Abstract
Pneumococcus is one of the most important human pathogens that causes life-threatening invasive diseases, especially at the extremities of age. Capsular polysaccharides (CPSs) are known to induce protective antibodies; however, it is not feasible to develop CPS-based vaccines that cover all of the 90 disease-causing serotypes. We applied a genomic approach and described the antibody repertoire for pneumococcal proteins using display libraries expressing 15–150 amino acid fragments of the pathogen's proteome. Serum antibodies of exposed, but not infected, individuals and convalescing patients identified the ANTIGENome of pneumococcus consisting of ∼140 antigens, many of them surface exposed. Based on several in vitro assays, 18 novel candidates were preselected for animal studies, and 4 of them showed significant protection against lethal sepsis. Two lead vaccine candidates, protein required for cell wall separation of group B streptococcus (PcsB) and serine/threonine protein kinase (StkP), were found to be exceptionally conserved among clinical isolates (>99.5% identity) and cross-protective against four different serotypes in lethal sepsis and pneumonia models, and have important nonredundant functions in bacterial multiplication based on gene deletion studies. We describe for the first time opsonophagocytic killing activity for pneumococcal protein antigens. A vaccine containing PcsB and StkP is intended for the prevention of infections caused by all serotypes of pneumococcus in the elderly and in children.
Streptococcus pneumoniae (pneumococcus) is responsible for >1.5 million deaths worldwide and kills more people in the United States and Europe than any other vaccine-preventable disease (1). Besides life-threatening invasive infections (meningitis, bacteremia, and pneumonia), it also causes millions of cases of otitis media in children. The highest incidence of invasive disease is detected in children (mainly <2 yr of age) and in the elderly (mainly >65 yr of age). High mortality caused by pneumococcal infections occurs in the elderly as well as in young children living in developing countries, where every sixth child death is related to this pathogen (2).
Because of this high medical need, great efforts are being made to develop effective vaccines for the prevention of pneumococcal diseases in both industrialized and developing countries. However, none of the current vaccines can address the needs of both the elderly and children in all parts of the world. The challenges are diverse. Although capsular polysaccharides (CPSs) are proven to be effective vaccine antigens, the existence of >90 different serotypes hinders the development of full-coverage vaccines. In addition, CPSs are not immunogenic enough in young children.
The 23-valent vaccine approved for adults shows efficacy only against bacteremia and meningitis in the elderly population, but not against pneumonia, the most prevalent pneumococcal disease of this age group (3, 4). The conjugated 7-valent vaccine, Prevnar, shows excellent efficacy against bacteremia and meningitis, which is the most frequent form of invasive pneumococcal disease in this age group, and also some effect on pneumonia, otitis media, and colonization when caused by the seven included CPS serotypes (5, 6, 7). However, pneumococcal serotype distribution varies from region to region in different parts of the world. In certain developing countries, Prevnar covers only one fourth of all disease-causing strains (8, 9). Most importantly, serotype replacement induced by vaccination has already been clearly demonstrated in several clinical and surveillance studies (10, 11, 12, 13). Thus, certain serotypes rarely detected in diseased children before vaccination now have major contributions, indicating that immune escape is taking place. Newer generations of pediatric conjugate vaccines including up to 13 different CPSs are in late-stage clinical development. However, these higher valency vaccines still only partially cover serotypes in developing countries, and the high manufacturing costs make it unaffordable for those with the greatest need. In the elderly, no clinical studies have shown improved efficacy and benefits of conjugate vaccines so far.
A promising alternative approach for new-generation vaccines is the use of nonpolysaccharide antigens that are conserved among pneumococcal strains (14). Thus, attention has focused recently on the development of recombinant protein–based subunit vaccines. Traditional approaches targeted single candidate proteins based on their roles in bacterial physiology and pathogenicity, such as PspA, PspC, and pneumolysin (15, 16, 17, 18). Although these antigens show protective effects in animal models of pneumococcal disease, the high sequence variability of PspA and PspC as well as the highly toxic nature of pneumolysin limit their use as vaccine candidates. Recently, more comprehensive technologies have been applied to identify novel antigens, taking advantage of complete bacterial genome sequences (19, 20) (for review see reference 21).
In general, vaccine candidate antigens can be selected in silico by bioinformatic prediction, as demonstrated by reverse vaccinology (22, 23, 24), but it is a labor-intensive endeavor and necessitates the expression of hundreds of recombinant proteins. Moreover, 25–40% of annotated genes in individual pathogen genomes are classified as hypothetical or with unknown function that could result in excluding them as potential vaccine candidates. In contrast, proteomic approaches select fewer antigens and have been shown to be highly successful in selecting protective antigens (for review see references 21, 25). However, the selection relies heavily on the expression of proteins in vitro, which may differ significantly from that under in vivo conditions with a requirement for virulence.
In our efforts to comprehensively describe the human immune response to pneumococcal protein antigens and, at the same time, select novel vaccine candidates, we aim to obtain crucial information about the natural human immune response during exposure to or infection by pneumococcus using sera from patients recovering from invasive disease, as well as healthy noncarrier adults exposed to the pathogen. Antigens that were expressed in vivo and induced antibodies in these donors were identified by using genomic surface display libraries, as previously described (26, 27, 28). The process of preselecting vaccine candidates from the ANTIGENome by in vitro epitope-based validation assays was described previously (29). In earlier studies, novel antigens from the human pathogen Staphylococcus aureus were identified (27), and one of those, IsdB, was selected for clinical testing based on animal protection data (30) and is currently aimed for phase II studies for the prevention of nosocomial infections.
During these studies, we rediscovered the majority of the known protective pneumococcal proteins, but most importantly, we identified two novel types of pneumococcal vaccine antigens. These serve as lead candidates for the development of a vaccine that has the potential to address the needs of both the elderly and children by being exceptionally conserved among all serotypes, immunogenic in young children and the elderly, and capable of inducing bactericidal antibodies that can be used as in vitro markers of protection during future clinical studies.
RESULTS
Selection of human antibodies for antigen identification
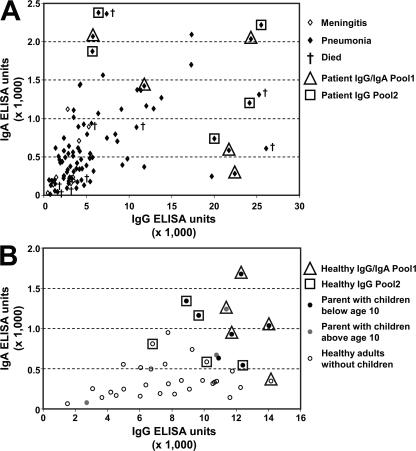
Human serum samples for antigen screening were collected from 97 patients convalescing from invasive pneumococcal diseases, with an average age of 54 yr, and from 40 healthy adults between 20 and 45 yr of age. Sera were characterized for antipneumococcal IgG and IgA levels by ELISA using total lysates and culture supernatant fractions of a capsule-negative mutant S. pneumoniae TIGR4 strain. Individual samples showed a wide range of IgG and IgA reactivity against both bacterial lysate and culture supernatant fractions. For antigen discovery, we selected sera with high antipneumococcal titer from donors who recovered successfully (Fig. 1 A). Analysis of sera from healthy donors revealed that the highest antipneumococcal antibody levels were mainly associated with adults who had young children living in the same household (Fig. 1 B). Sera from parents (n = 9) showed significantly (P < 0.05) higher ELISA reactivity than those from nonparents (n = 31), with median values of 10,236 ± 1,066 and 7,802 ± 561 U for IgG, and 980 ± 156 and 373 ± 38 U for IgA, respectively. None of these serum donors were colonized by pneumococcus in the nasopharynx based on two independent swab tests (taken 1 mo apart).
Figure 1.
Characterization and selection of human sera for antigen identification. Total anti–S. pneumoniae IgG and IgA antibody levels were measured by ELISA using total bacterial lysate prepared from the TIGR4 capsule-negative strain as a coating antigen. 97 serum samples from (A) convalescing patients with invasive diseases and (B) 40 healthy adults were analyzed; data are expressed as ELISA units calculated from absorbance at 405 nm. Sera selected for Ig purification and pooling are framed (triangle, Ig Pool1 for IgG and IgA; square, Ig Pool2 for IgG). Patients that suffered from meningitis and pneumonia are indicated by open and closed symbols; those who died during the convalescence phase are marked with a cross. Healthy adults with children <10 yr of age in the household are shown with black dots, those with children between 10–12 yr of age are in gray, and those without children are circled.
The ELISA reactivity of individual sera determined with lysate of the capsule-negative TIGR4 strain correlated well with the surface-staining intensity of encapsulated TIGR4 cells based on FACS analysis, demonstrating that the human sera had significant reactivity with pneumococcal surface antigens (unpublished data). Sera with the highest reactivity from both serum donor groups were further analyzed by Western blotting to facilitate selection based on reactivity against multiple proteinaceous antigens (unpublished data). Finally, 20 sera were chosen for antigen identification from both donor groups. Four IgG (P1IgG, P2IgG, H1IgG, and H2IgG) and two IgA pools (P1IgA and H1IgA) were generated by affinity purification of pooled sera from five different donors (Fig. 1).
Identification of the ANTIGENome of pneumococcus
The complete genome of S. pneumoniae TIGR4 was randomly fragmented into average sizes of ∼70 or 300 bp either by DNase I treatment or sonication, respectively, as reported previously (26). Two frame-selected libraries were generated by ligation of these fragments first into a frame-selection vector (26, 27), followed by their transfer into display plasmids expressing the corresponding peptides by the Escherichia coli outer membrane proteins LamB (for inserts <150 bp) and FhuA (150–600 bp), as described previously (28). Fusion of S. pneumoniae peptides to either of these two platform proteins allows recognition of the surface-exposed peptides by cognate antibodies.
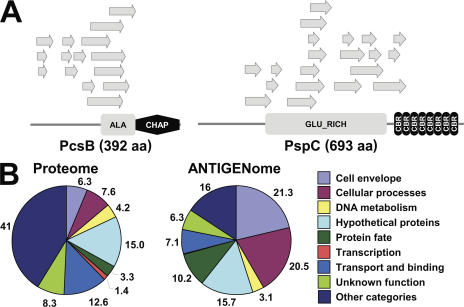
The surface display libraries were screened with six different biotinylated Ig pools by MACS, as described previously (27). 12 MACS screens were performed, 6 each with the LamB and FhuA libraries. Of these, eight screens were performed with IgGs (four with each platform) and four were performed with IgAs (two with each platform). From the 12 library screens, we collected ∼10,000 E. coli clones, as selected by human antibodies, and determined the DNA sequences of the pneumococcal genome-derived inserts. These sequences allowed localization of the epitopes within annotated genes as well as in nonannotated regions of the TIGR4 genome. The insert sequences were aligned (examples shown in Fig. 2 A), and all regions selected by at least two independent clones were verified for antibody reactivity by Western blot analysis of a representative clone using the screening Ig pools. Through these analyses we identified 97 predicted open reading frames (ORFs) and 45 peptides encoded by nonannotated genomic regions (Table S1, available at http://www.jem.org/cgi/content/full/jem.20071168/DC1).
Figure 2.
Selection of antigenic pneumococcal proteins. (A) Antigens of interest were identified by the reactivity of human serum antibodies with E. coli clones containing pneumococcal epitopes. Examples of antigenic regions are shown for two of the most frequently identified pneumococcal antigens, PcsB and PspC. Protein domains were predicted using the PROSITE database (reference 61). ALA- and GLU-rich domains are rich in the amino acids alanine and glutamate, respectively. The CHAP domain is proposed to function in peptidoglycan hydrolysis; cysteine and histidine form part of the putative active site of the CHAP domain. Choline-binding repeat (CBR) is found in Gram-positive bacteria. (B) The S. pneumoniae TIGR4 proteome consists of all annotated proteins assigned to role categories. The ANTIGENome encompasses all antigenic proteins identified by bacterial surface display screens and confirmed to be immunogenic. Numbers refer to the percentage contribution of each category.
By comparing the ANTIGENome with the complete proteome of S. pneumoniae TIGR4, we found an enrichment of antigens in the major cellular role categories: cell surface, cellular processes (virulence and pathogenicity), and protein fate (e.g., proteases; Fig. 2 B). These three categories contributed to >50% of all identified annotated antigens. Importantly, one fifth of the immunogenic pneumococcal proteins did not have predictable signatures and were annotated as hypothetical or unknown function genes.
Importantly, we rediscovered the majority of known protective pneumococcal antigens—such as PspA (15), PspC (16), serine protease (20), IgA1 protease (31), and histidine triad proteins SP1003, SP1004, SP1174, SP1175 (20, 32), NanA (33), LytC (20), and LytA (34)—with high frequency (Table I). SP2216, a novel antigen that was annotated as a secreted 45-kD protein (TIGR4 strain) or protein required for cell wall separation of group B streptococcus (PcsB; R6 strain), was identified as the most immunogenic protein, with 25% of all selected clones representing epitopes within this ORF. PcsB and two other proteins, PspA and PspC, which have been described previously as highly immunogenic antigens (15, 16), were selected in all 12 screens performed. Most of the highly immunogenic antigens (selected by at least 6 out of 12 screens) contained multiple epitopes in different regions of the proteins (Fig. 2 A). Moreover, these were identified by both IgG and IgA antibodies from both patients and healthy exposed noncolonized individuals, a strong indication for in vivo expression under disease and exposure at mucosal surfaces (Table I). Several antigens—such as LytC, SP0667, SP1527, SP1891, and SP2027—were identified mainly by serum pools obtained from parents of young children, whereas others—such as PcpA, CbpC, CbpI, and SP0107—were selected mainly by serum pools from patients in convalescent phase.
Table I.
Frequency of clone selection in genomic library screens
| ORF | Common name | Screens | Hits | FhuA | LamB | IgG | IgA | P | H |
|---|---|---|---|---|---|---|---|---|---|
| SP2216 | PcsB, secreted 45-kD protein (Usp45) | 12 | 1766 | 1606 | 160 | 1374 | 392 | 662 | 1104 |
| SP2190 | PspC/CbpA | 12 | 642 | 71 | 571 | 343 | 299 | 402 | 240 |
| SP0117 | PspA | 12 | 272 | 75 | 197 | 158 | 114 | 183 | 89 |
| SP0641 | Serine protease, subtilase family | 11 | 525 | 237 | 288 | 475 | 50 | 271 | 254 |
| SP0664 | Zinc metalloprotease ZmpB, putative | 11 | 253 | 145 | 108 | 158 | 95 | 184 | 69 |
| SP0071 | IgA1 protease (Iga1) | 10 | 164 | 26 | 138 | 70 | 94 | 105 | 59 |
| SP1154 | IgA1 protease (Iga2) | 10 | 61 | 26 | 35 | 46 | 15 | 21 | 40 |
| SP1174 | Conserved domain protein | 9 | 86 | 71 | 15 | 75 | 11 | 54 | 32 |
| SP1527 | Oligopeptide ABC transporter | 9 | 64 | 7 | 57 | 37 | 27 | 4 | 60 |
| SP1693 | NanA | 9 | 47 | 12 | 35 | 15 | 32 | 33 | 14 |
| SP2201 | CbpD | 9 | 46 | 16 | 30 | 18 | 28 | 36 | 10 |
| SP1004 | Conserved hypothetical protein | 9 | 43 | 38 | 5 | 29 | 14 | 26 | 17 |
| SP1772 | Cell wall surface anchor family protein | 9 | 32 | 24 | 8 | 30 | 2 | 17 | 15 |
| SP2136 | PcpA | 8 | 302 | 4 | 298 | 101 | 201 | 230 | 72 |
| SP2099 | Pbp1B | 8 | 34 | 29 | 5 | 31 | 3 | 9 | 25 |
| SP1175 | Conserved domain protein | 8 | 28 | 22 | 6 | 24 | 4 | 18 | 10 |
| SP0107 | LysM domain protein | 7 | 219 | 218 | 1 | 215 | 4 | 185 | 34 |
| SP1573 | Lysozyme (LytC) | 7 | 111 | 99 | 12 | 57 | 54 | 9 | 102 |
| SP1003 | Conserved hypothetical protein | 7 | 38 | 36 | 2 | 31 | 7 | 23 | 15 |
| SP0648 | β-galactosidase (BgaA) | 7 | 11 | 3 | 8 | 10 | 1 | 4 | 7 |
| SP0667 | Pneumococcal surface protein, putative | 6 | 327 | 327 | 0 | 235 | 92 | 85 | 242 |
| SP0082 | Cell wall surface anchor family protein | 6 | 112 | 15 | 97 | 112 | 0 | 77 | 35 |
| SP0377 | CbpC | 6 | 109 | 0 | 109 | 16 | 93 | 99 | 10 |
| SP2027 | Conserved hypothetical protein | 6 | 54 | 2 | 52 | 34 | 20 | 6 | 48 |
| SP0069 | Choline-binding protein I (CbpI) | 6 | 52 | 0 | 52 | 7 | 45 | 48 | 4 |
| SP1891 | Oligopeptide ABC transporter | 6 | 28 | 7 | 21 | 13 | 15 | 2 | 26 |
| SP1937 | Autolysin (LytA) | 6 | 19 | 4 | 15 | 4 | 15 | 16 | 3 |
| SP0057 | β-N-acetylhexosaminidase (StrH) | 6 | 7 | 4 | 3 | 5 | 2 | 3 | 4 |
Screens represent the number of screens in which a given antigen was selected. Hits describe the total number of E. coli clones selected for one ORF; FhuA and LamB describe the total number of E. coli clones selected from the FhuA or LamB library, respectively; IgG and IgA describe the total number of E. coli clones selected with IgGs or IgAs; and P and H describe the total number of E. coli clones selected with antibodies derived from patients or healthy adult donors, respectively.
Prioritization among pneumococcal antigens by in vitro validation
The large number of antigens identified by the ANTIGENome technology prompted us to preselect for animal protection studies. Several in vitro assays that did not necessitate the generation of recombinant proteins but focused on important qualities of vaccine candidates were applied. The following criteria were addressed in these assays: (a) immunogenicity in humans, (b) surface exposure and/or secretion, (c) capability of inducing bactericidal antibodies, and (d) sequence conservation. In addition, further selection was made based on intellectual property claims.
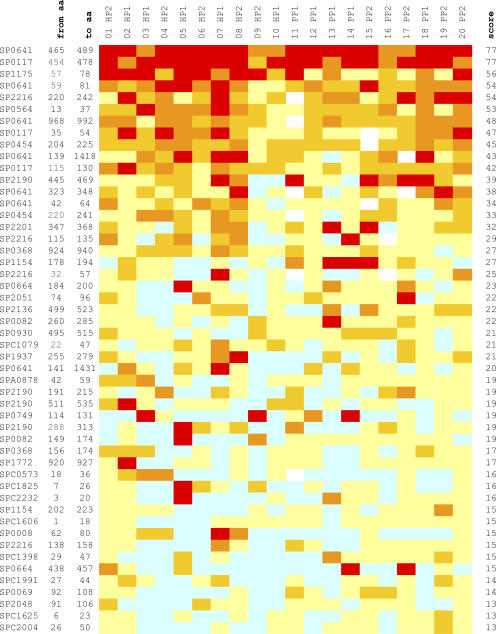
We performed an extensive serological analysis of those identified epitopes that were devoid of significant homologies to human sequences. For this purpose, the immune reactivity of 222 synthetic peptides, ranging in size from 9 to 26 aa and representing 136 S. pneumoniae epitopes derived from 64 annotated and 40 nonannotated ORFs, was determined by ELISA with the 20 individual human sera comprising the four Ig screening pools. The obtained results correlated well with the library screen data, as the most immunogenic peptides belonged to the most frequently selected antigens (e.g., PspA, SP0641, SP1175, PspC, and PcsB; Fig. 3). In addition, highly immunogenic epitopes from less frequently selected proteins were also detected (e.g., SP0564 and SP0454). In rare cases, frequently selected long epitopes did not detect comparably high antibody levels with overlapping synthetic peptides, most probably because of the presence of conformational, discontinuous epitopes. A more detailed peptide ELISA study with the 50 most immunogenic peptides showed, for example, an age-dependent increase in antibody levels among children and less reactivity in patients with unfavorable disease outcome (unpublished data).
Figure 3.
Peptide ELISA with human sera. Immune reactivities with individual human sera are shown for individual peptides representing the selected antigenic regions. Serum samples correspond to the donors indicated in Fig. 1. Peptides are named by their corresponding ORF based on the S. pneumoniae TIGR4 genome. Localization of epitopes is indicted by the position of the first and last amino acid residues of the peptides (“from aa” and “to aa”). ORF names are as follows: SP, annotated; SPA, potential ORF in alternative reading-frame; and SPC, potential ORF on complementary strand. Scores for reactivity based on ELISA Units are as follows: <50 U = 0 (blue); 50–99 U = 1 (light yellow); 100–199 U = 2 (mustard yellow); 200–499 U = 3 (orange); ≥500 U = 4 (red); and not determined (white cells). The total reactivity score for each peptide is calculated with the 20 serum samples analyzed. HP, healthy adult pool; PP, patient pool.
Further characterization of antigens was facilitated by the generation of epitope-specific hyperimmune sera in mice using total bacterial lysates prepared from E. coli clones displaying immunogenic pneumococcal epitopes. 90 hyperimmune sera were generated by immunization with single or pooled lysates of altogether 120 E. coli clones derived from 90 different antigens. Although it was possible to induce epitope-specific antibodies with 75% of the injected E. coli clones based on peptide ELISA and Western blot analysis, not all positive mouse sera detected the corresponding protein by Western blot analysis of S. pneumoniae total lysate, suggesting that some proteins were not expressed under in vitro growth conditions (unpublished data). Surface staining of encapsulated S. pneumoniae TIGR4 cells by FACS analysis was observed with 43 sera (Table S2, available at http://www.jem.org/cgi/content/full/jem.20071168/DC1). The 43 FACS-positive sera were further analyzed in an in vitro opsonophagocytic killing (OPK) assay using S. pneumoniae serotype 4 and 6B strains. Antibody- and complement-dependent killing was demonstrated with 31 sera (Table S2). Importantly, well-known protective pneumococcal antigens, such as PspA and PspC, were strongly positive, confirming the validity of this analysis. Both surface staining and OPK activity were considered as positive criteria for an antigen to be included among the list of candidates to be tested in animal models.
To select conserved antigens, we performed gene distribution studies by PCR using gene-specific primers and genomic DNA isolated from 50 different clinical strains collected from patients with bacteremia and representing the 22 most important serotypes of S. pneumoniae (1, 2, 3, 4, 5, 6A, 6B, 7F, 8, 9N, 9V, 12F, 14, 17F, 18C, 19A, 19F, 22F, 23F, 33F, 35A, and 35F). Based on this analysis, 10% of analyzed genes were absent in at least 15% of relevant clinical strains (e.g., SP0069, SP1154, and SP1330) and were not selected for in vivo studies.
Based on the in vitro validation results, 10 pneumococcal antigens fulfilling at least four out of the five validation criteria were identified as the most promising vaccine candidates: SP0082, PspA, SP0368, SP0454, SP0609, SP0749, SP1891, SP2108, PspC, and PcsB. An additional 10 novel conserved candidates were selected for animal testing in spite of the lack of positive FACS data, as this could be attributed to insufficient epitope-specific antibodies in hyperimmune mouse sera or a lack of in vitro expression. These were SP0107, SP0564, SP0667, SP1374, SP1522, SP1527, serine/threonine protein kinase (StkP)/SP1732, SP2048, SP2051, and SP2092. With the exception of PspA and PspC, the known protective pneumococcal antigens were excluded from this preselection.
Identification and characterization of protective vaccine candidates
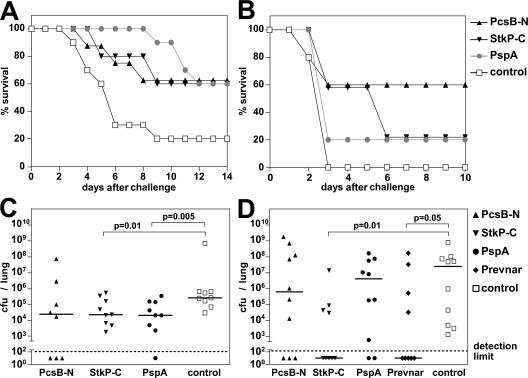
The 20 genes encoding the selected antigens (full-length ORFs) were cloned from the S. pneumoniae TIGR4 strain and expressed as His-tagged proteins. For protection studies, mice were immunized with recombinant proteins and challenged i.v. or i.p. with a 200× lethal dose of S. pneumoniae serotype 6B. Among the 20 antigens, 6 showed significant protection in these experiments: the 2 known protective antigens (PspA and PspC) and 4 novel ones (SP0368, SP0667, StkP, and PcsB), whereas the other antigens, such as SP0082, were clearly negative in these experiments (Fig. 4 A; and Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20071168/DC1). Importantly, we were able to show that protection was mainly afforded by antibodies, because passive immunization of naive animals with hyperimmune sera was effective (Fig. S1 B). Using the i.p. challenge model with this S. pneumoniae serotype 6B strain (against which Prevnar provides complete protection), we did not observe significant protection with the remaining antigens; however, these are currently being tested in further animal models of pneumococcal disease (pneumonia and otitis media).
Figure 4.
Protection by PcsB and StkP in models of pneumococcal sepsis and pneumonia. Mice were immunized with recombinant PcsBN and StkPC (with 1% ALUM) and challenged with S. pneumoniae as follows: (A) 105 CFU of the PJ-1259 strain i.p.; (B) 5 × 106 CFU of the 6301 strain i.n.; (C) 105 CFU of the WU2 strain i.n.; or (D) 6 × 107 CFU of the EF3030 strain i.n. (A and B) Survival of animals was monitored for 10–14 d after challenge. Numbers of surviving mice are plotted as a percentage of the total. In A, P = 0.04 for StkP and PcsB, and P = 0.006 for PspA comparing time to death relative to the control group; in B, P > 0.05. (C and D) Lung colonization of individual mice is shown at day 3 after challenge, indicating the median CFU per lung (horizontal lines). Statistically significant differences based on the Mann-Whitney two-sample rank test are indicated. For all experiments, adjuvant-immunized mice served as negative controls.
Characterization of the lead vaccine candidate antigens, PcsB and StkP
The four novel protective antigens were further characterized in a detailed gene conservation analysis. The SP0368 and SP0667 genes were missing from 8 and 10% of the 50 clinical strains tested, and the respective proteins showed variability in size (unpublished data). Thus, further animal testing focused on the two highly conserved candidates, StkP and PcsB. Because the N-terminal region of StkP contains the eukaryotic-type serine threonine kinase domain with ∼35% identity to the corresponding human protein, we generated deletion mutants of StkP and used the C-terminal 314 aa residues for further protection studies (StkP-C). The C-terminal hydrophobic cysteine, histidine-dependent amidohydrolase/peptidase CHAP domain was removed from PcsB, which made expression and purification of the protein easier (PcsB-N). We observed that the two subdomains afforded the same level of protection as the respective full-length antigens (unpublished data). Importantly, all epitopes identified in library screens for these antigens localized to the same regions, and the deleted parts were immunologically inert with human sera (even in recombinant forms). Interestingly, the recombinant StkP-C was much more immunogenic with individual sera from patients and parents than could be concluded from the low number of hits in the primary library screen with pooled human serum antibodies (unpublished data).
In an intranasal (i.n.) sepsis model, PcsB protected mice against death caused by an S. pneumoniae serotype 1 strain, whereas StkP immunization showed variable benefits (at least delaying the time of death; Fig. 4 B). No protection was observed with PspA in this model, possibly as a consequence of low amino acid identity (∼35% not considering the C-terminal region with the invariant choline binding domain) between the PspA variants used for immunization (TIGR4, family 2/clade 3) and expressed by the challenge strain 6301 (family 1/clade 1; PspA families and clades are according to Hollingshead et al. [35]). The good protection seen with the PJ-1259 6B strain is well supported by the 95% identity to the TIGR4- and 6B-derived PspA proteins (both family 2/clade 3).
To asses the effect of immunization on lung colonization, two pneumonia models were set up with two different serotypes. In the WU2 challenge model (serotype 3), both StkP and PcsB were capable of reducing the bacterial load in the lung 10 times relative to mock-immunized mice, similar to PspA (Fig. 4 C). The two antigens were also effective in reducing lung infection by the EF3030 (serotype 19) strain. Immunization with PcsB typically resulted in 10 times reduced median bacterial load, whereas StkP was reproducible as effective as Prevnar (containing serotype 19 CPS) and eliminated bacteria from the lung (below the detection limit) in 50–60% of animals (both significantly different from the adjuvant control group; P < 0.05; Fig. 4 D). The partial protection seen for PspA might be again explained by its serovariability in the two challenge strains relative to the one used for immunization (PspA family 1/clade 2 in WU2 and family 1/clade 1 in EF3030) (36, 37).
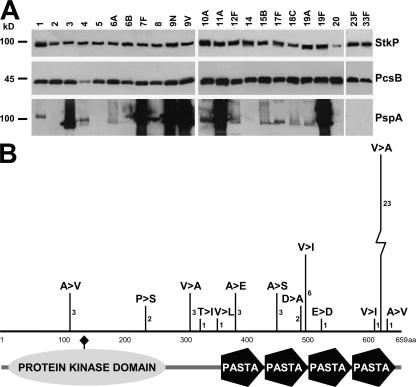
Detailed gene distribution analysis demonstrated that both genes were present in all of ∼100 clinical isolates that we collected from patients with different invasive diseases or from nasopharyngeal carriers as analyzed by PCR (unpublished data). Similarly, the corresponding proteins were detected in all of the 60 tested clinical isolates representing 48 different serotypes based on Western blot analysis using hyperimmune mouse sera generated with recombinant proteins (Fig. 5 A). Importantly, the electrophoretic mobility, the strength of immune reactivity, and the size of the protein products were uniform for both StkP and PcsB, unlike for PspA. DNA sequence analyses of both genes from up to 60 strains with 48 distinct pneumococcal serotypes revealed exceptionally high conservation. We detected only a single amino acid substitution for PcsB in only 1 out of 48 strains with different serotypes (A243S). StkP was sequenced from 60 strains encompassing the same 48 serotypes and was found to be highly conserved in 58 strains, with only up to three amino acid substitutions within one gene. Most of the amino acid variations occurred as substitutions with similar residues (Fig. 5 B). The two strains carrying a variant stkP gene were lacking the third repeat of the penicillin-binding protein and serine/threonine kinase–associated (PASTA) domain that had high sequence similarity to the second one.
Figure 5.
PcsB and StkP are highly conserved antigens. (A) Immunoblot analysis of pneumococcal isolates representing the serotypes contained in the 23-valent CPS vaccine. Bacterial lysates and culture supernatants were analyzed with hyperimmune mouse sera induced by recombinant PspA, StkP, and PcsB. (B) The amino acid sequence of StkP was determined in 58 S. pneumoniae strains representing 48 different serotypes. The number and type of amino acid exchanges compared with a serotype 6B isolate are indicated. Protein domains were predicted using the PROSITE database (reference 61). The active site residue of the predicted kinase domain of StkP is indicated by a diamond.
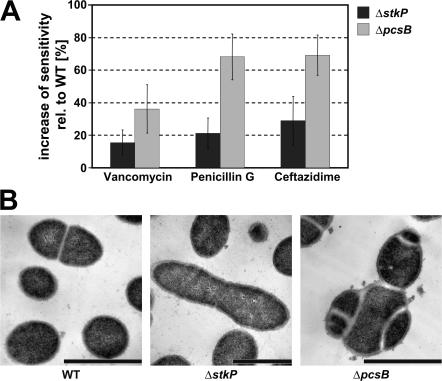
The importance of PcsB and StkP in pneumococcal disease was demonstrated with gene deletion mutant pneumococci. The lack of either gene resulted in a strong reduction of bacterial growth (Fig. S2 A, available at http://www.jem.org/cgi/content/full/jem.20071168/DC1) and increased antibiotic sensitivity to penicillin, cephalosporins (e.g., ceftazidime), and vancomycin in vitro (Fig. 6 A). We observed greatly reduced or no disease-causing potential for ΔstkP and ΔpcsB S. pneumoniae strains, respectively, in a lethal challenge model when compared with the parental 6B WT strain (Fig. S2 B). In this model, deletion of pspA did not have an effect on survival.
Figure 6.
Characterization of ΔpcsB and ΔstkP gene deletion mutant strains. (A) Antibiotic sensitivity was determined by the disc diffusion method in three independent experiments. The average increase in growth inhibition zone was expressed as a percentage relative to the WT; error bars represent standard deviations. (B) Transmission electron microscopic analyses of gene deletion strains ΔstkP and ΔpcsB KO and TIGR4 WT are shown. Bars, 1 μm.
The obvious effect on bacterial growth prompted us to study the morphology of the gene deletion mutant cells. Electron microscopy revealed a dramatic change in ΔpcsB cells demonstrating a defect in cell separation and/or septum formation. The normal diplococcus morphology disappeared; the daughter cells could not separate, resulting in cell conglomerates with division septa formed in different planes. The ΔstkP cells demonstrated an elongated shape and very few division septa separating the daughter cells (Fig. 6 B).
PcsB and StkP are immunogenic during pneumococcal infections and induce opsonophagocytic/bactericidal antibodies
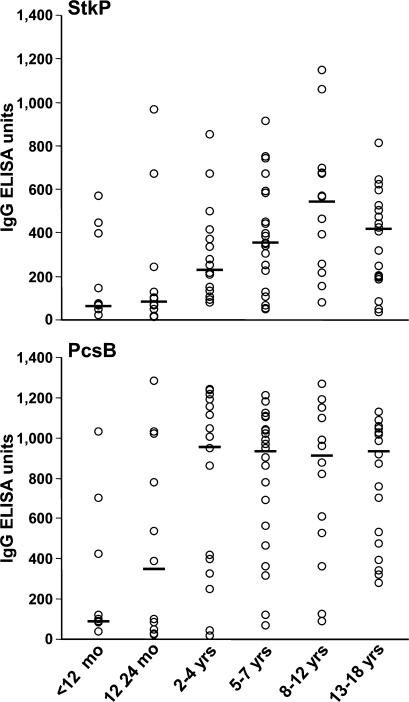
Because these two antigens were identified using antibodies from adult donors, it was interesting for us to detect natural antibody responses against them in healthy children and follow the development of specific antibody levels as a function of age (repeated exposure). Using 88 sera obtained from healthy children aged 2 mo to 18 yr, we detected a wide range of IgG titers from almost undetectable to high levels (comparable with the highest values obtained with high titer sera from patients and parents). In sera from infants (<12 mo old) and very young children (12–24 mo old), the median ELISA values for PcsB were significantly lower (107 and 388 ELISA U, respectively) than in older age groups (∼900 U; Fig. 7, bottom). The StkP-specific antibody levels showed a more continuous increase, with peak median value in the 8–12-yr-old group (Fig. 7, top).
Figure 7.
StkP and PcsB antigens are immunogenic in young children. 88 sera obtained from children not suffering from any infectious disease at the time of sampling were analyzed for antipneumococcal IgG levels by ELISA using recombinant proteins as coating antigens. Individual data points are indicated in six age subgroups (n = 8–21) for StkP (top) and PcsB (bottom). Data are expressed as ELISA units (OD, 405 nm) at 1:1,000 serum dilutions; horizontal lines represent medians.
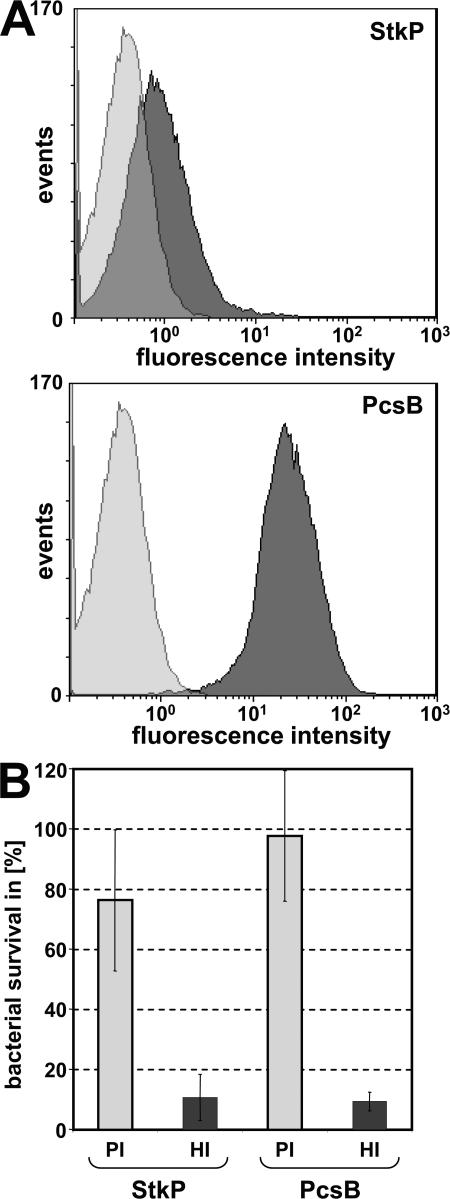
It has been well documented that specific antipneumococcal antibody levels induced by vaccination of children with polysaccharide-based vaccines correlate well with opsonophagocytic/bactericidal antibodies, whereas in elderly people, antibody levels overestimate the protection. Therefore, it was of paramount importance to develop a reliable in vitro functional assay to measure the bactericidal activity of antibodies induced by the novel vaccine candidate antigens. The in vitro OPK assay used for the prioritization of antigens (Table S2) was further optimized to make it suitable for future clinical studies as a surrogate marker. We have developed a reproducible and reliable assay with the human HL60 cell line, baby rabbit complement, and rabbit and human sera. We were able to detect surface staining with rabbit hyperimmune sera specific for PcsB and StkP by FACS analysis (Fig. 8 A) and OPK activity (Fig. 8 B). Importantly, OPK activity could also be shown for affinity-purified antigen-specific human antibodies induced during exposure to pneumococcus (unpublished data).
Figure 8.
Antigen-specific immune sera show surface staining and OPK activity. (A) Surface staining of S. pneumoniae serotype 4 DS2341-94 cells was performed with hyperimmune rabbit sera specific for StkP and PcsB and PE-conjugated secondary antibodies. The signal resulting from staining with preimmune sera is shown in light gray; StkP and PcsB specific signals are shown in dark gray. (B) Bactericidal activity of protein-specific antibodies as determined in an in vitro OPK assay. S. pneumoniae serotype 4 DS2341-94 cells were preincubated with rabbit hyperimmune (HI) or with their respective preimmune (PI) sera at a dilution of 1:1,000 in the presence of baby rabbit complement. HL60 phagocytic and pneumococcal cells were used at a 400:1 ratio. Data are expressed as the percentage of bacterial survival relative to the initial CFU at time 0. The average of three experiments is shown; error bars represent standard deviations.
DISCUSSION
To our knowledge, this is the most comprehensive study describing the antibody repertoire induced in the human host during pneumococcal disease and exposure. There are many reports on antipolysaccharide antibodies in vaccinated or diseased individuals, or in healthy children, but very few on species common antigens. The immunogenicity studies investigating protein-specific antibodies have focused on virulence factors and/or vaccine candidates (38, 39, 40, 41, 42). For the rational design of a novel protein-based vaccine, we first studied the human immune response against pneumococcal proteins on a global scale using genomic surface display libraries that served as tools for the expression of all potential epitopes. The serum antibodies from patients successfully recovering from invasive pneumococcal diseases or from parents of young children allowed us to identify those bacterial proteins that were expressed in the host during disease or mucosal exposure, respectively. To avoid bias from abundant anti-CPS antibodies, a capsule-negative mutant strain was used for immune characterization of our serum collection. We found a good correlation between antipneumococcal nonpolysaccharide-specific antibody levels and disease severity among patients with invasive disease. In the healthy adult group, high antibody levels were indicative of exposure to pneumococcus by small children living in the same household. Because none of these high titer parents were carriers, the antibodies could be considered potentially protective against nasopharyngeal colonization, the first step toward disease.
By fingerprinting of the pneumococcal proteome with preselected pooled IgG and IgA antibodies, several hundred B cell epitopes were identified that belonged to ∼140 ORFs and genomic regions not annotated as coding sequences. We rediscovered the majority of known protective proteins, such as PspA, PspC, and histidine triad proteins (15, 16, 20, 34). Half of the annotated proteins of the pneumococcal ANTIGENome belong to predicted or known surface proteins, virulence factors, extracellular matrix binding proteins, and proteases. However, every fifth of them was encoded by hypothetical or unknown function genes that could have been missed by approaches merely driven by prediction algorithms. Further analysis is needed to asses the value of nonannotated antigens in correcting gene assignment and discovery of novel short reading frames. Importantly, as this antigen selection is not biased by the often artificial abundance or lack of protein expression in bacteria grown under in vitro conditions (25), we also detected immunogenic epitopes that were derived from proteins not expressed by in vitro grown pneumococci.
At the same time, some of the current vaccine candidates, such as PsaA and pneumolysin, were not among the ANTIGENome. In the case of PsaA, this could be explained by the low antibody titers against this protein present in sera selected for antigen identification (unpublished data). It is also possible that certain protective antigens are not immunogenic in the human host during infection or contain mainly structural epitopes that cannot be expressed in the genomic surface expression libraries.
We found a large overlap between proteins identified by the two different serum donor groups, suggesting that in general the same antigens were expressed during invasive disease and mucosal exposure. Additional detailed serological analysis of the epitopes with immune reagents from different cohorts of donors may identify “missing” antibodies in susceptible individuals (e.g., very young children or acutely diseased elderly people) and further characterize host immune responses to pneumococcus under different disease conditions and exposure.
A series of in vitro epitope-based validation assays were applied to preselect vaccine candidates for animal testing. 6 out of 20 pneumococcal antigens (cloned from TIGR4) showed significant cross-protection in a stringent mouse model of lethal sepsis (with a human serotype 6B isolate). These were the two best-characterized pneumococcal vaccine candidates, PspA and PspC, and four novel antigens, SP0368, SP0667, StkP, and PcsB. The functions of SP0368, a choline-binding protein, and SP0667, pneumococcal surface protein, have not been revealed yet. PcsB in S. pneumoniae and its homologues from Streptococcus agalactiae and mutans were previously shown to be involved in important bacterial survival mechanisms, maintenance of cell morphology, and growth (43, 44, 45, 46). We substantiated these findings by successfully creating gene deletion strains for the first time and, thus, demonstrating that pcsB is not an essential gene for pneumococcus. We observed greatly reduced in vitro growth and complete loss of virulence that might be caused by a lack of in vivo growth. Electron microscopic analysis confirmed previous light microscopy findings with strains expressing low levels of PcsB (43, 44) and demonstrated drastic changes in cell morphology and aberrant division septum formation. The predicted CHAP domain at the C terminus of PcsB is indicative for its involvement in hydrolysis of peptidoglycan, whereas no such activity could be shown for any of the homologues nor by us with recombinant or native PcsB (unpublished data). We delineated the protective epitopes to the N-terminal immunogenic region. Interestingly, glucan-binding protein B, the homologue from S. mutans, was shown to protect from caries (47, 48).
StkP possesses amino acid sequence homology to serine/threonine kinases with a suggested important role in cell–cell signaling and competence triggering, as well as resistance to various stress conditions, by acting as a transcriptional regulator (49, 50, 51 52). Several activities are associated with this protein, such as autophosphorylation and phosphorylation of phosphoglucosamine mutase, which is important for cell wall synthesis (50). In accordance with our findings, gene deletion D39 and 23477 mutant strains were found to be less virulent in mice. However, no in vitro growth defect was observed, in contrast to our data generated with two different strains (TIGR4 and PJ-1259 6B). Our electron microscopic studies revealed an altered cell shape and a defect in forming division septa. StkP contains a characteristic structural signature: four copies of the PASTA domain that is also present in penicillin-binding proteins (53). Although StkP does not contain predictable signal peptide sequences, it is expressed on the bacterial surface (Fig. 8 A) (49, 53). We concluded that the C-terminal PASTA domains are the immunogenic region of the protein, which induces protective immune responses in animals.
PspA, PspC, and PcsB were the most frequently selected antigens in the genomic library screens. Highly immunogenic proteins are suspected to be fake targets of the immune response, with protective ones being prone to genetic variability as exploited by pathogens to escape host defense. Indeed, PspA and PspC are highly variable, which is thought to be a result of immune pressure (35, 54). In this context, it is a very significant finding that the most immunogenic pneumococcal protein in our screens, PcsB, is exceptionally conserved among the different serotypes, with almost 100% amino acid identity. A similar high level of sequence conservation was found for StkP. The extent of sequence diversity of PspA and PspC (35, 54), as well as of SP0368 and SP0667 (unpublished data), would necessitate several clade variants to be included in a broad coverage vaccine. Because it is likely that a combination of several different antigens is necessary to develop an effective antipneumococcal protein-based vaccine (55), the selection of highly conserved, species-common proteins seems to be crucial.
Genetic variability of antigens under immune pressure by the host is not limited to epitope diversity but can also lead to the complete loss of an antigen or phase variants. Targeting bacterial proteins that fulfill important nonredundant roles in in vivo survival and growth, instead of virulence factors with often redundant functions, minimizes the possibility of negative selection and strain replacement induced by vaccination. Our studies suggested important functions for PcsB and StkP, but not for PspA, in bacterial growth, antibiotic sensitivity, and in vivo survival that could also explain the high degree of gene conservation observed.
A major requirement for viable vaccine candidates for the prevention of pneumococcal disease is the demonstration of protective effects against several of the major disease causing serotypes. The limitation of the different animal models that try to mimic human disease can be compensated for by testing multiple in vivo models. Moreover, results generated in widely used models allow comparison of different candidates considered for vaccine development. In addition to the 6B serotype–induced i.p. sepsis model, the lead vaccine antigens PcsB and StkP were also tested by i.n. challenge of mice with the sepsis-causing serotype 1 and pneumonia-inducing serotype 3 (WU2) or 19F (EF3030) strains (37). Both proteins were effective in all models with equal or superior protection relative to PspA (TIGR4 derived). The most striking observation was the highly significant protection by StkP immunization in the EF3030-induced pneumonia model that was comparable with the effect of the conjugated Prevnar vaccine.
A further very important aspect of developing novel protein-based antibacterial vaccines is to apply in vitro functional antibody assays and define correlates of protection that can be used as surrogate markers in clinical development. Based on the characteristics of PcsB and StkP (surface localization, important in vivo function, and a high level of conservation), both opsonophagocytic and neutralizing antibodies are expected to contribute to cross-protection, mainly because of antibodies based on results of serum transfer experiments. Our detection of OPK activity induced by PcsB and StkP immunization represents the first in vitro functional antibody assay for pneumococcal protein antigens. It remains to be established in additional preclinical and clinical studies whether OPK antibodies induced by protein antigens will suffice as surrogate markers of protection. However, it is firmly established that vaccine efficacy against pneumococcus can be correlated with serum levels of opsonizing antibodies induced by polysaccharide antigens (56).
Based on the results presented in this study, a subunit vaccine using a combination of antigens is being developed. Because the lead vaccine candidates, StkP and PcsB, are immunogenic in both the elderly and very young children and expressed during invasive disease, as well as during colonization and exposure, it is possible that the same vaccine can address the needs of both target populations. The serotype-independent expression of the antigens suggests that the broad spectrum of disease-causing serotypes detected in the elderly, the distinct distribution of pediatric strains in developing countries, and the emergence of nonvaccine strains in developed countries can be also addressed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Clinical isolates were collected by the Swedish Institute for Infectious Disease Control. The WU2 (serotype 3) and EF3030 (serotype 19) strains were provided by D. Briles (University of Alabama at Birmingham, Birmingham, AL), the 6301 (serotype 1) strain was provided by I. Jonsdottir (Landspitali University Hospital, Reykjavik, Iceland), the DS2341-94 strain was provided by E. Ades (Centers for Disease Control and Prevention, Atlanta, GA), and the serotype 4 unencapsulated strain was provided by E. Tuomanen (St. Jude Children's Research Hospital, Memphis, TN). Bacteria were grown in Todd-Hewitt broth (BBL; Becton Dickinson) plus 0.5% yeast extract (THY) or in brain-heart infusion broth for S. pneumoniae 6301 at 37°C in an atmosphere of 5% CO2.
Human serum samples and their characterization.
Sera from 96 patients were obtained at the Karolinska University Hospital in a study approved by the Ethical Committees of the Karolinska University Hospital and the Karolinska Institute. Sera from healthy adults were collected during routine occupational health checks (leftover samples were used with informed consent) or were from existing serum collections (Austrian Red Cross, Vienna, Austria). 88 sera were collected from children while they were hospitalized for noninfectious disorders (samples were saved after completed routine blood tests and handled according to institutional regulations at the Bethesda Childrens's Hospital, Budapest, Hungary).
Antibodies against E. coli proteins were removed from the pooled sera as described previously (26). For screening, Igs were purified by affinity chromatography and labeled with biotin (27). Peptide serology was performed with biotin-labeled synthetic peptides coated on streptavidin-covered ELISA plates. Serum antibodies were detected by horseradish peroxidase (HRPO)–conjugated anti–human IgG secondary antibodies (SouthernBiotech). Antigen-specific antibodies in sera of children were analyzed by recombinant protein–coated ELISA plates and HRPO-conjugated anti–human secondary antibodies.
Gene distribution and sequence analysis of pneumococcal antigens.
Oligonucleotides were designed for selected ORFs to amplify PCR products of approx. 1,000 bp. For gene distribution, PCR with genomic DNA of S. pneumoniae isolates was performed using Taq polymerase (Invitrogen). The full-length stkP and pcsB genes were amplified with primers matching the sequences in the S. pneumoniae TIGR4 genome flanking the corresponding ORFs, using the Expand High Fidelity PCR kit (Roche).
Surface staining of bacteria.
Bacteria from the early logarithmic growth phase were collected and washed twice in HBSS. Approximately 105 CFU in 100 μl HBSS, 0.5% BSA were incubated with mouse or rabbit sera at 0.5 and 2% final concentrations for 60 min at 4°C before detection with FITC-labeled goat anti–mouse IgGs (F(ab′)2 fragment specific; Dako) or PE-conjugated goat anti–rabbit IgG (H+L; Beckman Coulter) antibodies. After fixation with 2% paraformaldehyde, surface staining was detected by a flow cytometer (Cytomics FC500; Beckman Coulter), and data were analyzed using analysis software (CXP; Beckman Coulter).
OPK assays.
For the analysis of epitope-specific mouse hyperimmune sera (27), a mouse macrophage cell line (RAW264.7) and S. pneumoniae strains TIGR4 or PJ-1259 (human isolate from a nasopharyngeal carrier, serotype 6B; ratio of phagocytes to bacteria = 20:1) were used in the presence of guinea pig complement. For the analysis of rabbit and human sera, DS2341-94 and the human HL60 cell line (differentiated for 5–6 d with 100 mM dimethylformamide [57]) were used at a ratio of 400:1 in the presence of baby rabbit complement. Bacteria were washed in HBSS and ∼105 CFU/40 μl (for RAW264.7) and 5 × 103 CFU/80 μl (for HL60) incubated with 50 μl of mouse serum and 10 μl of guinea pig complement or with 2 μl of 1:10 diluted rabbit/human serum and 10 μl of baby rabbit complement in a total volume of 100 μl for 60 min at 4–6°C. Serum-opsonized bacteria were mixed with 100 μl of phagocytic cells and incubated for 60 min at 37°C. An aliquot of each sample was diluted in sterile water and incubated for 5 min at room temperature, and serial dilutions were plated onto blood agar plates (BioMerieux). After overnight incubation at 37°C, plates were analyzed with a colony counter (Countermat Flash [IUL Instruments]; Easy Count 2 [AES Laboratories]), and data were expressed as the percentage of bacterial survival relative to the CFU at time 0.
Cloning and expression of recombinant pneumococcal proteins.
Genes of interest were cloned into pET28b(+) vector (EMD). Subfragments of PcsB and StkP were generated by PCR with internal primers, amplifying the N-terminal two thirds of PcsB (PcsB-N, aa 28–278) and the C-terminal half of StkP (StkP-C, aa 345–659). Proteins were expressed in E. coli BL21 Star cells (Invitrogen), and His-tagged proteins were purified by binding to Ni-sepharose beads (Ni Sepharose 6 Fast Flow; GE Healthcare). Proteins from the insoluble fraction were first solubilized in 8 M urea in 50 mM Tris-HCl, pH 8.
Immunization and challenge of animals.
All animal experiments were performed according to Austrian Law (BGB1 Nr. 501/1989, approved by Magistratsabteilung 58, Landeskultur und Wasserrecht). Female C3H/HeNHsd mice (Harlan Winkelmann) at 6–10 wk of age were immunized three times at 14-d intervals s.c. (flank) with 50 μg of protein or PBS mixed with CFA/IFA or 1% ALUM in groups of 5–10. 1 wk after the last immunization, hyperimmune sera were taken from the tail vein, and 3–7 d thereafter, animals were challenged either i.p. with a 200× LD50 dose of S. pneumoniae PJ-1259 (∼104–105 CFU) or i.n. with an LD90-100 dose of a serotype 1 strain (6301; ∼5 × 106 CFU) in a volume of 40 μl under injection anesthesia (Ketamin/Rompun). Antigen-specific total IgG levels were determined for all sera of immunized mice. Only experimental groups with sufficiently high antibody levels (end-point ELISA titer >1:10,000) were evaluated. In serum transfer experiments, mice received 150 μl of mouse serum i.p. 1–3 h before challenge. In the pneumonia model, CD-1 mice were challenged with strains WU2 (∼105 CFU) or EF3030 (∼5 × 107 CFU), and the lungs were removed at day 3 after challenge and homogenized in PBS. Samples were serially diluted and plated on blood agar plates, and viable counts were determined after overnight incubation. A nonparametric test (Mann-Whitney) was used to compare numbers of CFU (log10) or time to death between groups. P ≤ 0.05 was considered statistically significant.
Generation of pneumococcal gene deletion strains.
Gene deletion strains were generated by competence stimulatory peptide–induced transformation of S. pneumoniae strains TIGR4 and PJ-1259 (58, 59). The gene replacement cassettes—the kanamycin gene (60) surrounded by 1-kb flanking regions of the target genes—were generated by ligation-mediated PCR. For transformation, 2 × 106 pneumococcal logarithmic growing cells were incubated in competence medium with 0.5 μg of PCR-generated linear DNA for 3 h at 37°C. Transformants were selected on blood agar containing 250 μg/ml kanamycin.
Electron microscopy.
WT, ΔstkP, and ΔpcsB mutant cells of S. pneumoniae strains TIGR4 and PJ-1259 were fixed with 2.5% glutaraldehyde in PBS (pH 7.4) for 2 h on ice and for 1 h at room temperature, filled into cellulose capillary tubes (0.2 mm in diameter), fixed with 2% OsO4 in Sorensen's buffer for 90 min, and dehydrated with increasing concentrations of ethanol. Samples were embedded in epoxy resin (Agar 100), and thin sections (60–80 nm) were cut with an ultramicrotome (Ultracut S; Leica) mounted on copper grids, contrasted by uranyl acetate and lead citrate, and examined at 80 kV in an electron microscope (JEM-1210; JEOL). Images were acquired using a digital camera (Morada) for the wide-angle port of the TEM and analySIS FIVE software (all from Soft Imaging System).
Online supplemental material.
Table S1 lists all pneumococcal antigens (TIGR4 strain annotation) that were identified by genomic library screening, indicating the total number of clones identified in the different screens with IgG and IgA pools as well as their immunogenic regions. Table S2 summarized the results of surface staining and OPK assay obtained with epitope-specific mouse sera. Fig. S1 depicts the selection of novel protective pneumococcal vaccine candidates in a mouse sepsis model. Fig. S2 shows the characterization of ΔpcsB and ΔstkP gene deletion mutant strains regarding their in vitro growth and in vivo virulence. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071168/DC1.
Supplemental Material
Acknowledgments
The authors would like to thank Christine Triska, Ulrike Stierschneider, Markus Horky, Sonja Prustomersky, Birgit Noiges, Barbara Maierhofer, and Ingrid Andersson for technical help; Siegfried Reipert for electron microscopic analysis; and David Briles, Ingileif Jonsdottir, Eddie Ades, and Elaine Tuomanen for providing bacterial strains for in vivo and in vitro studies.
The authors declare a potential conflict of financial interest as employees and/or consultants of Intercell AG, a biotechnology company. The authors have no other competing financial interests.
Abbreviations used: CHAP, cysteine, histidine-dependent amidohydrolase/peptidase; CPS, capsular polysaccharide; i.n., intranasal(ly); OPK, opsonophagocytic killing; ORF, open reading frame; PASTA, penicillin-binding protein and serine/threonine kinase associated; PcsB, protein required for cell wall separation of group B streptococcus; StkP, serine/threonine protein kinase.
C. Giefing and A.L. Meinke contributed equally to this work.
T. Henics' present address is Max F. Perutz Laboratories, 1030 Vienna, Austria.
D.B. Minh's present address is Icon Genetics GmbH, 06120 Halle/Saale, Germany.
A. Habel's present address is Berna Biotech AG, 3018 Bern, Switzerland.
References
- 1.Centers for Disease Control and Prevention. 2007. Pneumococcal disease. In Epidemiology and Prevention of Vaccine-Preventable Diseases (The Pink Book). Tenth edition. W. Atkinson, J. Hamborsky, L. McIntyre, and S. Wolfe, editors. Public Health Foundation, Washington, DC. 257–270.
- 2.Cutts, F.T., S.M. Zaman, G. Enwere, S. Jaffar, O.S. Levine, J.B. Okoko, C. Oluwalana, A. Vaughan, S.K. Obaro, A. Leach, et al. 2005. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet. 365:1139–1146. [DOI] [PubMed] [Google Scholar]
- 3.Mangtani, P., F. Cutts, and A.J. Hall. 2003. Efficacy of polysaccharide pneumococcal vaccine in adults in more developed countries: the state of the evidence. Lancet Infect. Dis. 3:71–78. [DOI] [PubMed] [Google Scholar]
- 4.Jackson, L.A., K.M. Neuzil, O. Yu, P. Benson, W.E. Barlow, A.L. Adams, C.A. Hanson, L.D. Mahoney, D.K. Shay, and W.W. Thompson. 2003. Effectiveness of pneumococcal polysaccharide vaccine in older adults. N. Engl. J. Med. 348:1747–1755. [DOI] [PubMed] [Google Scholar]
- 5.Black, S., H. Shinefield, B. Fireman, E. Lewis, P. Ray, J.R. Hansen, L. Elvin, K.M. Ensor, J. Hackell, G. Siber, et al. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr. Infect. Dis. J. 19:187–195. [DOI] [PubMed] [Google Scholar]
- 6.Black, S.B., H.R. Shinefield, J. Hansen, L. Elvin, D. Laufer, and F. Malinoski. 2001. Postlicensure evaluation of the effectiveness of seven valent pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 20:1105–1107. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher, M.A., and B. Fritzell. 2007. Brief review of the clinical effectiveness of PREVENAR against otitis media. Vaccine. 25:2507–2512. [DOI] [PubMed] [Google Scholar]
- 8.Hill, P.C., A. Akisanya, K. Sankareh, Y.B. Cheung, M. Saaka, G. Lahai, B.M. Greenwood, and R.A. Adegbola. 2006. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian villagers. Clin. Infect. Dis. 43:673–679. [DOI] [PubMed] [Google Scholar]
- 9.Adegbola, R.A., P.C. Hill, O. Secka, U.N. Ikumapayi, G. Lahai, B.M. Greenwood, and T. Corrah. 2006. Serotype and antimicrobial susceptibility patterns of isolates of Streptococcus pneumoniae causing invasive disease in The Gambia 1996-2003. Trop. Med. Int. Health. 11:1128–1135. [DOI] [PubMed] [Google Scholar]
- 10.Singleton, R.J., T.W. Hennessy, L.R. Bulkow, L.L. Hammitt, T. Zulz, D.A. Hurlburt, J.C. Butler, K. Rudolph, and A. Parkinson. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA. 297:1784–1792. [DOI] [PubMed] [Google Scholar]
- 11.Beall, B., M.C. McEllistrem, R.E. Gertz Jr., S. Wedel, D.J. Boxrud, A.L. Gonzalez, M.J. Medina, R. Pai, T.A. Thompson, L.H. Harrison, et al. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44:999–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez, B.E., K.G. Hulten, L. Lamberth, S.L. Kaplan, and E.O. Mason Jr. 2006. Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr. Infect. Dis. J. 25:301–305. [DOI] [PubMed] [Google Scholar]
- 13.Pai, R., M.R. Moore, T. Pilishvili, R.E. Gertz, C.G. Whitney, and B. Beall. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192:1988–1995. [DOI] [PubMed] [Google Scholar]
- 14.Bogaert, D., P.W. Hermans, P.V. Adrian, H.C. Rumke, and R. de Groot. 2004. Pneumococcal vaccines: an update on current strategies. Vaccine. 22:2209–2220. [DOI] [PubMed] [Google Scholar]
- 15.McDaniel, L.S., J.S. Sheffield, P. Delucchi, and D.E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenow, C., P. Ryan, J.N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H.R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819–829. [DOI] [PubMed] [Google Scholar]
- 17.Paton, J.C., R.A. Lock, and D.J. Hansman. 1983. Effect of immunization with pneumolysin on survival time of mice challenged with Streptococcus pneumoniae. Infect. Immun. 40:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogunniyi, A.D., M. Grabowicz, D.E. Briles, J. Cook, and J.C. Paton. 2007. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect. Immun. 75:350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barocchi, M.A., S. Censini, and R. Rappuoli. 2007. Vaccines in the era of genomics: the pneumococcal challenge. Vaccine. 25:2963–2973. [DOI] [PubMed] [Google Scholar]
- 20.Wizemann, T.M., J.H. Heinrichs, J.E. Adamou, A.L. Erwin, C. Kunsch, G.H. Choi, S.C. Barash, C.A. Rosen, H.R. Masure, E. Tuomanen, et al. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meinke, A., T. Henics, and E. Nagy. 2004. Bacterial genomes pave the way to novel vaccines. Curr. Opin. Microbiol. 7:314–320. [DOI] [PubMed] [Google Scholar]
- 22.Mora, M., D. Veggi, L. Santini, M. Pizza, and R. Rappuoli. 2003. Reverse vaccinology. Drug Discov. Today. 8:459–464. [DOI] [PubMed] [Google Scholar]
- 23.Pizza, M., V. Scarlato, V. Masignani, M.M. Giuliani, B. Arico, M. Comanducci, G.T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, et al. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 287:1816–1820. [DOI] [PubMed] [Google Scholar]
- 24.Maione, D., I. Margarit, C.D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E.T. Iacobini, R. Rosini, et al. 2005. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 309:148–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Ortega, M.J., N. Norais, G. Bensi, S. Liberatori, S. Capo, M. Mora, M. Scarselli, F. Doro, G. Ferrari, I. Garaguso, et al. 2006. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24:191–197. [DOI] [PubMed] [Google Scholar]
- 26.Henics, T., B. Winkler, U. Pfeifer, S.R. Gill, M. Buschle, A. von Gabain, and A.L. Meinke. 2003. Small-fragment genomic libraries for the display of putative epitopes from clinically significant pathogens. Biotechniques. 35:196–202, 204, 206 passim. [DOI] [PubMed] [Google Scholar]
- 27.Etz, H., D.B. Minh, T. Henics, A. Dryla, B. Winkler, C. Triska, A.P. Boyd, J. Sollner, W. Schmidt, U. von Ahsen, et al. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 99:6573–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etz, H., D.B. Minh, C. Schellack, E. Nagy, and A. Meinke. 2001. Bacterial phage receptors, versatile tools for display of polypeptides on the cell surface. J. Bacteriol. 183:6924–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meinke, A., T. Henics, M. Hanner, D.B. Minh, and E. Nagy. 2005. Antigenome technology: a novel approach for the selection of bacterial vaccine candidate antigens. Vaccine. 23:2035–2041. [DOI] [PubMed] [Google Scholar]
- 30.Kuklin, N.A., D.J. Clark, S. Secore, J. Cook, L.D. Cope, T. McNeely, L. Noble, M.J. Brown, J.K. Zorman, X.M. Wang, et al. 2006. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74:2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanello, V., M. Marcacci, F. Dal Molin, M. Moschioni, S. Censini, A. Covacci, A.G. Baritussio, C. Montecucco, and F. Tonello. 2006. Cloning, expression, purification, and characterization of Streptococcus pneumoniae IgA1 protease. Protein Expr. Purif. 45:142–149. [DOI] [PubMed] [Google Scholar]
- 32.Adamou, J.E., J.H. Heinrichs, A.L. Erwin, W. Walsh, T. Gayle, M. Dormitzer, R. Dagan, Y.A. Brewah, P. Barren, R. Lathigra, et al. 2001. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69:949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lock, R.A., J.C. Paton, and D. Hansman. 1988. Comparative efficacy of pneumococcal neuraminidase and pneumolysin as immunogens protective against Streptococcus pneumoniae. Microb. Pathog. 5:461–467. [DOI] [PubMed] [Google Scholar]
- 34.Lock, R.A., D. Hansman, and J.C. Paton. 1992. Comparative efficacy of autolysin and pneumolysin as immunogens protecting mice against infection by Streptococcus pneumoniae. Microb. Pathog. 12:137–143. [DOI] [PubMed] [Google Scholar]
- 35.Hollingshead, S.K., R. Becker, and D.E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roche, H., B. Ren, L.S. McDaniel, A. Hakansson, and D.E. Briles. 2003. Relative roles of genetic background and variation in PspA in the ability of antibodies to PspA to protect against capsular type 3 and 4 strains of Streptococcus pneumoniae. Infect. Immun. 71:4498–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briles, D.E., S.K. Hollingshead, J.C. Paton, E.W. Ades, L. Novak, F.W. van Ginkel, and W.H. Benjamin Jr. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J. Infect. Dis. 188:339–348. [DOI] [PubMed] [Google Scholar]
- 38.Goldblatt, D., M. Hussain, N. Andrews, L. Ashton, C. Virta, A. Melegaro, R. Pebody, R. George, A. Soininen, J. Edmunds, et al. 2005. Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. J. Infect. Dis. 192:387–393. [DOI] [PubMed] [Google Scholar]
- 39.McCool, T.L., T.R. Cate, E.I. Tuomanen, P. Adrian, T.J. Mitchell, and J.N. Weiser. 2003. Serum immunoglobulin G response to candidate vaccine antigens during experimental human pneumococcal colonization. Infect. Immun. 71:5724–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linder, A., S. Hollingshead, R. Janulczyk, B. Christensson, and P. Akesson. 2007. Human antibody response towards the pneumococcal surface proteins PspA and PspC during invasive pneumococcal infection. Vaccine. 25:341–345. [DOI] [PubMed] [Google Scholar]
- 41.Baril, L., D.E. Briles, P. Crozier, J. King, M. Punar, S.K. Hollingshead, and J.B. McCormick. 2004. Characterization of antibodies to PspA and PsaA in adults over 50 years of age with invasive pneumococcal disease. Vaccine. 23:789–793. [DOI] [PubMed] [Google Scholar]
- 42.Simell, B., M. Korkeila, H. Pursiainen, T.M. Kilpi, and H. Kayhty. 2001. Pneumococcal carriage and otitis media induce salivary antibodies to pneumococcal surface adhesin a, pneumolysin, and pneumococcal surface protein a in children. J. Infect. Dis. 183:887–896. [DOI] [PubMed] [Google Scholar]
- 43.Ng, W.L., K.M. Kazmierczak, and M.E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161–1175. [DOI] [PubMed] [Google Scholar]
- 44.Ng, W.L., G.T. Robertson, K.M. Kazmierczak, J. Zhao, R. Gilmour, and M.E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647–1663. [DOI] [PubMed] [Google Scholar]
- 45.Mattos-Graner, R.O., K.A. Porter, D.J. Smith, Y. Hosogi, and M.J. Duncan. 2006. Functional analysis of glucan binding protein B from Streptococcus mutans. J. Bacteriol. 188:3813–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinscheid, D.J., B. Gottschalk, A. Schubert, B.J. Eikmanns, and G.S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, D.J., W.F. King, L.A. Barnes, Z. Peacock, and M.A. Taubman. 2003. Immunogenicity and protective immunity induced by synthetic peptides associated with putative immunodominant regions of Streptococcus mutans glucan-binding protein B. Infect. Immun. 71:1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, D.J., W.F. King, and R. Godiska. 2001. Passive transfer of immunoglobulin Y antibody to Streptococcus mutans glucan binding protein B can confer protection against experimental dental caries. Infect. Immun. 69:3135–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Echenique, J., A. Kadioglu, S. Romao, P.W. Andrew, and M.C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72:2434–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novakova, L., L. Saskova, P. Pallova, J. Janecek, J. Novotna, A. Ulrych, J. Echenique, M.C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 272:1243–1254. [DOI] [PubMed] [Google Scholar]
- 51.Pallova, P., K. Hercik, L. Saskova, L. Novakova, and P. Branny. 2007. A eukaryotic-type serine/threonine protein kinase StkP of Streptococcus pneumoniae acts as a dimer in vivo. Biochem. Biophys. Res. Commun. 355:526–530. [DOI] [PubMed] [Google Scholar]
- 52.Saskova, L., L. Novakova, M. Basler, and P. Branny. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 189:4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeats, C., R.D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27:438. [DOI] [PubMed] [Google Scholar]
- 54.Brooks-Walter, A., D.E. Briles, and S.K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogunniyi, A.D., R.L. Folland, D.E. Briles, S.K. Hollingshead, and J.C. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson, S.E., L. Rubin, S. Romero-Steiner, J.K. Dykes, L.B. Pais, A. Rizvi, E. Ades, and G.M. Carlone. 1999. Correlation of opsonophagocytosis and passive protection assays using human anticapsular antibodies in an infant mouse model of bacteremia for Streptococcus pneumoniae. J. Infect. Dis. 180:133–140. [DOI] [PubMed] [Google Scholar]
- 57.Romero-Steiner, S., D. Libutti, L.B. Pais, J. Dykes, P. Anderson, J.C. Whitin, H.L. Keyserling, and G.M. Carlone. 1997. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin. Diagn. Lab. Immunol. 4:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L.S. Havarstein, L. Piccoli, D. Simon, and D.A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whatmore, A.M., V.A. Barcus, and C.G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banai, M., and D.J. LeBlanc. 1983. Genetic, molecular, and functional analysis of Streptococcus faecalis R plasmid pJH1. J. Bacteriol. 155:1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. De Castro, P.S. Langendijk-Genevaux, M. Pagni, and C.J. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.