Abstract
Suppressor of cytokine signaling 3 (SOCS3) down-regulates several signaling pathways in multiple cell types, and previous data suggest that SOCS3 may shut off cytokine activation at the early stages of liver regeneration (Campbell, J.S., L. Prichard, F. Schaper, J. Schmitz, A. Stephenson-Famy, M.E. Rosenfeld, G.M. Argast, P.C. Heinrich, and N. Fausto. 2001.J. Clin. Invest. 107:1285–1292). We developed Socs3 hepatocyte-specific knockout (Socs3 h-KO) mice to directly study the role of SOCS3 during liver regeneration after a two-thirds partial hepatectomy (PH). Socs3 h-KO mice demonstrate marked enhancement of DNA replication and liver weight restoration after PH in comparison with littermate controls. Without SOCS3, signal transducer and activator of transcription 3 (STAT3) phosphorylation is prolonged, and activation of the mitogenic extracellular signal-regulated kinase 1/2 (ERK1/2) is enhanced after PH. In vitro, we show that SOCS3 deficiency enhances hepatocyte proliferation in association with enhanced STAT3 and ERK activation after epidermal growth factor or interleukin 6 stimulation. Microarray analyses show that SOCS3 modulates a distinct set of genes, which fall into diverse physiological categories, after PH. Using a model of chemical-induced carcinogenesis, we found that Socs3 h-KO mice develop hepatocellular carcinoma at an accelerated rate. By acting on cytokines and multiple proliferative pathways, SOCS3 modulates both physiological and neoplastic proliferative processes in the liver and may act as a tumor suppressor.
Cytokines are secreted proteins that regulate multiple processes, including growth and differentiation, cell survival, hematopoiesis, and immunological functions. Many cytokine effects are transduced through the JAK–STAT pathway. JAK proteins, when bound to cytokine receptors, assemble in phosphorylated receptor complexes that create docking sites for proteins such as the STATs, which contain Src homology 2 domains. STATs are activated through phosphorylation by JAKs, and the activated STATs can dimerize and bind to DNA to activate transcription of target genes. Important STAT targets include the suppressor of cytokine signaling (Socs) genes, which encode eight proteins that inhibit a variety of signaling pathways (1). SOCS proteins appear to inhibit cytokine signaling by targeting different components of the signaling complex either (a) by directly binding to activated, tyrosine phosphorylated cytokine receptors or JAKs via their Src homology 2 domains, or (b) by targeting receptor complexes for proteosomal degradation via the SOCS box (1). Thus, stimulation of Socs transcription by STATs establishes a negative feedback loop that inhibits ongoing activation of cytokine signaling. SOCS proteins have been shown to play an important role in regulating cytokine activity at several levels, including modulating cytokine production and by inhibiting downstream signaling cascades (1, 2).
Both in the hematopoietic system and in the liver, SOCS3 is a critical inhibitor of IL-6 signaling mediated through the gp130 receptor (2). Mice deficient in the gp130 receptor in the liver do not produce STAT3 in response to IL-6 (3), and Yang et al. showed that IL-6 has a crucial role in the expression of Socs3 during inflammatory responses (4). Inhibition of this pathway involves the binding of SOCS3 to phosphotyrosine 759 of the activated gp130 receptor (5). IL-6 is one of the main mediators of the acute-phase response, which is induced by inflammatory stimuli in the liver. By regulating the activity of the IL-6–gp130 pathway, SOCS3 could have a major effect on the acute-phase response to liver injury or inflammation.
One of the most interesting findings regarding the mechanisms that initiate liver regeneration after two-thirds partial hepatectomy (PH) is the demonstration that several components of the innate immune system may be involved in the initiation process. These components, which include cytokines such as IL-6, proteins of the complement system, and lymphotoxins, are particularly active during the first 12–18 h after PH, a time period during which hepatocytes transition from a quiescent state into the cell cycle. Through work with animal models, we and others have shown that IL-6 is a component of a cytokine pathway activated very shortly after PH that involves signaling through TNFR1 and the activation of the transcription factors NF-κB and STAT3 (6–8). It appears that the components of this pathway are involved in both cell survival and hepatocyte replication (6, 8). Data from our laboratory showed that Socs3 is highly expressed between 2 and 8 h after PH, which partially overlaps with the time frame of STAT3 expression, and that Socs3 expression in the regenerating liver is IL-6 dependent (9). We concluded from these experiments that in the initial stages of liver regeneration, SOCS3 might shut off the early cytokine response, perhaps to protect liver cells against the cytotoxic effects of prolonged cytokine expression. However we did not have direct evidence either to support this hypothesis or to determine whether SOCS3 might have other functions in the regenerating liver. Because Socs3 KO mice die during embryogenesis, we developed mice in which Socs3 was specifically deleted in hepatocytes (Socs3fl/fl, Alb-Cre+; termed Socs3 hepatocyte-specific knockout [h-KO] mice) and used these animals to directly investigate the role of SOCS3 during liver regeneration.
Our main expectation was that Socs3 h-KO mice would show a prolonged acute-phase response, and that excessive cytokine signaling would lead to toxicity and a decrease in cell proliferation after PH. Contrary to these expectations, we demonstrate that in the absence of SOCS3, (a) hepatocyte DNA replication and progression through the cell cycle are markedly enhanced after PH, leading to an acceleration of liver regeneration; (b) hepatocytes isolated from Socs3 h-KO mice have an increased replication capacity; and (c) Socs3-deficient mice develop hepatocellular carcinoma (HCC) at an accelerated rate. These data suggest that, in addition to its role in the control of cytokine expression in the regenerating liver, SOCS3 coordinates the responses of innate immune system components with that of proliferative pathways. Coordination between these systems may be required for the precise regulation and synchronization of hepatocyte proliferation during liver regeneration and for the prevention of tumorigenesis in such a highly proliferative environment.
RESULTS
Liver regeneration is enhanced in Socs3 h-KO mice
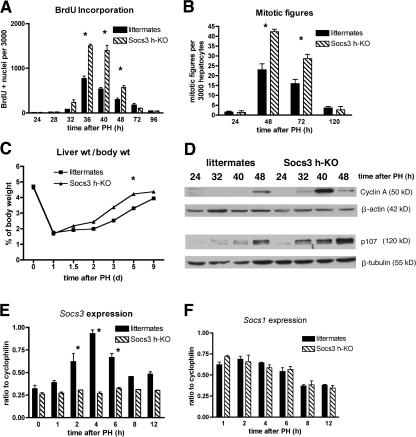
Socs3 is robustly induced during the first few hours after PH (9), suggesting that SOCS3 might act as a negative regulator of liver regeneration. To determine whether Socs3 deficiency altered regeneration, we generated Socs3 h-KO mice (2). In the absence of an operative or chemical stimulus, Socs3 h-KO livers were identical to those of control littermates in weight, histology, and proliferative and apoptotic indices (unpublished data). To determine whether the lack of SOCS3 would have an effect on DNA replication after PH, we evaluated the proliferative response to PH in Socs3 h-KO mice compared with that of littermates with intact Socs3. Surprisingly, Socs3 h-KO mice display marked enhancement of hepatocyte DNA replication, as shown by increased BrdU incorporation (Fig. 1 A). Nuclear hepatocyte BrdU labeling in Socs3 h-KO mice is significantly higher than in control littermates from 32 to 48 h after PH, and is 90–160% higher at the peak of DNA replication between 36 and 40 h after PH. Additionally, the number of hepatocyte mitoses is 85 and 89% higher in Socs3 h-KO mice than that of controls at 48 and 72 h, respectively (Fig. 1 B). As a consequence of the increased hepatocyte replication in Socs3 h-KO mice, these animals restore their liver weights after PH 2 d earlier than do controls (Fig. 1 C). To further demonstrate negative regulation by SOCS3 of the progression of hepatocytes through the cell cycle in the regenerating liver, we performed immunoblotting for the cell-cycle proteins cyclin A and p107, which are known to be up-regulated during liver regeneration (10). Lysates harvested between 24 and 48 h after PH showed that Socs3 h-KO mice had both earlier and increased expression of these proteins during liver regeneration, particularly p107, which is strongly expressed in Socs3 h-KO mice from 32 to 48 h after PH (Fig. 1 D).
Figure 1.
Liver regeneration is enhanced in Socs3 h-KO mice. (A) Staining for BrdU indicates increased DNA synthesis in Socs3 h-KO mice. (B) Increased mitotic figures in hepatocytes lacking Socs3. (C) More rapid return to preoperative liver weight in the absence of Socs3. (D) Increased expression of cyclin A and p107 in Socs3 h-KO mice after PH, demonstrated by immunoblotting. β-Actin and β-tubulin are shown as loading controls. (E) Confirmation of lack of Socs3 induction after PH in Socs3 h-KO mice by Northern blotting, with cyclophilin as a loading control. (F) Northern blotting demonstrates no compensatory up-regulation of Socs1 in the absence of Socs3. Data shown are representative of three to six mice per genotype per time point and are presented as mean ± SEM. *, P < 0.05.
Though previous work has demonstrated that 90–95% of hepatocyte genomic Socs3 is excised in Socs3 h-KO mice (2), we wanted to be certain that the strong physiological stimulus of PH (9) would not lead to significant expression from residual copies of the gene and that expression by nonparenchymal cells (NPCs) was very low. We thus performed Northern blotting for Socs3 on RNA isolated from Socs3 h-KO and control littermates at various times after PH. We found virtually no induction of Socs3 after PH in the KO mice at any of the times examined (Fig. 1 E) and, similarly, did not observe a compensatory up-regulation of Socs1 (Fig. 1 F). In summary, the data presented in this section clearly demonstrate that SOCS3 deficiency increases hepatocyte replication and accelerates liver regeneration after PH.
Activation of STAT3 and extracellular signal-regulated kinase (ERK) signaling pathways in Socs3 h-KO mice after PH
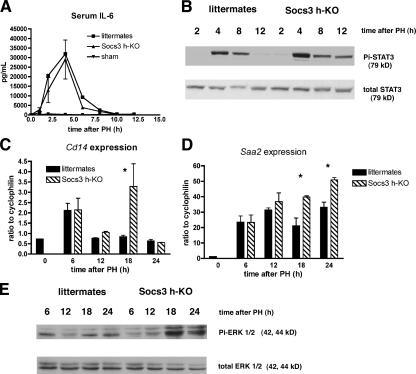
After PH, IL-6 is released by Kupffer cells (11) and subsequently binds its specific receptor on the surface of hepatocytes. Receptor binding activates JAK to phosphorylate and activate STAT3, which then dimerizes and translocates to the nucleus. As we have previously shown that Socs3 expression after PH is largely dependent on the IL-6–STAT3 signaling pathway (9), we examined the activation of this pathway during liver regeneration in Socs3 h-KO mice. Serum IL-6 levels were determined by ELISA from 30 min to 12 h after PH and do not significantly differ between Socs3 h-KO and control littermates (Fig. 2 A). This result is not unexpected, because the production of IL-6 by NPCs would not be altered by SOCS3 deficiency in hepatocytes. However, there is a marked difference in the phosphorylation of STAT3 between Socs3 h-KO and control littermates. STAT3 phosphorylation is still present 12 h after PH in Socs3 h-KO mice, whereas it is no longer detectable in control littermates, as shown by immunoblotting (Fig. 2 B). These data confirm the observation that SOCS3 acts in a negative feedback loop to arrest STAT3 activation in hepatocytes during liver regeneration. To confirm that this prolonged activation of STAT3 would lead to differences in gene expression, we examined the induction Cd14 and Saa2, which are two acute-phase response genes and known STAT3 targets in the liver. Northern blot analysis shows that the expression of these genes after PH is prolonged in Socs3 h-KO mice compared with littermates (Fig. 2, C and D). We measured IL-6 levels at 18 and 24 h after PH to confirm that this cytokine is not expressed later in Socs3 h-KO mice, thus contributing to prolonged expression of these genes, and did not observe elevated levels of IL-6 in either Socs3 h-KO or control littermates at these times (unpublished data). We also determined whether either STAT1 or STAT5 are abnormally activated in the absence of SOCS3 by immunoblotting for phospho-STAT1 and -STAT5 on liver lysates prepared from Socs3 h-KO mice and littermates from 0 to 12 h after PH. We did not observe any activation of either STAT1 or STAT5 at any of the times examined (unpublished data).
Figure 2.
Activation of STAT3 and ERK1/2 are enhanced in Socs3 h-KO mice. (A) Serum IL-6 levels after PH are unchanged in Socs3 h-KO mice (ELISA). (B) Lack of SOCS3 in hepatocytes leads to prolonged phosphorylation of STAT3 (Y705) after PH, demonstrated by immunoblotting. (C) Northern blot analysis indicates prolonged expression of Cd14 Socs3 h-KO mice. (D) Saa2 expression is also prolonged in Socs3 h-KO mice. (E) Socs3 deficiency leads to enhanced phosphorylation of ERK1/2 after PH, demonstrated by immunoblotting. All demonstrated data are representative of three to six mice per genotype per time point and are presented as mean ± SEM. *, P < 0.05.
TNF and IL-6 participate in both the initiation of liver regeneration and the induction of the acute-phase response, which are very different biological processes (12). We show that in SOCS3-deficient mice, both cell proliferation and the expression of some acute-phase response genes are enhanced, suggesting that both processes may be negatively regulated by SOCS3. Phosphorylation of the mitogen-activated protein kinases (MAPKs) ERK1/2 has been shown to be a key event in the regenerative response to PH and is believed to occur via activation of the epidermal growth factor receptor (EGFR) by a variety of ligands (13). We found that the increased proliferative activity in Socs3 h-KO mice after PH is associated with a marked increase in phospho-ERK1/2, particularly at 18 and 24 h after PH (Fig. 2 E). Thus, SOCS3 deficiency after PH accelerates liver regeneration, prolongs STAT3 activation and the acute-phase response, and enhance ERK1/2 activation.
Socs3-deficient hepatocytes exhibit enhanced proliferation in vitro, associated with increased phosphorylation of STAT3 and ERK1/2
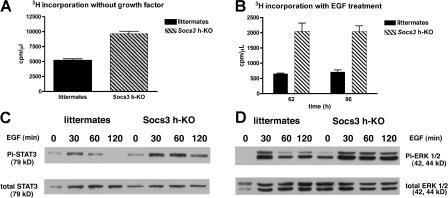
After demonstrating the marked enhancement of both cytokine signaling and hepatocyte proliferation during liver regeneration, an in vivo growth process that restores liver mass after removal of two thirds of the liver, we asked whether primary hepatocytes isolated from Socs3 h-KO mice also display enhanced proliferative activity in culture. For these experiments, we perfused the livers of Socs3 h-KO and control mice with collagenase, Percoll-purified the hepatocytes, plated them in media containing 5% serum for attachment, and maintained the cells in the absence of serum or growth factors. We found that the incorporation of [3H]thymidine in hepatocytes lacking SOCS3 is approximately double of that of hepatocytes with intact SOCS3 (Fig. 3 A). These data indicate that SOCS3 deficiency appears to result in autocrine mechanisms that lead to enhanced hepatocyte replication. The difference between the proliferative response of hepatocytes from Socs3 h-KO mice and their littermates in culture was further accentuated by exposure of the cells to EGF (Fig. 3 B). After 62 and 86 h in culture, EGF-stimulated hepatocytes from SOCS3-deficient mice incorporate [3H]thymidine at a rate that is almost fourfold higher than the hepatocytes from control littermates. Similar to our findings in the regenerating liver in vivo, SOCS3-deficient hepatocytes in culture displayed increased activation of STAT3 and ERK1/2 in response to EGF (Fig. 3, C and D). Thus, the deficiency in SOCS3 not only increases the intrinsic replicative capacity of hepatocytes but also makes them more responsive to the proliferative effects of growth factors such as EGF.
Figure 3.
SOCS3 down-regulates hepatocyte proliferation and growth factor signaling in vitro. (A) [3H]Thymidine incorporation by primary hepatocytes in the absence of growth factors 36 h after isolation and initiation of cell culture. (B) Hepatocyte proliferation in response to 20 ng/ml EGF. (C) Pi-STAT3Y705 is enhanced in response to EGF in Socs3-deficient hepatocytes. (D) Similar enhancement of Pi-ERK1/2 in response to EGF in Socs3 KO hepatocytes. All data are representative of at least three separate experiments, with four to six replicates in each 3H experiment, and are presented as mean ± SEM. *, P < 0.05.
JAK inhibition by AG490 and MAPK/ERK kinase (MEK) inhibition by U0126 inhibit hepatocyte proliferation in vitro
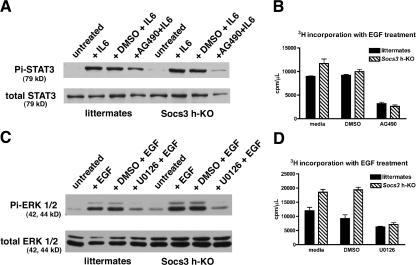
The data presented suggest that enhanced signaling through STAT3 and ERK1/2 may be responsible for the heightened proliferative state of SOCS3-deficient cells. We thus used small molecule inhibitors of the upstream kinases JAK (upstream of STAT3) and MEK (upstream of ERK1/2) to determine whether we could decrease the proliferation of Socs3 KO cells to the level of control hepatocytes. As expected, AG490, a JAK inhibitor, inhibits STAT3 phosphorylation in response to IL-6 in cultured hepatocytes, whereas the vehicle control (DMSO) has no effect on IL-6–stimulated STAT3 phosphorylation (Fig. 4 A). When added to culture medium 1 h before the addition of EGF, AG490 also inhibits hepatocyte proliferation, as measured by [3H]thymidine incorporation (Fig. 4 B). Addition of U0126, a selective MEK inhibitor, inhibits both ERK1/2 activation (Fig. 4 C) and hepatocyte proliferation (Fig. 4 D) in response to EGF. These results indicate that SOCS3 can modulate hepatocyte replication in vitro through effects on both the STAT3 and MAPK signaling pathways, similar to our in vivo observations.
Figure 4.
Inhibition of JAK or MEK in culture inhibits hepatocyte proliferation. (A) AG490 inhibits tyrosine phosphorylation of STAT3. (B) AG490 inhibits hepatocyte proliferation ([3H]thymidine assay). (C) U0126 inhibits ERK activation in response to EGF. (D) U0126 inhibits hepatocyte proliferation ([3H]thymidine assay). Data are representative of three independent experiments and are presented as mean ± SEM. *, P < 0.05.
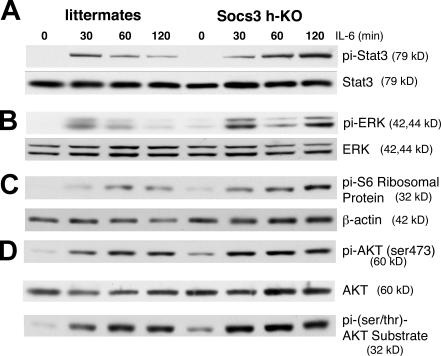
Socs3-deficient hepatocytes exhibit enhanced activation of multiple IL-6–dependent signaling pathways
The observations that both STAT3 and ERK1/2 activation are prolonged in vivo after PH in Socs3-deficient livers and in vitro after EGF stimulation of Socs3 deficient hepatocytes suggested that SOCS3 may also inhibit signaling pathways downstream of IL-6. To determine whether IL-6 stimulation of Socs3-deficient hepatocytes in culture would alter the response of downstream pathways, hepatocytes isolated from Socs3 h-KO mice and control littermates were exposed to IL-6. Protein lysates were prepared at the indicated times, and phosphorylation of STAT3, ERK1/2, Akt, and S6 protein was analyzed by phosphoimmunoblotting. IL-6–induced STAT3 phosphorylation is prolonged in Socs3 h-KO hepatocytes compared with controls (Fig. 5 A). ERK1/2 activation is biphasic, peaking at 30 and 120 min after IL-6 treatment (Fig. 5 B), and although the initial activation of ERK1/2 appears to be similar in hepatocytes from Socs3 h-KO mice and littermates, Socs3 KO hepatocytes demonstrate prolonged activation of ERK1/2 compared with control cells.
Figure 5.
Activation of STAT3, ERK1/2, and Akt are prolonged in Socs3 KO hepatocytes after IL-6 treatment. (A) Pi-STAT3Y705 is prolonged in response to IL-6 in Socs3 KO hepatocytes. (B) Activation of ERK1/2 is enhanced in response to IL-6 in Socs3 KO hepatocytes. (C) Prolonged activation of Pi-S6 S240/244 ribosomal protein in Socs3 KO versus littermate hepatocytes. β-Actin is shown as a loading control. (D) No difference in activation of Akt in Socs3 versus control hepatocytes. Data are representative of three to five independent experiments.
The protein kinase target of rapamycin (TOR) complexes 1/2, which regulate mammalian TOR (mTOR), are involved in multiple cellular processes, such as protein translation, nutrient sensing, and cell growth (14). To determine whether Socs3 could regulate IL-6–stimulated mTOR activity, we examined the levels of phospho-S6 ribosomal protein, a downstream target of mTOR. We observed enhanced and prolonged phosphorylation of S6 protein in Socs3 KO cells (Fig. 5 C), indicating that S6 kinase is activated to a greater extent, and thus, the mTOR pathway may also be enhanced. In contrast, Akt phosphorylation and activation, as determined by phospho-S473–Akt and a phospho-Akt substrate antibody, is similar in Socs3 KO and control hepatocytes after IL-6 treatment (Fig. 5 D). These results confirm that IL-6 stimulates multiple signaling pathways, such as those mediated by STAT3, ERK1/2, Akt, and S6 kinase in primary hepatocytes, and that SOCS3 deficiency results in prolonged and enhanced activation of some of these pathways.
The absence of Socs3 results in profound changes in gene expression after PH
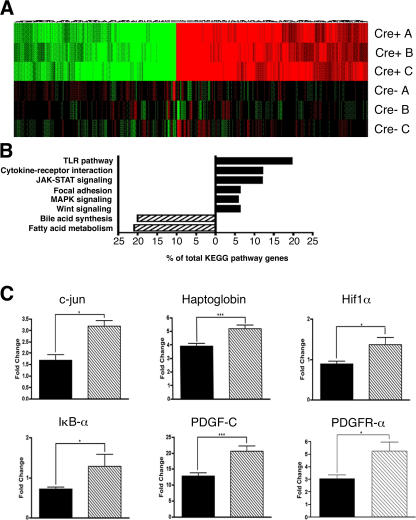
Given the significant enhancement of liver regeneration observed in Socs3 h-KO mice and our findings in vitro, we hypothesized that diverse cellular pathways contribute to the proliferative advantage of these cells. To determine whether the lack of SOCS3 has broad effects on gene expression during liver regeneration, we performed complementary DNA (cDNA) microarray analysis on RNA prepared from Socs3 h-KO mice and control littermates 18 h after PH. Three pooled samples per genotype were applied to Affymetrix oligonucleotide arrays, and data were analyzed as described in Supplemental materials and methods (available at http://www.jem.org/cgi/content/full/jem.20070820/DC1). The heatmap shown in Fig. 6 A represents a global view of differential gene expression patterns in Socs3 h-KO mice versus control littermates at 18 h after PH. The map demonstrates uniformity of expression among the pools for each genotype and reveals striking differences in gene expression profiles between Socs3 h-KO mice and control littermates. The Affymetrix data were subjected to National Institutes of Health (NIH) database for annotation, visualization, and integrated discovery (DAVID) analysis and Kyoto Encyclopedia of Genes and Genomes annotation. NIH DAVID analysis of differentially regulated genes revealed that several pathways thought to be important in liver regeneration are enhanced in Socs3 h-KO mice (Fig. 6 B). In addition to the JAK–STAT and MAPK signaling pathways, which we had already shown to be enhanced in the absence of SOCS3, we found that Toll-like receptor (TLR) signaling and cytokine–cytokine receptor interaction, focal adhesion, and Wnt signaling pathways are similarly up-regulated. These pathways have been shown by multiple investigators to be critical to normal regeneration (13, 15, 16), and in some cases may be involved in the development of HCC (8, 17, 18). Our microarray data support the view that the enhancement of multiple intracellular signaling pathways in Socs3 h-KO mice allows them to regenerate more efficiently than control littermates. Interestingly, DAVID analysis revealed that bile acid synthesis and fatty acid metabolism were down-regulated in Socs3 h-KO mice in comparison with control littermates, suggesting that SOCS3 may enhance rather than inhibit these functions. Recent data suggest that these pathways are themselves necessary for optimal liver regeneration (19, 20). Our results do not necessarily contradict these studies, as the multiple changes created by SOCS3 deficiency may alter the liver metabolic requirements during regeneration.
Figure 6.
Oligonucleotide microarrays demonstrate broad and diverse effects of SOCS3 on gene expression after PH. (A) Heatmap demonstrating a global picture of differential gene expression in Socs3 h-KO mice versus control littermates at 18 h after PH. (B) NIH DAVID analysis of a differentially regulated gene list reveals that multiple pathways are differentially regulated during regeneration in Socs3 h-KO mice. (C) Confirmed up-regulation of c-jun, Haptoglobin, Hif1α, IκBα, Pdgf-c, and Pdgfrα in Socs3 h-KO mice versus control littermates at 18 h after PH, demonstrated by real-time RT-PCR. Data are presented as fold change using the values from nonoperated mice as the denominator and are presented as mean ± SEM. *, P < 0.05; ***, P < 0.001.
To validate our microarray gene expression data, we performed real-time RT-PCR on several genes that had been shown to be up-regulated in Socs3 h-KO mice. Work by several investigators has shown that c-jun, a subunit of the transcription factor activator protein 1, is critical to the proliferative response after PH (21), and data from our laboratory suggest that AP-1 activity may be necessary for induction of heparin-binding EGF (unpublished data; Mitchell, C., personal communication), a member of the EGF family that participates in the linkage between cytokine activity and cell-cycle progression during regeneration (22). In the present study, we found that the expression of c-jun messenger RNA is approximately twofold higher in Socs3 h-KO mice compared with littermates (Fig. 6 C).
We also observed increased expression of haptoglobin, an acute-phase response protein, further supporting our observation of extended STAT3 activation in Socs3 h-KO mice (Fig. 2). IκBα is rapidly resynthesized after it is phosphorylated and degraded, which results in the release and activation of NF-κB (23, 24). We observed increased expression of Iκbα in Socs3 h-KO mice, suggesting that NF-κB was active at 18 h after PH in these animals. Increased expression of Iκbα is also consistent with the enrichment of genes in the TLR pathway (Fig. 6 B).
Hypoxia-inducible factor 1α (Hif1α) is induced under hypoxic conditions and transcribes factors that are important to angiogenesis, and has been reported to increase after PH (25). Hif1α expression was significantly increased in Socs3 h-KO mice compared with littermates after PH. Both platelet-derived growth factor C (Pdgfc) and PDGF receptor α (Pdgfrα) transcribe potent angiogenic factors (26), and were significantly up-regulated in Socs3 h-KO mice. The increased expression of three angiogenic factors in the livers of Socs3 h-KO mice suggests that initiation of liver architecture remodeling seen after PH may occur earlier in these mice. Collectively, our real-time RT-PCR results both validate our microarray data and provide additional insight into potential mechanisms behind the enhancement in liver regeneration seen in Socs3 h-KO mice.
Promoter analysis after PH in Socs3 h-KO mice
We were also interested in identifying the potential regulatory networks that might account for the changes in messenger RNA expression identified by the microarray and, thus, used transcriptional regulatory network analysis (TRNA) using the promoter analysis and interaction network tool (PAINT), as described in Materials and methods (27). TRNA was performed for genes found to be up-regulated >1.5-fold in Socs3 h-KO mice to identify transcription factor binding sites or transcriptional regulatory elements (TREs) in the 5′ flanking regions. The most enriched TREs in the gene set are listed by consensus sequence, along with their associated transcription factors, categorized, and ranked by frequency of presence in Table I. Most notably, TREs for several members of the NF-κB promoter family (p65, NF-κB, and c-Rel) are among the most overrepresented, consistent with the enrichment of cytokine signaling and TLR pathways determined by DAVID analysis (28). Additionally, the presence of the TRE for Elk-1, a downstream target of the MAPK pathway, supports our findings of extended and increased ERK1/2 activation in Socs3 h-KO mice (Fig. 2 E). Collectively, these results corroborate our in vivo and in vitro data demonstrating the increased proliferative capacity in Socs3 h-KO mice after PH, in the context of enhanced signaling via multiple pathways.
Table I.
TREs enriched in Socs3 h-KO mice 18 h after PH
| Candidate binding factor for TRE | TRE consensus sequence | Counts | p-value |
|---|---|---|---|
| NF-κB superfamily | |||
| c-Rel | SGGRNTTTCC | 87 | 0.05 |
| NF-κB | NGGGGAMTTTCCNN | 20 | 0.25 |
| NF-κB | GGGAMTTYCC | 15 | 0.02 |
| NF-κB (p65) | GGGRATTTCC | 12 | 0.01 |
| Cell growth and proliferation | |||
| Elk-1 | NAAACMGGAAGTNCVH | 71 | 0.02 |
| c-Jun/CRE-BP1 | TGACGTYA | 42 | 0.28 |
| CREB | TGACGTMA | 39 | 0.09 |
| Evi-1 | WGAYAAGATAGATAA | 25 | 0.19 |
| E2F | TTTSGCGC | 20 | 0.04 |
| ATF-2/CRE-BP1 | VGTGACGTMACN | 14 | 0.28 |
| CREB | NSTGACGTAANN | 11 | 0.07 |
| Egr-1 | WTGCGTGGGCGK | 1 | 0.39 |
| Egr-3 | NTGCGTGGGCGK | 1 | 0.28 |
| Differentiation and hepatocyte specificity | |||
| USF | GYCACGTGNC | 49 | 0.09 |
| Pax-6 | NNNNTTCACGCWTGANTKNNN | 34 | 0.28 |
| USF | NCACGTGN | 31 | 0.28 |
| CP2 | GCHCDAMCCAG | 23 | 0.17 |
| Oct-1 | CWNAWTKWSATRYN | 22 | 0.28 |
| HNF-4/COUP-TF | TGAMCTTTGMMCYT | 12 | 0.05 |
| Oct-1 | MKVATTTGCATATT | 7 | 0.41 |
| MEF-2 | NNTGTTACTAAAAATAGAAMNN | 2 | 0.21 |
Enriched TREs (biologically relevant transcription factor binding sites) were identified by comparing promoter regions of genes up-regulated >1.5-fold in Socs3 h-KO mice to all M. musculus promoter regions in the PAINT database. To identify TREs overrepresented, 5 kb of upstream sequence of each gene from the list of significantly up-regulated genes was analyzed by PAINT. TRE consensus sequence is the consensus binding site for the indicated binding factor (known transcription factor), provided as the International Union of Pure and Applied Chemistry 15-letter code. Multiple listings of candidate binding factors are based on differences in consensus binding sites. Count refers to the total number of times the TRE was found in the gene set. p-values were calculated as described in Materials and methods.
Accelerated development of N-nitrosodiethylamine (DEN)–induced HCC in Socs3 h-KO mice
Recent work on human HCCs demonstrated that the JAK–STAT and/or Ras-Raf–MAPK pathways are virtually always up-regulated in these cancers (17). Because we have shown that SOCS3 is a critical negative regulator of these pathways during the physiological regenerative response to PH, we wondered whether a lack of SOCS3 would promote neoplastic proliferative processes as well. To test this hypothesis, we used a model of DEN-induced hepatocarcinogenesis, in which Socs3 h-KO mice and control littermates were injected with a single dose of DEN at 12–14 d of life. The mice received no other treatment and were killed between 3 and 12 mo of age. Foci of altered hepatocytes were observed by 6 mo of age in both Socs3 h-KO mice and littermates but were larger in size in Socs3 h-KO mice (1.2 vs. 0.5% of the area). Socs3 h-KO mice developed HCC significantly earlier and in greater numbers than did littermate controls (Table II). By 9 mo, four out of six SOCS3-deficient mice had developed tumors, whereas none were detected in the control littermates; by 12 mo, all Socs3 h-KO mice had developed HCC. The tumors were typical trabecular-type HCCs with a moderate degree of differentiation (unpublished data). Using immunoblot analysis, we observed increased levels of activated, phosphorylated ERK1/2 in the tumor tissue from 9-mo Socs3 h-KO mice compared with adjacent tissue, as well as from liver tissue from control littermates (unpublished data), suggesting that signaling pathways that control proliferation may be activated in Socs3-deficient tumors.
Table II.
Development of HCC in Socs3 h-KO and control littermates after DEN injection
| Strain | Time
|
|||
|---|---|---|---|---|
| 3 mo | 6 mo | 9 mo | 12 mo | |
| Control | 0/10 | 0/5 | 0/6 | 4/8 |
| Socs3 h-KO | 0/4 | 0/3 | 4/6 | 4/4 |
12–14-d-old pups were injected with a single dose of 5 mg/kg DEN. At the indicated times, male mice were killed and livers were examined for tumors. Macroscopic HCCs were confirmed by hematoxylin and eosin analysis, as described in Materials and methods. Tumor development was significantly different in Socs3 h-KO mice as determined by a log-rank test, with a p-value of 0.0027.
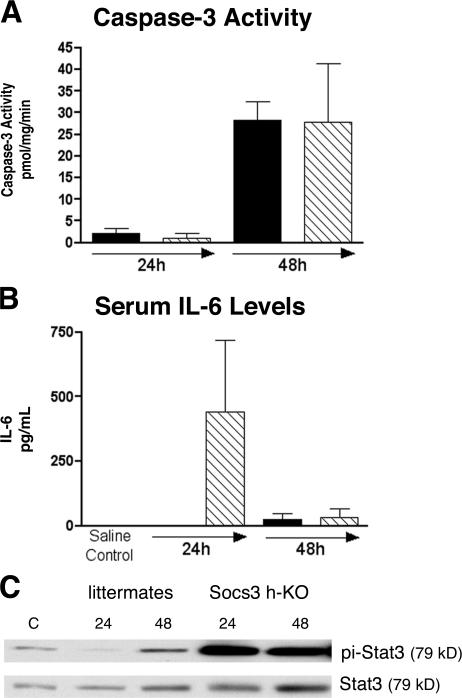
Previous work suggested that SOCS3 may decrease tumor formation in the liver by blocking apoptosis (29), so we examined the extent of apoptosis induced by DEN in our mice using a caspase 3 activity assay (30). We found no differences among caspase activities of Socs3 h-KO mice and control littermates 24 h after injection (Fig. 7 A), though caspase 3 is activated in both groups of mice 48 h after DEN injection. To determine whether DEN injection induces expression of the antiapoptotic protein Bcl-xL, we performed immunoblot analysis of liver lysates. We observed a modest induction of Bcl-xL at 24 and 48 h after DEN injection, but the levels were not different between Socs3 h-KO mice compared with littermate controls (unpublished data), suggesting that dysregulated apoptotic pathways did not result in the earlier development of HCC in the absence of SOCS3 in our model.
Figure 7.
Analysis of early response to DEN injection in Socs3 h-KO mice versus littermates. Mice were injected with 100 μg DEN per gram of body weight, and liver tissues were harvested 24 and 48 h later. (A) DEN injection induces caspase 3 activity at 48 h but similarly in Socs3 h-KO (hatched bars) and littermate (black bars) mice. (B) Serum IL-6 levels are elevated in Socs3 h-KO versus littermate mice 24 h after DEN injection. (C) Enhanced activation of STAT3 after DEN injection in Socs3 h-KO mice. Data presented are representative of six mice per genotype per time point and are presented as mean ± SEM. *, P < 0.05.
It has recently been demonstrated that IL-6 is rapidly released after DEN injection (31), and we found that serum IL-6 levels were dramatically elevated in Socs3 h-KO mice compared with littermates at 24 h after injection (Fig. 7 B). Elevated levels of phospho-STAT3 are observed at both 24 and 48 h after DEN injection in Socs3 h-KO mice (Fig. 7 C). These data support the hypothesis that SOCS3 may prevent DEN-induced HCC formation by altering the response to IL-6 rather than by inhibiting apoptotic pathways.
DISCUSSION
Liver regeneration after PH is a unique growth process in which the hepatic mass is rapidly restored after surgical removal of two thirds of the liver. The regenerative process after PH is dependent on the replication of hepatocytes, which are completely differentiated and normally quiescent cells, and does not rely on the activation of a compartment of liver stem cells. Hepatocyte replication does require the involvement of NPCs, the major source of hepatic cytokines and growth factors, and the remodeling of extracellular matrix (6, 8, 32). A large number of components of the innate immune system, including TNF, complement proteins, and the IL-6–STAT3 pathway, participate in the initiation of liver regeneration (6, 8, 33). Additionally, studies have shown that growth factors such as HGF, members of the EGF family, and stem cell factor participate in the replicative response of hepatocytes after PH (32, 34). Although it may appear that the cytokine and growth factor pathways in the regenerating liver may have distinct functions, these networks may overlap in some cases. For instance, it has been demonstrated that STAT3 activates multiple genes and contributes to hepatocyte replication during liver regeneration (16). Moreover, IL-6 has multiple functions, including cell survival, cell proliferation, and the acute-phase response to inflammation (35–37), and the role of IL-6 in the regenerating liver remains a controversial issue. Thus, it is unclear how cytokines and growth factors may interact to produce the precisely orchestrated liver growth response that occurs after PH.
SOCS proteins were originally described as negative regulators of cytokine signaling. More recent studies using specifically targeted mouse models reveal that, in the hematopoietic system, SOCS1 and SOCS3 inhibit myeloid signaling pathways and may influence conditions such as inflammation, autoimmunity, and malignancy (17, 38–41). In the liver, SOCS3 regulates the activity of the cytokine pathway stimulated by the IL-6 family, which signals through the IL-6R–gp130 to activate the STAT3 transcription factor. IL-6 is a key mediator of the acute-phase response to inflammation, and by controlling the IL-6–STAT3 pathway, SOCS3 functions as a regulator of this response. Previous work from our laboratory showed that Socs3 expression is greatly induced during the first 12 h after PH (9). Our interpretation of these results was that SOCS3 halts STAT3 activation and terminates the early phase of liver regeneration in which cytokines are major participants, leading to the growth factor–regulated progression of hepatocytes through the cell cycle and, ultimately, DNA replication. Thus, SOCS3 could function at the interface between cytokine expression and growth factor activity during the regenerative response. To directly analyze the role of SOCS3 during liver regeneration after PH, we studied a variety of proliferative processes in Socs3 h-KO mice. The main findings of this work were that (a) Socs3 h-KO mice show an enhancement of hepatocyte DNA synthesis and mitosis after PH; (b) Socs3 KO hepatocytes are highly proliferative in primary culture, even in the absence of growth factors; (c) SOCS3 deficiency enhances multiple pathways related to both cytokine activity and cell proliferation; and (d) Socs3 h-KO mice develop HCC at an accelerated rate.
The enhancement of liver regeneration caused by SOCS3 deficiency was an unexpected result. Based on existing data on the role of SOCS3 in blocking STAT3 expression, it was expected that SOCS3 deficiency after PH would lead to an enhanced and prolonged acute-phase reaction that might inhibit the proliferative response. However, SOCS3 deficiency led to both an enhancement of the expression of acute-phase response genes and the up-regulation of multiple pathways related to cell proliferation. These data are consistent with reports in other systems showing that SOCS3 has a broader role than the control of cytokine activity, which include the enhancement of the proliferation of myeloid cells in the hematopoietic system (38), islet cell hyperplasia (42), inhibition of insulin receptor phosphorylation associated with the metabolic syndrome in the liver (43, 44), and the promotion of liver fibrosis through TGF-β production (45). Sun et al. reported that STAT3 activation was enhanced after PH in Socs3 heterozygous (+/−) mice, but that hepatocytes isolated from these mice and exposed to IL-6 in culture showed a decrease in cell proliferation because of increased expression of p21 (46). We suggest that the difference in our results from those of Sun et al. may be explained by the use of Socs3 h-KO mice in our experiments.
As expected, IL-6 levels after PH were not altered by hepatocyte Socs3 deficiency because IL-6 is produced by liver NPCs, which were not genetically targeted in our mice. The data we obtained with hepatocytes isolated from Socs3 h-KO mice and placed in primary culture demonstrate that these cells have a high proliferative activity even when maintained in medium without growth factors and are highly sensitive to EGF stimulation. Similar to the experiments performed in regenerating livers, the increased proliferation of cultured hepatocytes from Socs3 h-KO mice is associated with enhanced activation of STAT3 and ERK1/2 after IL-6 or EGF stimulation. These effects can be blocked by inhibitors of the JAK–STAT (AG490) or MEK–ERK1/2 (U0126) pathways. Our results are consistent with other work demonstrating that SOCS3 (as well as other SOCS proteins) can regulate signaling through the EGFR (47, 48).
Calvisi et al. reported that the JAK–STAT pathway is enhanced in human HCC compared with nonneoplastic liver and is associated with the down-regulation of various suppressors of this pathway (17). We wondered whether the enhanced hepatocyte proliferation in the regenerating liver and the high proliferative capacity of Socs3 KO hepatocytes in culture would predispose these mice to liver carcinogenesis. We found that tumor development is accelerated in Socs3 h-KO mice that are injected with DEN, a known hepatocarcinogen. These data are consistent with the observation that SOCS3 deficiency promotes cell growth in human HCC by enhancing the JAK–STAT and focal adhesion kinase signaling pathways (18). Our microarray analysis of post-PH liver RNA using DAVID and the Kyoto Encyclopedia of Genes and Genomes annotation identified both of these pathways as being activated in Socs3 h-KO mice. Recently, Ogata et al. reported that Socs3 hepatic-deficient mice developed a greater number of hepatic tumors that were larger than those of control mice when injected with DEN for 6 wk or DEN in combination with 6 mo of a choline-deficient, l–amino acid diet (29). Ogata et al. concluded that in the setting of inflammation-induced tumorigenesis, STAT3 was activated and induced expression of antiapoptotic proteins such as Bcl-2 and Bcl-xL (29). Our findings included accelerated HCC development in Socs3 h-KO mice after a single DEN injection (Table II), but we did not observe a difference in DEN-induced apoptosis 24 or 48 h after DEN injection as measured by caspase 3 activation or Bcl-xL expression. However we did observe increased release of IL-6 in Socs3 h-KO mice and subsequent enhanced phosphorylation of STAT3. It is possible that the elevated levels of IL-6 provide a cell proliferative or survival advantage to tumor cells in Socs3 h-KO mice (35–37, 49). Regardless of mechanisms, our results and those of Ogata et al. demonstrate that SOCS3 deficiency increases the risk of HCC development.
Despite the fact that individual pathways involving various cytokines and growth factors during liver regeneration have been described in detail, there is little information regarding the mechanisms that coordinate these events and lead to a precisely regulated and synchronized growth process. Our work demonstrates that, in the regenerating liver, SOCS3 regulates not only cytokine expression through various pathways involving TLR receptors and the IL-6–STAT3 pathway but also controls the expression of multiple genes involved in proliferative pathways that require ERK activation. We suggest that SOCS3 expression at the early stages of liver regeneration is an essential element that coordinates the termination of the main cytokine response with the activation of growth factors that regulate cell-cycle progression. In the absence of SOCS3, hepatocytes acquire an enhanced proliferative capacity, both in vivo and in culture. Thus, hepatic SOCS3 can function both as an antiinflammatory agent and a tumor suppressor. SOCS3 may be a suitable target for the regulation of acute-phase responses to inflammation and for the prevention or treatment of HCC.
MATERIALS AND METHODS
Animals and operative technique.
The generation of mice in which the Socs3 gene has been flanked by loxP sequences (Socs3fl/fl mice) has been previously described (2). Hepatocyte-specific excision of the Socs3 gene was achieved by breeding Socs3fl/fl mice with mice expressing the Cre recombinase transgene under control of the albumin promoter (Alb-Cre+), yielding Socs3 h-KO mice. Socs3fl/fl, Alb-Cre− littermates were used as controls for all experiments and are henceforth referred to as littermates. All mice (C57BL/6) were free of Helicobacter species and were housed in a specific pathogen-free facility with 12-h light/dark cycles with free access to standard food and water. PH and sham operations were performed as previously described (n = three to six mice per genotype per time point) (15, 50). Liver remnants were weighed after removal of necrotic stumps and sutures and compared with postoperative body weight. For HCC experiments, a single i.p. injection of 5 mg DEN per kilogram (Sigma-Aldrich) was performed 12–14 d after birth. For short time points, a single injection of 100 mg DEN per kilogram (31) was given to 4-wk-old mice. At the times indicated in the figures, mice were killed by CO2 inhalation. All animal studies were performed under approved institutional animal care and use committee protocols at the University of Washington.
Histology.
For all mice harvested 24 h or later after PH, 50 μg BrdU per gram of body weight was injected i.p. 2 h before harvest. Liver lobes were fixed and stained as previously described (15, 22). For DEN-induced HCC experiments, macroscopic tumors were counted at necropsy. Liver sections were stained with hematoxylin and eosin and scored for foci of altered hepatocytes and microscopic HCC using previously described criteria (51, 52).
ELISA.
Serum IL-6 concentration was determined using an ELISA, according to the manufacturer's instructions (R&D Systems), as described previously (15, 22).
RNA isolation, Northern blotting, and real-time RT-PCR analysis.
RNA was isolated from whole liver or primary hepatocytes using TRIzol (Invitrogen) as previously described (9, 15). Northern blot analyses were done as previously described (9, 15) using 32P-labeled cDNA probes for Socs3, Socs1, Cd14, or Saa2 (15), or a 32P-labeled riboprobe for cyclophilin (Ambion) as a loading control. Bands were quantified using a phosphorimager (Storm; Molecular Dynamics). For cDNA synthesis, 2 μg RNA from 18-h PH samples were reverse transcribed using the Retroscript kit (Invitrogen), and real-time RT-PCR was performed on the resultant cDNA using FAM-labeled primers and reagents (Applied Biosystems). ΔΔCt values were calculated by subtracting Ct values for the gene of interest from the Ct values for 18S (a housekeeping gene) and then subtracting the ΔCt value obtained for each gene from nonoperated control mice. Fold change was calculated by normalizing all values to those of nonoperated wild-type mice.
Immunoblotting and caspase 3 activity assay.
Whole liver or primary hepatocyte protein lysates were prepared in a Triton X-100 lysis buffer and quantified as previously described (15). Caspase 3 activity was determined using a fluorogenic substrate as previously described (30). 20 or 50 μg of protein from each sample were separated using SDS-PAGE, and immunoblotting was performed using standard procedures with the following primary antibodies from Cell Signaling Technology (phosphotyrosine 705 STAT3, total STAT3, phospho-ERK1/2, phospho-S473–Akt, total Akt, phospho-S/T Akt substrates, phospho-S6 S240/244 protein, phosphotyrosine 701 STAT1, total STAT1, and phosphotyrosine 649 STAT5), Santa Cruz Biotechnology, Inc. (cyclin A and p107), Sigma-Aldrich (β-actin), and Zymed (total STAT5a/b), and total ERK1/2 (53) as previously described (9, 22).
Primary hepatocyte culture, [3H]thymidine incorporation, and kinase inhibitor treatment.
Primary hepatocytes were isolated from Socs3 h-KO mouse and control livers by collagenase perfusion and Percoll gradients, as previously described (54). To measure DNA replication in cultured hepatocytes, [3H]thymidine was added to media at a final concentration of 1 mCi/ml for 4 h, and incorporation was measured as previously described (55). Where indicated in the figures, 20 ng/ml IL-6 or 30 ng/ml EGF was added to media. For kinase inhibitor experiments, 50 μM AG490, 5 μM U0126, or DMSO (vehicle control) was added 1 h before IL-6 or EGF stimulation. Total RNA and protein lysates were prepared from hepatocyte cell cultures as described.
Microarray analysis of 18-h PH samples.
To perform statistically relevant gene expression profiling at 18 h after PH, additional PHs were performed so that there were 9–11 samples for each genotype at this time point. Total RNA was isolated as described in Materials and methods and then purified using the RNeasy kit (QIAGEN). Equal amounts of each sample were pooled, resulting in three groups per genotype. All samples were of good quality, as assessed by a bioanalyzer (model 2100; Agilent Technologies). A detailed description of microarray methods can be found in Supplemental materials and methods. Microarray data have been deposited in the Gene Expression Omnibus under accession no. GSE9549.
TRNA.
Promoter analysis was performed using PAINT (version 3.5, available at http://www.dbi.tju.edu/dbi/tools/paint) (27). The input file contained the Locuslink identifiers for 583 Affymetrix probe sets determined by microarray to be up-regulated >1.5-fold in Socs3 h-KO mice 18 h after PH. PAINT retrieved 5 kb of 5′ flanking sequence for 524 genes and identified 473 unique promoters. Using a TRE core similarity threshold of 1, 132 TREs were found for known promoters within the input sequences. PAINT tested for enrichment of specific TREs and transformed the results into a candidate interaction matrix based on a p-value threshold of 0.05 and a false discovery rate threshold of 0.3. For the group of genes analyzed, the p-values for significance of enrichment of each TRE were calculated using hypergeometric distribution, with the abundance of each TRE compared with the default PAINT reference set of randomly selected Mus musculus promoters. For additional details, please refer to Vadigepalli et al. (27) and Addya et al. (56).
Statistical analyses.
For all in vivo experiments, three to six mice per genotype per time point were used, except for the microarray analyses done on 18-h post-PH samples. All in vitro experiments were performed at least three times. Data presented are either representative of the replicates, or quantitation and statistical analysis of all replicates. Statistical analysis was done by nonparametric analysis, using the Mann-Whitney test or an unpaired t test with Welch's correction. Data are presented as mean ± SEM, with P < 0.05 considered statistically significant. Prism software (GraphPad) was used for all statistical analyses except for microarray data.
Online supplemental material.
Supplemental materials and methods provides detailed methods for the generation and analysis of microarray data (Fig. 6, A and B), as performed by the Center for Ecogenetics and Environmental Health at the University of Washington. Table S1 provides a complete list of the genes and identifiers presented in Fig. 6 A. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070820/DC1.
Supplemental Material
Acknowledgments
The authors would like to thank Doug Hilton, Warren Alexander, and Ben Croker for developing and providing the Socs3fl/fl mice. The authors also wish to thank John Brooling and Renay Bauer for their technical assistance.
This work was supported by NIH grants CA-23226 (to J.S. Campbell) and CA-023226 and CA-074131 (to N. Fausto), an American College of Surgeons Resident Research Scholarship (to K.J. Riehle), and University of Washington National Institute of Environmental Health Sciences–sponsored Center for Ecogenetics and Environmental Health grant P30ES07033.
The authors have no conflicting financial interests.
Abbreviations used: cDNA, complementary DNA; DAVID, database for annotation, visualization, and integrated discovery; DEN, N-nitrosodiethylamine; EGF, epidermal growth factor; ERK1/2, extracellular signal-regulated kinase 1/2; h-KO, hepatocyte-specific knockout; HCC, hepatocellular carcinoma; HIF1α, hypoxia-inducible factor 1 α; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; mTOR, mammalian target of rapamycin; NPC, nonparenchymal cell; PAINT, promoter analysis and interaction network tool; PDGF, platelet-derived growth factor; PH, two-thirds partial hepatectomy; SOCS, suppressor of cytokine signaling; TLR, Toll-like receptor; TRE, transcriptional regulatory element; TRNA, transcriptional regulatory network analysis.
References
- 1.Alexander, W.S., and D.J. Hilton. 2004. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 22:503–529. [DOI] [PubMed] [Google Scholar]
- 2.Croker, B.A., D.L. Krebs, J.G. Zhang, S. Wormald, T.A. Willson, E.G. Stanley, L. Robb, C.J. Greenhalgh, I. Forster, B.E. Clausen, et al. 2003. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 4:540–545. [DOI] [PubMed] [Google Scholar]
- 3.Streetz, K.L., T. Wustefeld, C. Klein, K.J. Kallen, F. Tronche, U.A. Betz, G. Schutz, M.P. Manns, W. Muller, and C. Trautwein. 2003. Lack of gp130 expression in hepatocytes promotes liver injury. Gastroenterology. 125:532–543. [DOI] [PubMed] [Google Scholar]
- 4.Yang, X.P., F. Schaper, A. Teubner, F. Lammert, P.C. Heinrich, S. Matern, and E. Siewert. 2005. Interleukin-6 plays a crucial role in the hepatic expression of SOCS3 during acute inflammatory processes in vivo. J. Hepatol. 43:704–710. [DOI] [PubMed] [Google Scholar]
- 5.Schaper, F., C. Gendo, M. Eck, J. Schmitz, C. Grimm, D. Anhuf, I.M. Kerr, and P.C. Heinrich. 1998. Activation of the protein tyrosine phosphatase SHP2 via the interleukin-6 signal transducing receptor protein gp130 requires tyrosine kinase Jak1 and limits acute-phase protein expression. Biochem. J. 335:557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fausto, N., J.S. Campbell, and K.J. Riehle. 2006. Liver regeneration. Hepatology. 43:S45–S53. [DOI] [PubMed] [Google Scholar]
- 7.Koniaris, L.G., I.H. McKillop, S.I. Schwartz, and T.A. Zimmers. 2003. Liver regeneration. J. Am. Coll. Surg. 197:634–659. [DOI] [PubMed] [Google Scholar]
- 8.Taub, R. 2004. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 5:836–847. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, J.S., L. Prichard, F. Schaper, J. Schmitz, A. Stephenson-Famy, M.E. Rosenfeld, G.M. Argast, P.C. Heinrich, and N. Fausto. 2001. Expression of suppressors of cytokine signaling during liver regeneration. J. Clin. Invest. 107:1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrecht, J.H., B.M. Rieland, C.J. Nelsen, and C.L. Ahonen. 1999. Regulation of G(1) cyclin-dependent kinases in the liver: role of nuclear localization and p27 sequestration. Am. J. Physiol. 277:G1207–G1216. [DOI] [PubMed] [Google Scholar]
- 11.Aldeguer, X., F. Debonera, A. Shaked, A.M. Krasinkas, A.E. Gelman, X. Que, G.A. Zamir, S. Hiroyasu, K.K. Kovalovich, R. Taub, and K.M. Olthoff. 2002. Interleukin-6 from intrahepatic cells of bone marrow origin is required for normal murine liver regeneration. Hepatology. 35:40–48. [DOI] [PubMed] [Google Scholar]
- 12.Greenwel, P., M.J. Iraburu, M. Reyes-Romero, N. Meraz-Cruz, E. Casado, J.A. Solis-Herruzo, and M. Rojkind. 1995. Induction of an acute phase response in rats stimulates the expression of alpha 1(I) procollagen messenger ribonucleic acid in their livers. Possible role of interleukin-6. Lab. Invest. 72:83–91. [PubMed] [Google Scholar]
- 13.Talarmin, H., C. Rescan, S. Cariou, D. Glaise, G. Zanninelli, M. Bilodeau, P. Loyer, C. Guguen-Guillouzo, and G. Baffet. 1999. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signalling pathway involved in the regulation of G(1) phase progression in proliferating hepatocytes. Mol. Cell. Biol. 19:6003–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaskar, P.T., and N. Hay. 2007. The two TORCs and Akt. Dev. Cell. 12:487–502. [DOI] [PubMed] [Google Scholar]
- 15.Campbell, J.S., K.J. Riehle, J.T. Brooling, R.L. Bauer, C. Mitchell, and N. Fausto. 2006. Proinflammatory cytokine production in liver regeneration is Myd88-dependent, but independent of Cd14, Tlr2, and Tlr4. J. Immunol. 176:2522–2528. [DOI] [PubMed] [Google Scholar]
- 16.Li, W., X. Liang, C. Kellendonk, V. Poli, and R. Taub. 2002. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J. Biol. Chem. 277:28411–28417. [DOI] [PubMed] [Google Scholar]
- 17.Calvisi, D.F., S. Ladu, A. Gorden, M. Farina, E.A. Conner, J.S. Lee, V.M. Factor, and S.S. Thorgeirsson. 2006. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 130:1117–1128. [DOI] [PubMed] [Google Scholar]
- 18.Niwa, Y., H. Kanda, Y. Shikauchi, A. Saiura, K. Matsubara, T. Kitagawa, J. Yamamoto, T. Kubo, and H. Yoshikawa. 2005. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 24:6406–6417. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez, M.A., C. Albor, M. Ingelmo-Torres, S.J. Nixon, C. Ferguson, T. Kurzchalia, F. Tebar, C. Enrich, R.G. Parton, and A. Pol. 2006. Caveolin-1 is essential for liver regeneration. Science. 313:1628–1632. [DOI] [PubMed] [Google Scholar]
- 20.Shteyer, E., Y. Liao, L.J. Muglia, P.W. Hruz, and D.A. Rudnick. 2004. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology. 40:1322–1332. [DOI] [PubMed] [Google Scholar]
- 21.Stepniak, E., R. Ricci, R. Eferl, G. Sumara, I. Sumara, M. Rath, L. Hui, and E.F. Wagner. 2006. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 20:2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell, C., M. Nivison, L.F. Jackson, R. Fox, D.C. Lee, J.S. Campbell, and N. Fausto. 2005. Heparin-binding epidermal growth factor-like growth factor links hepatocyte priming with cell cycle progression during liver regeneration. J. Biol. Chem. 280:2562–2568. [DOI] [PubMed] [Google Scholar]
- 23.Brown, K., S. Park, T. Kanno, G. Franzoso, and U. Siebenlist. 1993. Mutual regulation of the transcriptional activator NF-kappa B and its inhibitor, I kappa B-alpha. Proc. Natl. Acad. Sci. USA. 90:2532–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacker, H., and M. Karin. 2006. Regulation and function of IKK and IKK-related kinases. Sci. STKE. 2006:re13. [DOI] [PubMed] [Google Scholar]
- 25.Hickey, M.M., and M.C. Simon. 2006. Regulation of angiogenesis by hypoxia and hypoxia-inducible factors. Curr. Top. Dev. Biol. 76:217–257. [DOI] [PubMed] [Google Scholar]
- 26.Maeno, H., T. Ono, D.K. Dhar, T. Sato, A. Yamanoi, and N. Nagasue. 2005. Expression of hypoxia inducible factor-1alpha during liver regeneration induced by partial hepatectomy in rats. Liver Int. 25:1002–1009. [DOI] [PubMed] [Google Scholar]
- 27.Vadigepalli, R., P. Chakravarthula, D.E. Zak, J.S. Schwaber, and G.E. Gonye. 2003. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS. 7:235–252. [DOI] [PubMed] [Google Scholar]
- 28.Dennis, G., Jr., B.T. Sherman, D.A. Hosack, J. Yang, W. Gao, H.C. Lane, and R.A. Lempicki. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- 29.Ogata, H., T. Kobayashi, T. Chinen, H. Takaki, T. Sanada, Y. Minoda, K. Koga, G. Takaesu, Y. Maehara, M. Iida, and A. Yoshimura. 2006. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology. 131:179–193. [DOI] [PubMed] [Google Scholar]
- 30.Chaisson, M.L., J.T. Brooling, W. Ladiges, S. Tsai, and N. Fausto. 2002. Hepatocyte-specific inhibition of NF-kappaB leads to apoptosis after TNF treatment, but not after partial hepatectomy. J. Clin. Invest. 110:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naugler, W.E., T. Sakurai, S. Kim, S. Maeda, K. Kim, A.M. Elsharkawy, and M. Karin. 2007. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 317:121–124. [DOI] [PubMed] [Google Scholar]
- 32.Michalopoulos, G.K., and M.C. DeFrances. 1997. Liver regeneration. Science. 276:60–66. [DOI] [PubMed] [Google Scholar]
- 33.Strey, C.W., M. Markiewski, D. Mastellos, R. Tudoran, L.A. Spruce, L.E. Greenbaum, and J.D. Lambris. 2003. The proinflammatory mediators C3a and C5a are essential for liver regeneration. J. Exp. Med. 198:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren, X., C. Hogaboam, A. Carpenter, and L. Colletti. 2003. Stem cell factor restores hepatocyte proliferation in IL-6 knockout mice following 70% hepatectomy. J. Clin. Invest. 112:1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wuestefeld, T., C. Klein, K.L. Streetz, U. Betz, J. Lauber, J. Buer, M.P. Manns, W. Muller, and C. Trautwein. 2003. Interleukin-6/glycoprotein 130-dependent pathways are protective during liver regeneration. J. Biol. Chem. 278:11281–11288. [DOI] [PubMed] [Google Scholar]
- 36.Blindenbacher, A., X. Wang, I. Langer, R. Savino, L. Terracciano, and M.H. Heim. 2003. Interleukin 6 is important for survival after partial hepatectomy in mice. Hepatology. 38:674–682. [DOI] [PubMed] [Google Scholar]
- 37.Jin, X., T.A. Zimmers, E.A. Perez, R.H. Pierce, Z. Zhang, and L.G. Koniaris. 2006. Paradoxical effects of short- and long-term interleukin-6 exposure on liver injury and repair. Hepatology. 43:474–484. [DOI] [PubMed] [Google Scholar]
- 38.Croker, B.A., D. Metcalf, L. Robb, W. Wei, S. Mifsud, L. DiRago, L.A. Cluse, K.D. Sutherland, L. Hartley, E. Williams, et al. 2004. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 20:153–165. [DOI] [PubMed] [Google Scholar]
- 39.Wong, P.K., P.J. Egan, B.A. Croker, K. O'Donnell, N.A. Sims, S. Drake, H. Kiu, E.J. McManus, W.S. Alexander, A.W. Roberts, and I.P. Wicks. 2006. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J. Clin. Invest. 116:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, T., H. Ogata, M. Kamio, A. Joo, H. Shiraishi, Y. Tokunaga, M. Sata, H. Nagai, and A. Yoshimura. 2004. SOCS1 is a suppressor of liver fibrosis and hepatitis-induced carcinogenesis. J. Exp. Med. 199:1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanada, T., T. Kobayashi, T. Chinen, K. Saeki, H. Takaki, K. Koga, Y. Minoda, T. Sanada, T. Yoshioka, H. Mimata, et al. 2006. IFN-γ–dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J. Exp. Med. 203:1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindberg, K., S.G. Ronn, D. Tornehave, H. Richter, J.A. Hansen, J. Romer, M. Jackerott, and N. Billestrup. 2005. Regulation of pancreatic beta-cell mass and proliferation by SOCS-3. J. Mol. Endocrinol. 35:231–243. [DOI] [PubMed] [Google Scholar]
- 43.Ueki, K., T. Kondo, and C.R. Kahn. 2004. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol. Cell. Biol. 24:5434–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard, J.K., and J.S. Flier. 2006. Attenuation of leptin and insulin signaling by SOCS proteins. Trends Endocrinol. Metab. 17:365–371. [DOI] [PubMed] [Google Scholar]
- 45.Ogata, H., T. Chinen, T. Yoshida, I. Kinjyo, G. Takaesu, H. Shiraishi, M. Iida, T. Kobayashi, and A. Yoshimura. 2006. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene. 25:2520–2530. [DOI] [PubMed] [Google Scholar]
- 46.Sun, R., B. Jaruga, S. Kulkarni, H. Sun, and B. Gao. 2005. IL-6 modulates hepatocyte proliferation via induction of HGF/p21cip1: regulation by SOCS3. Biochem. Biophys. Res. Commun. 338:1943–1949. [DOI] [PubMed] [Google Scholar]
- 47.Xia, L., L. Wang, A.S. Chung, S.S. Ivanov, M.Y. Ling, A.M. Dragoi, A. Platt, T.M. Gilmer, X.Y. Fu, and Y.E. Chin. 2002. Identification of both positive and negative domains within the epidermal growth factor receptor COOH-terminal region for signal transducer and activator of transcription (STAT) activation. J. Biol. Chem. 277:30716–30723. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson, S.E., D. Metcalf, N.S. Sprigg, R. Columbus, F. Walker, A. Silva, D. Cary, T.A. Willson, J.G. Zhang, D.J. Hilton, et al. 2005. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc. Natl. Acad. Sci. USA. 102:2328–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovalovich, K., W. Li, R. DeAngelis, L.E. Greenbaum, G. Ciliberto, and R. Taub. 2001. Interleukin-6 protects against Fas-mediated death by establishing a critical level of anti-apoptotic hepatic proteins FLIP, Bcl-2, and Bcl-xL. J. Biol. Chem. 276:26605–26613. [DOI] [PubMed] [Google Scholar]
- 50.Greene, A.K., and M. Puder. 2003. Partial hepatectomy in the mouse: technique and perioperative management. J. Invest. Surg. 16:99–102. [PubMed] [Google Scholar]
- 51.Maronpot, R.R., C.A. Montgomery Jr., G.A. Boorman, and E.E. McConnell. 1986. National Toxicology Program nomenclature for hepatoproliferative lesions of rats. Toxicol. Pathol. 14:263–273. [DOI] [PubMed] [Google Scholar]
- 52.Pierce, R.H., M.E. Vail, L. Ralph, J.S. Campbell, and N. Fausto. 2002. Bcl-2 expression inhibits liver carcinogenesis and delays the development of proliferating foci. Am. J. Pathol. 160:1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seger, R., D. Seger, A.A. Reszka, E.S. Munar, H. Eldar-Finkelman, G. Dobrowolska, A.M. Jensen, J.S. Campbell, E.H. Fischer, and E.G. Krebs. 1994. Overexpression of mitogen-activated protein kinase kinase (MAPKK) and its mutants in NIH 3T3 cells. Evidence that MAPKK involvement in cellular proliferation is regulated by phosphorylation of serine residues in its kinase subdomains VII and VIII. J. Biol. Chem. 269:25699–25709. [PubMed] [Google Scholar]
- 54.Wu, J.C., G. Merlino, and N. Fausto. 1994. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. USA. 91:674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Argast, G.M., J.S. Campbell, J.T. Brooling, and N. Fausto. 2004. Epidermal growth factor receptor transactivation mediates tumor necrosis factor-induced hepatocyte replication. J. Biol. Chem. 279:34530–34536. [DOI] [PubMed] [Google Scholar]
- 56.Addya, S., M.A. Keller, K. Delgrosso, C.M. Ponte, R. Vadigepalli, G.E. Gonye, and S. Surrey. 2004. Erythroid-induced commitment of K562 cells results in clusters of differentially expressed genes enriched for specific transcription regulatory elements. Physiol. Genomics. 19:117–130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.