg (−/− TOX-Tg) thymocytes gated on CD5 and TCR is shown. (C and D) The frequency of thymocyte populations in Tox+/− (+/−) ΔHMG-Tg and Tox−/− (−/−) ΔHMG-Tg mice, stained as indicated.
Other TOX-dependent T cell lineages
CD1d-dependent NKT, many T reg cells, and γδT require the thymic environment for development. Mature T cells of these cell lineages differ in coreceptor expression; mature γδT cells are DN or CD8αα+ (37), NKT cells are CD4+ or DN (38), and T reg cells are mostly CD4+ (39). In addition, γδT cells develop from DN thymocytes (40), whereas NKT and T reg cells develop from DP thymocytes as a result of a positive selection process (38, 41). Thus, we asked how loss of TOX would affect development of these distinct T cell lineages.
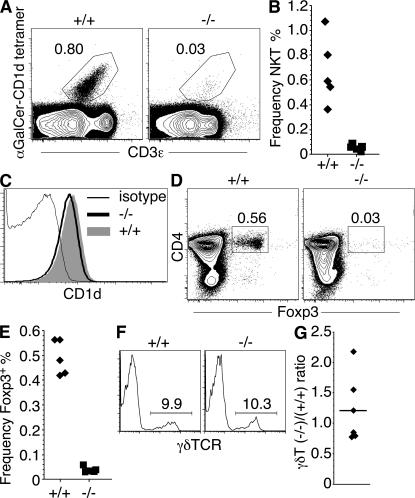
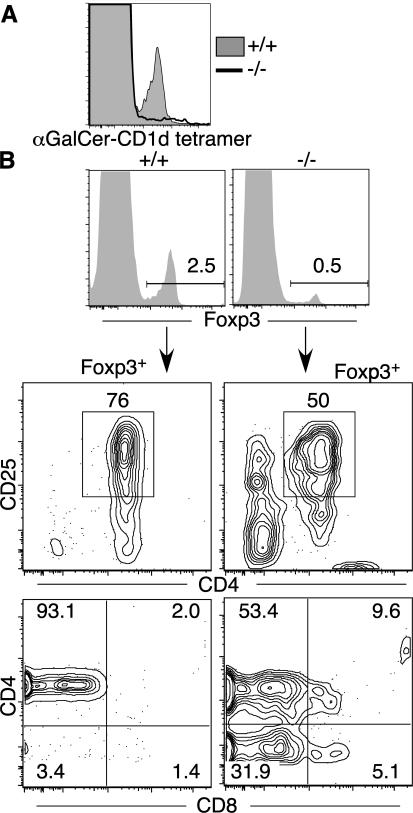
NKT cell precursors, defined as CD3int α-galactosylceramide (αGalCer)-CD1d tetramer+ thymocytes (42), were severely reduced in the thymuses (Fig. 9, A and B) and spleens (Fig. 10 A) of Tox−/− mice. This was not caused by the absence of the selecting MHC molecule because loss of TOX had no effect on expression of CD1d by DP thymocytes (Fig. 9 C). In wild-type mice, the majority of T reg cell precursor thymocytes, defined by expression of the Foxp3 transcription factor (41, 43), expressed CD4 (Fig. 9 D). In Tox−/− mice, few Foxp3+ cells were detected in the thymus (Fig. 9, D and E), whereas a low frequency of Foxp3+ T cells was present in the spleen (Fig. 10 B). Although wild-type T reg cells are almost entirely CD4+ and the majority express CD25, Foxp3+ splenocytes from Tox−/− mice contained a sizeable proportion of DN cells that were CD25− (Fig. 10 B). These results contrasted with γδT cell precursors, which were not reduced in the thymus of Tox−/− mice when compared with wild-type animals (Fig. 9, F and G).
Figure 9.
Development of γδT but not NKT or T reg cell precursor thymocytes in Tox−/− mice. (A) Inhibition of development of CD1d-dependent NKT cells in the thymus of Tox−/− mice as indicated by loss of CD3int αGalCer-CD1d tetramer+ thymocytes. (B) Compilation of data for individual animals generated as in A. (C) Expression of CD1d on +/+ (shaded histogram) and −/− (thick black line) DP thymocytes. Control isotype staining (thin black line) is shown on −/− DP thymocytes. (D) Inhibition of development of T reg cells in the thymus of Tox−/− mice as indicated by the loss of Foxp3+ thymocytes, the vast majority of which are CD4+ in wild-type mice. (E) Compilation of data for individual animals generated as in D. (F) Frequency (percentage) of γδTCR+ cells among otherwise lin− thymocytes in Tox−/− mice. (G) Compilation of analysis of six −/− and +/+ animal pairs expressed as the (−/−)/(+/+) ratio of the frequency of γδTCR+ thymocytes in each experiment. The frequency of γδTCR+ thymocytes was determined in total viable thymocytes, DN thymocytes, or lin− DN thymocytes in individual experiments. The mean ratio value is shown as a horizontal bar.
Figure 10.
Loss of CD4+ NKT and T reg mature cells in Tox−/− mice. (A) Few αGalCer-CD1d tetramer+ cells are present in the spleens of Tox−/− (−/−) mice. (B) Tox−/− mice have fewer Foxp3+ splenic T cells and an increased proportion of CD25−CD4−CD8− Foxp3+ cells. Shown is the staining pattern for CD25 and CD4 or CD4 and CD8 among gated Foxp3+ splenocytes from +/+ and −/− mice.
DISCUSSION
Our results have demonstrated that TOX plays a critical role in the development of T but not B lymphocytes, consistent with the expression pattern of this nuclear protein (22). Expression of TOX, however, is not T cell specific. Despite this, no gross abnormalities outside the immune system were observed in Tox−/− mice. The ability of thymocytes to undergo cell differentiation and expansion as a result of β selection was also largely unimpaired in Tox−/− mice. This was surprising because TOX is transiently up-regulated during β selection, and expression of transgene-encoded TOX is sufficient to induce Cd8 gene demethylation and DN to DP differentiation, although not cell expansion, of RAG-deficient thymocytes (23). Whether the failure to detect a role for TOX during β selection is due to compensation from other TOX family members (44), however, remains to be determined.
The most striking phenotype of Tox−/− mice was initiation but not completion of positive selection of CD4 lineage T cells, leading to severe CD4 T lymphopenia. This was caused by a block at the DD to CD4+8lo transition in the thymus, consistent with the high level of TOX in these cell subpopulations in wild-type mice (22).
The TOX dependence of the DD to CD4+8lo transition provides the first molecular evidence that this subset represents a distinct stage of positive selection. In this regard, the phenotype of Tox−/− mice differs from that of mice deficient in GATA-3 or functional Zbtb7b in several critical characteristics. GATA-3 is also required for development or survival of conventional CD4 but not CD8 T cells (31, 32). However, loss of TOX does not prevent Gata3 mRNA up-regulation in DD thymocytes, suggesting that the Tox−/− phenotype is not due to loss of GATA-3. Moreover, CD69+CD4+8lo transitional but not CD4SP thymocytes are present in CD4-Cre Gata3fl/fl mice (32). Thus, GATA-3 may play a role at a later stage of development than TOX, possibly mediating survival of lineage-committed CD4 T cells. In addition, loss of GATA-3 inhibits development or survival of CD4+ but not DN NKT cells in the thymus (45). This is in contrast to TOX deficiency that leads to a near-complete loss of all CD1d tetramer+ thymocytes.
The helper-deficient mutation in the Zbtb7b gene also prevents CD4 T cell development but, in contrast to the results reported here, does not inhibit development of CD4+8lo thymocytes (46). This is consistent with the relatively late expression of this lineage commitment factor during positive selection (10, 11). The failure to detect high-level expression of Zbtb7b in Tox−/− thymocytes, then, likely reflects the inability of the cells to differentiate to the CD4+8lo developmental stage. However, even in DD thymocytes, low-level Zbtb7b expression was not detected in Tox−/− mice. Therefore, TOX could play a more direct role in Zbtb7b up-regulation. Because TOX is expressed during both CD4 and CD8 lineage development, an additional CD4 lineage–specific factor(s) would be required to induce Zbtb7b in CD4-destined cells.
Interestingly, the integration of lineage commitment with positive selection was largely intact in mice lacking TOX. Thus, the majority of CD8αβ+ T cells that developed in Tox−/− mice were class I MHC specific, as assessed by loss of most CD8 T cells in β2M−/− Tox−/− mice. The minor population of CD8 T cells that did develop in these animals, however, may be comprised of a subset of cells with class II MHC–specific TCR. Similarly, OT-I (class I MHC–specific) but not DO11 (class II MHC–specific) TCR transgenes promoted the development of CD8 T cells in Tox−/− mice.
The absence of CD4+8lo thymocytes in Tox−/− mice demonstrates that this transitional population is not an absolute requirement for CD8 lineage development. Indeed, in mice that expressed a limited TCR repertoire, cells with certain TCR specificities were found to be enriched in the CD8SP but not CD4+8lo thymocyte populations (12). Thus, cells bearing some class I MHC specificities may skip the CD4+8lo stage altogether. This could account for the ability of CD8 T cells to develop in Tox−/− mice. The frequency of CD4+8lo cells varies appreciably between different class I MHC–specific TCR-Tg mice (47). However, even in OT-I TCR-Tg mice that normally contain a pronounced CD4+8lo cell population, CD8 T cell development was maintained in the absence of TOX and these transitional cells. Thus, under normal circumstances in the presence of TOX, many or most CD8 T cells may develop from the CD4+8lo stage via a “coreceptor reversal” pathway as has been proposed (8).
The question arises then of why class II MHC–specific T cells are not shunted into the CD8 lineage as they pass through the DD stage, as might be expected due to CD4 coreceptor down-regulation (48). We suggest that CD4-dependent selection of class II MHC specificities during the initiation of positive selection may involve lineage specification, although not fixed commitment. This may help maintain appropriate lineage commitment as cells undergo complex changes in coreceptor expression. In support of this, repositioning of the Cd8 gene locus to heterochromatin in class II MHC–specific AND TCR-Tg cells is detectable as early as the DD stage (7). Despite this apparent specification, AND TCR-Tg cells can undergo CD8 lineage commitment when CD4+8lo cells form in the absence of Zbtb7b (10, 11), indicating the maintenance of plasticity as late as the CD4+8lo stage.
This model predicts that normal lineage commitment would break down in the case of CD4-independent class II MHC specificities. Indeed, cells expressing class II MHC–specific TCR develop into CD8 or DN T cells in CD4-deficient mice (49, 50). Similarly, class II MHC–specific AND or DO11 TCR-Tg thymocytes have been reported to adopt a CD8 lineage fate in mice that lack CD4, although this requires expression of MHC molecules of sufficient affinity for these TCR to be borderline inducers of negative selection in the presence of CD4 (51). Thus, selection for cells that can undergo CD4-independent class II MHC–specific TCR activation during initiation of positive selection in these systems appears to eliminate lineage bias. This is in contrast to Tox−/− mice, where the initiation of positive selection and repertoire selection occurs with normal expression of CD4 and where normal fidelity of lineage commitment is largely maintained.
The development of Runx3+ CD8SP thymocytes and mature CD8 T cells in Tox−/− mice was puzzling given the phenotype of TOX-Tg mice. We have previously demonstrated that Runx3+ CD8SP thymocytes develop, although do not mature, in TOX-Tg mice, even in the absence of positive selection (23). Although these gain-of-function and loss-of-function results appear contradictory, we think they illustrate the context dependence of TOX function. This was evident when TOX-Tg mice were bred to Tox−/− mice. CD4 T cell development was restored in Tox−/− mice by transgene-encoded TOX, presumably in conjunction with appropriate positive selection signals. At the same time, early expression of TOX at the DP stage induced an abnormal population of CD5lo/− CD8SP thymocytes, likely formed in the absence of positive selection. The competition between these effects may also cause the observed reduction in CD4SP development in TOX-Tg Tox−/− mice. One intriguing possibility is that TOX plays a role not only in the DD to CD4+8lo transition, but also in the CD4+8lo to CD8SP coreceptor reversal. Ectopic expression of TOX in DP thymocytes could partially mimic this latter activity, potentially mediated by Runx3 (5, 23).
Additional T cell types formed in the thymus could be clearly split into TOX-dependent (NKT and T reg cell) and TOX-independent (γδT) lineages. The best overall correlation with TOX dependence is positive selection coupled with expression of CD4 on a sizeable fraction of the lineage. Most T reg cells are CD4+ cells. However, NKT cells can be DN as well as CD4+ in the thymus (for review see reference 39). Despite this, both subsets of NKT are derived from DP thymocytes (38). In addition, the appearance of a significant fraction of αGalCer-CD1d tetramer+ DD thymocytes in newborn animals may suggest that both CD4 and DN subsets of developing NKT pass through a DD stage (38). As with conventional CD4 T cells, therefore, progression from the DD stage of NKT cell development may be TOX dependent.
The shared role of TOX in the development of conventional CD4 T cells, CD1d-dependent NKT cells, and most thymically derived T reg cells may indicate that NKT and T reg cells are true CD4 sublineages. Because the initial CD4 and CD8 lineage split likely occurs at the CD4+8lo stage of positive section, a serial pathway in which development of other functional lineages branch off from the conventional CD4 T cell subset is possible. The fact that medullary signals may be involved in the induction of Foxp3 would be consistent with such a model (41). In the case of CD1d-dependent NKT cells, however, where the TCR-α is invariant and there are distinct signaling requirements for development (for review see reference 39), an earlier instructive signal could alternatively lead to a parallel pathway of TOX-dependent development of this nonconventional T cell subset.
MATERIALS AND METHODS
Mice.
All mice were bred at The Scripps Research Institute and kept under specific pathogen-free conditions. Tox genomic fragments were obtained from a screen of the RPCI-22 129/SvEvTACBr BAC library (The Centre for Applied Genomics, Canada) and cloned into the pFlox vector (52). Gene targeting and subsequent transient Cre expression was performed in CMTI-1 mouse ES cells (derived from 129S6/SvEv strain mice) using standard methodology. Heterozygous Tox+/− mice were bred to generate homozygous Tox−/− and control Tox+/+ littermates. The generation of TOX-Tg and ΔHMG-Tg mice has been described (21, 22). β2M−/− (53), OT-I TCR-Tg (54), and DO11 TCR-Tg (55) mice on either a wild-type or Tox−/− background and TAP-1−/− mice (56) were also used. Experiments were conducted in accordance with National Institutes of Health guidelines for the care and use of animals and with an approved animal protocol from The Scripps Research Institute Animal Care and Use Committee.
Antibodies, flow cytometry, and Western blotting.
All antibodies were purchased from eBioscience or BD Biosciences, except for anti-TOX and pan anti-Runx polyclonal antibodies (the latter were provided by M. Satake, Tohoku University, Sendai, Japan) that were described previously (23). α-GalCer–loaded CD1d tetramers were provided by L. Teyton (The Scripps Research Institute, La Jolla, CA). Flow cytometry and Western blotting were performed as described previously (23).
PCR.
Real-time quantitative RT-PCR analysis was performed using the standard curve method, where samples were normalized based on Gapdh expression, and analyzed using SDS 2.1 software (Applied Biosystems). Primers for real-time RT-PCR were purchased from QIAGEN. Primer sequences for Tox and ActB (β-actin) genomic screens are available upon request.
Signaling.
For calcium flux, total thymocytes were labeled at 2 × 107 cells/ml with 20 μM indo-1 (Invitrogen), followed by surface staining. TCR stimulations were performed by adding purified functional-grade anti-CD3ε mAb (clone 2C11; eBioscience) at the indicated concentrations and cross-linking bound antibodies with anti–hamster IgG at 55 μg/ml. The ratio of calcium-bound indo-1 to free indo-1 was measured as a function of time by flow cytometry. For ERK analysis, total thymocytes were prestained with surface markers and incubated with 10 μg/ml of purified functional-grade anti-CD3ε mAb for 30 min. TCR cross-linking was performed by adding anti–hamster IgG at 75 μg/ml to prewarmed cells for 2 min. Cells were immediately fixed in 2% paraformaldehyde for 10 min on ice, washed, and permeabilized in 100% ice-cold methanol for at least 30 min. Phospho-ERK1/2 was measured by intracellular staining (clone E10; Cell Signaling Technology). In some cases, 500 ng/ml PMA, with or without 50 μg/ml of the MEK inhibitor U0126 (EMD), was used instead of anti-CD3ε mAb stimulation.
T cell stimulation cultures.
DP thymocyte death curves were calculated as the frequency of live cells (negative for staining with annexin V and propidium iodide [PI]) after overnight incubation with 50 μg/ml of coimmobilized anti-CD28 mAb and various concentrations of anti-CD3ε mAb. In some experiments, purified DP thymocytes were activated with 10 μg/ml of coimmobilized anti-CD2 and 5 μg/ml anti–TCR-β antibodies for 14 h before analysis of CD4 and CD8 expression by flow cytometry.
Splenic CD8 T cells were purified by negative selection using magnetic beads (StemCell Technologies Inc.) and activated at 106 cells/ml in 200 μl in 96-well plates with 10 μg/ml of immobilized anti-CD3ε and 20 μg/ml of soluble anti-CD28 mAb in the absence or presence of 25 U/ml of recombinant human IL-2.
T cells were cultured for 72 h and pulsed with 1 μCi per well of [3H]-TdR for the last 16 h before TdR incorporation was determined. In other experiments, total splenocytes were cultured at 106 cells/ml in 2 ml with 50 ng/ml PMA and 500 ng/ml ionomycin for 4 h before cells were surface stained for CD40L or internally stained for IFN-γ. To generate cytotoxic effector cells, purified splenic H-2b CD8 T cells were cultured for 4–5 d at 106 cells/ml with 4 × 106 cells/ml irradiated (3,000 rad) red blood cell–lysed B10.BR splenocytes in the presence of 20 U/ml of recombinant human IL-2. Wild-type target spleen cells were labeled with a high (CFSEhi) or low (CFSElo) concentration of CFSE. Activated CD8 T cells were incubated for 20 h with a 1:1 mixture of CFSEhi B10.BR and CFSElo C57BL/6 target cells at the indicated E/T ratios. Loss of CFSEhi or CFSElo cells among the viable cell population was determined by flow cytometry. The percentage of specific lysis was determined by the following formulas: ratio = (percentage of CFSEhi/percentage of CFSElo); percentage of specific lysis = [1− (ratio in the presence of effector cells/ratio in the absence of effector cells) × 100].
BM chimeric mice.
Recipient CD90.1 mice were lethally irradiated (990 rads) to deplete hematopoietic cells. A 1:1 mixture of donor wild-type (CD90.2+CD45.1+) and Tox−/− (CD90.2+ CD45.2+) lin− BM cells was injected intravenously, and chimeras were analyzed 5–8 wk later. More than 98% of thymocytes were of donor origin in these mice. In other experiments, lethally irradiated β2M−/− or TAP-1−/− hosts were reconstituted with wild-type or Tox−/− BM.
Microscopy.
5-μm frozen sections from the thymuses of BM chimeric mice were immunostained and analyzed for the expression of CD45.1 and CD45.2 as well as binding of Ulex europaeus agglutinin (Vector Laboratories) and visualized by a MRC1024 laser scanning confocal microscope (BioRad Laboratories) at a magnification of 20.
Statistics.
The probability (P) associated with a Student's t test using a two-tailed distribution of equal variance is shown in some figures.
Online supplemental material.
Fig. S1 shows that loss of TOX does not affect B cell development. Fig. S2 shows initiation of positive selection in Tox−/− mice as analyzed by CD69 up-regulation. Fig. S3 shows class I MHC dependence of generation of splenic CD8 T cells in Tox−/− mice. Figs. S1–S3 are available at http://www.jem.org/cgi/content/full/jem.20071944/DC1.
Supplemental Material
Acknowledgments
We thank Olivia Garijo for expert technical assistance, Nayna Sanathara for help with genotyping, and Dr. Michelle Fung for help with microscopy and critical reading of the manuscript. We also thank the Nemazee laboratory for input on analysis of B cell development, the TSRI Mouse Genetics Core, especially Greg Martin, and the TSRI Microscopy and Flow Cytometry core facilities for services provided.
This work was supported by a predoctoral scholarship from Les Fonds de la Recherche en Santé du Québec (to P. Aliahmad) and National Institutes of Health grant AI054977 (to J. Kaye). This is manuscript 18463 from The Scripps Research Institute.
The authors have no conflicting financial interests.
Abbreviations used: αGalCer, α-galactosylceramide; β2M, β2-microglobulin; DD, CD4loCD8lo double dull; DP, CD4+CD8+ double positive; ERK, extracellular signal–regulated kinase; ES, embryonic stem; ETP, early T lineage precursor; NKT cell, natural killer T; PI, propidium iodide; SP, single positive; TAP, transporter associated with antigen processing; Tg, transgenic; TOX or Tox, thymocyte selection–associated HMG box protein or gene, respectively; T reg, T regulatory.
References
- 1.Lucas, B., and R.N. Germain. 1996. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 5:461–477. [DOI] [PubMed] [Google Scholar]
- 2.Sant'Angelo, D.B., B. Lucas, P.G. Waterbury, B. Cohen, T. Brabb, J. Goverman, R.N. Germain, and C.A. Janeway Jr. 1998. A molecular map of T cell development. Immunity. 9:179–186. [DOI] [PubMed] [Google Scholar]
- 3.Hogquist, K.A., A.J. Tomlinson, W.C. Kieper, M.A. McGargill, M.C. Hart, S. Naylor, and S.C. Jameson. 1997. Identification of a naturally occurring ligand for thymic positive selection. Immunity. 6:389–399. [DOI] [PubMed] [Google Scholar]
- 4.Lundberg, K., W. Heath, F. Kontgen, F.R. Carbone, and K. Shortman. 1995. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4−8+TCRhi thymocytes. J. Exp. Med. 181:1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato, T., S. Ohno, T. Hayashi, C. Sato, K. Kohu, M. Satake, and S. Habu. 2005. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 22:317–328. [DOI] [PubMed] [Google Scholar]
- 6.Kersh, G.J., and S.M. Hedrick. 1995. Role of TCR specificity in CD4 versus CD8 lineage commitment. J. Immunol. 154:1057–1068. [PubMed] [Google Scholar]
- 7.Merkenschlager, M., S. Amoils, E. Roldan, A. Rahemtulla, E. O'Connor, A.G. Fisher, and K.E. Brown. 2004. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J. Exp. Med. 200:1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brugnera, E., A. Bhandoola, R. Cibotti, Q. Yu, T.I. Guinter, Y. Yamashita, S.O. Sharrow, and A. Singer. 2000. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 13:59–71. [DOI] [PubMed] [Google Scholar]
- 9.Bosselut, R., T.I. Guinter, S.O. Sharrow, and A. Singer. 2003. Unraveling a revealing paradox: why major histocompatibility complex I–signaled thymocytes “paradoxically” appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J. Exp. Med. 197:1709–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, X., V.P. Dave, Y. Zhang, X. Hua, E. Nicolas, W. Xu, B.A. Roe, and D.J. Kappes. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833. [DOI] [PubMed] [Google Scholar]
- 11.Sun, G., X. Liu, P. Mercado, S.R. Jenkinson, M. Kypriotou, L. Feigenbaum, P. Galera, and R. Bosselut. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6:373–381. [DOI] [PubMed] [Google Scholar]
- 12.Correia-Neves, M., D. Mathis, and C. Benoist. 2001. A molecular chart of thymocyte positive selection. Eur. J. Immunol. 31:2583–2592. [DOI] [PubMed] [Google Scholar]
- 13.Adlam, M., and G. Siu. 2003. Hierarchical interactions control CD4 gene expression during thymocyte development. Immunity. 18:173–184. [DOI] [PubMed] [Google Scholar]
- 14.Sawada, S., and D.R. Littman. 1991. Identification and characterization of a T-cell-specific enhancer adjacent to the murine CD4 gene. Mol. Cell. Biol. 11:5506–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawada, S., and D.R. Littman. 1993. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol. Cell. Biol. 13:5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, Z., H. Xie, V. Ioannidis, W. Held, H. Clevers, M.S. Sadim, and Z. Sun. 2006. Transcriptional regulation of CD4 gene expression by T cell factor-1/beta-catenin pathway. J. Immunol. 176:4880–4887. [DOI] [PubMed] [Google Scholar]
- 17.Ellmeier, W., M.J. Sunshine, K. Losos, and D.R. Littman. 1998. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 9:485–496. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, X.L., R. Seong, R. Piracha, M. Larijani, M. Heeney, J.R. Parnes, and J.W. Chamberlain. 1998. Distinct stage-specific cis-active transcriptional mechanisms control expression of T cell coreceptor CD8 alpha at double- and single-positive stages of thymic development. J. Immunol. 161:2254–2266. [PubMed] [Google Scholar]
- 19.Garefalaki, A., M. Coles, S. Hirschberg, G. Mavria, T. Norton, A. Hostert, and D. Kioussis. 2002. Variegated expression of CD8 alpha resulting from in situ deletion of regulatory sequences. Immunity. 16:635–647. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki, H., J.A. Punt, L.G. Granger, and A. Singer. 1995. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 2:413–425. [DOI] [PubMed] [Google Scholar]
- 21.Aliahmad, P., and J. Kaye. 2006. Commitment issues: linking positive selection signals and lineage diversification in the thymus. Immunol. Rev. 209:253–273. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson, B., J.Y. Chen, P. Han, K.M. Rufner, O.D. Goularte, and J. Kaye. 2002. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat. Immunol. 3:272–280. [DOI] [PubMed] [Google Scholar]
- 23.Aliahmad, P., E. O'Flaherty, P. Han, O.D. Goularte, B. Wilkinson, M. Satake, J.D. Molkentin, and J. Kaye. 2004. TOX provides a link between calcineurin activation and CD8 lineage commitment. J. Exp. Med. 199:1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhandoola, A., and A. Sambandam. 2006. From stem cell to T cell: one route or many? Nat. Rev. Immunol. 6:117–126. [DOI] [PubMed] [Google Scholar]
- 25.Azzam, H.S., A. Grinberg, K. Lui, H. Shen, E.W. Shores, and P.E. Love. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendelac, A., P. Matzinger, R.A. Seder, W.E. Paul, and R.H. Schwartz. 1992. Activation events during thymic selection. J. Exp. Med. 175:731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neilson, J.R., M.M. Winslow, E.M. Hur, and G.R. Crabtree. 2004. Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity. 20:255–266. [DOI] [PubMed] [Google Scholar]
- 28.Cibotti, R., A. Bhandoola, T.I. Guinter, S.O. Sharrow, and A. Singer. 2000. CD8 coreceptor extinction in signaled CD4(+)CD8(+) thymocytes: coordinate roles for both transcriptional and posttranscriptional regulatory mechanisms in developing thymocytes. Mol. Cell. Biol. 20:3852–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouillet, P., J.F. Purton, D.I. Godfrey, L.C. Zhang, L. Coultas, H. Puthalakath, M. Pellegrini, S. Cory, J.M. Adams, and A. Strasser. 2002. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 415:922–926. [DOI] [PubMed] [Google Scholar]
- 30.Cante-Barrett, K., E.M. Gallo, M.M. Winslow, and G.R. Crabtree. 2006. Thymocyte negative selection is mediated by protein kinase C- and Ca2+-dependent transcriptional induction of bim. J. Immunol. 176:2299–2306. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez-Hoyos, G., M.K. Anderson, C. Wang, E.V. Rothenberg, and J. Alberola-Ila. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. [DOI] [PubMed] [Google Scholar]
- 32.Pai, S.Y., M.L. Truitt, C.N. Ting, J.M. Leiden, L.H. Glimcher, and I.C. Ho. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 19:863–875. [DOI] [PubMed] [Google Scholar]
- 33.Turka, L.A., D.G. Schatz, M.A. Oettinger, J.J. Chun, C. Gorka, K. Lee, W.T. McCormack, and C.B. Thompson. 1991. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 253:778–781. [DOI] [PubMed] [Google Scholar]
- 34.Taniuchi, I., M. Osato, T. Egawa, M.J. Sunshine, S.C. Bae, T. Komori, Y. Ito, and D.R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. [DOI] [PubMed] [Google Scholar]
- 35.Woolf, E., C. Xiao, O. Fainaru, J. Lotem, D. Rosen, V. Negreanu, Y. Bernstein, D. Goldenberg, O. Brenner, G. Berke, et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst, B., D.S. Lee, J.M. Chang, J. Sprent, and C.D. Surh. 1999. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 11:173–181. [DOI] [PubMed] [Google Scholar]
- 37.Lin, T., G. Matsuzaki, H. Kenai, and K. Nomoto. 1994. Progenies of fetal thymocytes are the major source of CD4-CD8+ alpha alpha intestinal intraepithelial lymphocytes early in ontogeny. Eur. J. Immunol. 24:1785–1791. [DOI] [PubMed] [Google Scholar]
- 38.Gapin, L., J.L. Matsuda, C.D. Surh, and M. Kronenberg. 2001. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2:971–978. [DOI] [PubMed] [Google Scholar]
- 39.Kronenberg, M., and A. Rudensky. 2005. Regulation of immunity by self-reactive T cells. Nature. 435:598–604. [DOI] [PubMed] [Google Scholar]
- 40.Petrie, H.T., R. Scollay, and K. Shortman. 1992. Commitment to the T cell receptor-alpha beta or -gamma delta lineages can occur just prior to the onset of CD4 and CD8 expression among immature thymocytes. Eur. J. Immunol. 22:2185–2188. [DOI] [PubMed] [Google Scholar]
- 41.Fontenot, J.D., J.P. Rasmussen, L.M. Williams, J.L. Dooley, A.G. Farr, and A.Y. Rudensky. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. [DOI] [PubMed] [Google Scholar]
- 42.Benlagha, K., A. Weiss, A. Beavis, L. Teyton, and A. Bendelac. 2000. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 191:1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fontenot, J.D., M.A. Gavin, and A.Y. Rudensky. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336. [DOI] [PubMed] [Google Scholar]
- 44.O'Flaherty, E., and J. Kaye. 2003. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim, P.J., S.Y. Pai, M. Brigl, G.S. Besra, J. Gumperz, and I.C. Ho. 2006. GATA-3 regulates the development and function of invariant NKT cells. J. Immunol. 177:6650–6659. [DOI] [PubMed] [Google Scholar]
- 46.Keefe, R., V. Dave, D. Allman, D. Wiest, and D.J. Kappes. 1999. Regulation of lineage commitment distinct from positive selection. Science. 286:1149–1153. [DOI] [PubMed] [Google Scholar]
- 47.Chan, S., M. Correia-Neves, C. Benoist, and D. Mathis. 1998. CD4/CD8 lineage commitment: matching fate with competence. Immunol. Rev. 165:195–207. [DOI] [PubMed] [Google Scholar]
- 48.Sarafova, S.D., B. Erman, Q. Yu, F. Van Laethem, T. Guinter, S.O. Sharrow, L. Feigenbaum, K.F. Wildt, W. Ellmeier, and A. Singer. 2005. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 23:75–87. [DOI] [PubMed] [Google Scholar]
- 49.Locksley, R.M., S.L. Reiner, F. Hatam, D.R. Littman, and N. Killeen. 1993. Helper T cells without CD4: control of leishmaniasis in CD4-deficient mice. Science. 261:1448–1451. [DOI] [PubMed] [Google Scholar]
- 50.Tyznik, A.J., J.C. Sun, and M.J. Bevan. 2004. The CD8 population in CD4-deficient mice is heavily contaminated with MHC class II–restricted T cells. J. Exp. Med. 199:559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matechak, E.O., N. Killeen, S.M. Hedrick, and B.J. Fowlkes. 1996. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 4:337–347. [DOI] [PubMed] [Google Scholar]
- 52.Priatel, J.J., M. Sarkar, H. Schachter, and J.D. Marth. 1997. Isolation, characterization and inactivation of the mouse Mgat3 gene: the bisecting N-acetylglucosamine in asparagine-linked oligosaccharides appears dispensable for viability and reproduction. Glycobiology. 7:45–56. [DOI] [PubMed] [Google Scholar]
- 53.Zijlstra, M., M. Bix, N.E. Simister, J.M. Loring, D.H. Raulet, and R. Jaenisch. 1990. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 344:742–746. [DOI] [PubMed] [Google Scholar]
- 54.Hogquist, K.A., S.C. Jameson, W.R. Heath, J.L. Howard, M.J. Bevan, and F.R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. [DOI] [PubMed] [Google Scholar]
- 55.Murphy, K.M., A.B. Heimberger, and D.Y. Loh. 1990. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 250:1720–1723. [DOI] [PubMed] [Google Scholar]
- 56.Van Kaer, L., P.G. Ashton-Rickardt, H.L. Ploegh, and S. Tonegawa. 1992. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 71:1205–1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.