Abstract
The single recombinant expressing the Streptomyces coelicolor minimal whiE (spore pigment) polyketide synthase (PKS) is uniquely capable of generating a large array of well more than 30 polyketides, many of which, so far, are novel to this recombinant. The characterized polyketides represent a diverse set of molecules that differ in size (chain length) and shape (cyclization pattern). This combinatorial biosynthetic library is, by far, the largest and most complex of its kind described to date and indicates that the minimal whiE PKS does not independently control polyketide chain length nor dictate the first cyclization event. Rather, the minimal PKS enzyme complex must rely on the stabilizing effects of additional subunits (i.e., the cyclase whiE-ORFVI) to ensure that the chain reaches the full 24 carbons and cyclizes correctly. This dramatic loss of control implies that the growing polyketide chain does not remain enzyme bound, resulting in the spontaneous cyclization of the methyl terminus. Among the six characterized dodecaketides, four different first-ring cyclization regiochemistries are represented, including C7/C12, C8/C13, C10/C15, and C13/C15. The dodecaketide TW93h possesses a unique 2,4-dioxaadamantane ring system and represents a new structural class of polyketides with no related structures isolated from natural or engineered organisms, thus supporting the claim that engineered biosynthesis is capable of producing novel chemotypes.
Engineered or combinatorial biosynthesis recently has emerged as a powerful technique to generate novel, structurally diverse molecules that are not available naturally nor generated readily through combinatorial synthesis. This approach has provided diverse chemical structures of the type required in the discovery of new lead-drug candidates (1), either directly or after synthetic modification to generate even greater structural diversity.
Recent progress has focused on the generation of hybrid polyketide metabolites through genetic manipulations of the encoding polyketide synthase (PKS) genes in a combinatorial fashion, either by “mixing and matching” the components of different gene sets or by exchanging parts of genes by genetic engineering (2). Polyketides, a large family of structurally diverse natural products possessing broad ranges of biological activities, are synthesized by successive Claisen condensations of extender units derived from (methyl)malonyl-CoA with an acyl-CoA starter unit in a manner reminiscent of fatty acid synthesis (3). Two types of bacterial PKSs have been recognized through molecular genetics (4) and are exemplified by prototype products, the macrolide antibiotic erythromycin (5, 6) (type I) and the aromatic polyketides actinorhodin (7) and tetracenomycin (8) (type II). In the modular type I system, the multifunctional PKS contains a separate, active site for each enzyme-catalyzed reaction, and the product thus is dictated strictly by the number and arrangement of these sites. The aromatic or type II system, on the other hand, contains a single set of iteratively used individual proteins for the construction of a linear poly-β-ketoacyl thioester intermediate, which then undergoes enzyme-catalyzed, regiospecific cyclizations to give an aromatic product.
A set of programming rules for the prediction of potential structures produced by engineered type II PKSs recently has been proposed (9, 10). Each PKS contains a “minimal” set of three protein subunits [the two β-ketoacyl/ACP synthase subunits, KSα and KSβ (also referred to as the chain length factor; ref. 11), and an acyl carrier protein (ACP)], which is required for in vivo polyketide biosynthesis (12). Additional PKS subunits, including ketoreductases (KRs), cyclases (CYCs), and aromatases (AROs), are responsible for modification of the nascent chain to form the specific cyclized polyketide product. A large series of novel polyketides has been generated by manipulating type II PKS systems that express a variety of combinations of a minimal PKS with KRs, CYCs, and AROs (for an overview on combinatorial biosynthesis with PKSs, see ref. 4).
Although engineered biosynthesis can generate just one or only a very few structures per recombinant, this is not always the case. Here, we describe an array of engineered aromatic polyketides that were generated by the single recombinant Streptomyces coelicolor YU105/pIJ4193 (13). The expression plasmid pIJ4193, a derivative of pSEK4 (11) that contains the minimal whiE (spore pigment) PKS (ORFs III–V) from S. coelicolor (14), was introduced via transformation into the genetically engineered S. coelicolor host YU105 (15) from which the natural chromosomal copies of the act and whiE gene clusters have been deleted. This strain is an ideal host for engineering recombinant polyketides because it displays a very low level of background polyketide synthesis. We analyzed an encoded library of well more than 30 polyketides from S. coelicolor YU105/pIJ4193 by liquid chromatography–electrospray MS (LC-ESMS) and isolated and fully characterized eight novel polyketides. Importantly, the characterized polyketides represent a diverse set of molecules that differ in size (chain length) and shape (cyclization pattern), indicating that the minimal whiE PKS, when separated from the other PKS components, does not independently control polyketide chain length or dictate the first cyclization event.
MATERIALS AND METHODS
Materials.
S. coelicolor YU105/pIJ4293 has been described elsewhere (13). SEK4 and SEK4b were a generous gift from R. J. Cox (University of Bristol), and SEK15 and SEK15b were from R. McDaniel (Kosan Biosciences, Burlingame, CA).
Spectral Analysis.
The 1H and 13C NMR spectra were obtained on Bruker DRX-499 and WM-500 spectrometers. 1H and 13C chemical shifts are referenced to dimethyl sulfoxide (DMSO)-d6 at 2.50 and 39.5 ppm, respectively. Proton data are reported as follows: chemical shift [multiplicity, coupling constants in hertz, assignment, integration, heteronuclear multiple bond correlations (HMBCs)]. One-bond and multiple-bond 1H-13C connectivities were determined, respectively, by gradient-enhanced heteronuclear multiple quantum correlation (HMQC) (16) and HMBC (17) experiments. Homonuclear 1H nuclear Overhauser effects (NOEs) were obtained by difference NOE experiments using a 0.7-s irradiation period.
Culture Conditions and Isolation of TW93e–h from S. coelicolor YU105/pIJ4293.
The construction and culture conditions of S. coelicolor YU105/pIJ4293 have been described earlier (13). YU105/pIJ4293 was grown on R5 agar plates (2 liter) and extracted with 80:20:1 EtOAc/MeOH/AcOH as described previously (13). The concentrated crude extract was subjected to C-18 silica gel flash chromatography and eluted with increasing amounts of acetone (10–30%) in H2O containing 1% AcOH. Fractions of 15 ml were collected and combined into six mixed fractions based on RP-18 TLC analysis (30:70:1 CH3CN/H2O/AcOH).
The first three flash-chromatography fractions contained polyketide pigments as determined by 1H NMR. These fractions were chromatographed by repetitive reversed-phase HPLC (Alltech Ultrasphere C18, 5 μ, 250 × 4.6 mm) with 30% CH3CN in 1% AcOH at flow rates between 3 and 6 ml/min and monitored at 280 nm. Fraction 1 yielded TW93a (8 mg), fraction 2 gave TW93b (6 mg), TW93c (20 mg), TW93d (10 mg), and TW93 g (7 mg), and fraction 3 provided TW93e (25 mg), TW93f (20 mg), and TW93h (10 mg). TW93f was purified further on Sephadex LH-20 using 7:3:0.1 MeOH/H2O/AcOH as eluent.
TW93e (5).
High-resolution fast atom bombardment MS (HRFABMS) m/z 465.0839 (calculated for C24H17O10, 1.7 mmu error); 1H NMR δ 2.18 (s, H-24, 3H, C-14, C-22, C-23), 3.85 (s, H-6, 2H, C-4, C-5, C-7, C-8, C-12), 5.15 (d, J = 2.5, H-2, 1H, C-1, C-3, C-4), 5.68 (d, J = 2.5, H-4, 1H, C-2, C-3, C-5, C-6), 6.09 (s, H-18, 1H, C-16, C-17, C-19, C-20), 6.21 (d, J = 2.5, H-10, 1H, C-8, C-9, C-11), 6.28 (d, J = 2.5, H-8, 1H, C-6, C-9, C-10, C12), 7.38 (s, H-22, 1H, C-13, C-14, C-16, C-17, C-20, C-24); 13C NMR δ 19.0 (q, C-24), 37.3 (t, C-6), 87.9 (d, C-2), 99.6 (d, C-4), 101.3 (d, C-10), 109.9 (d, C-18), 111.3 (s, C-16), 111.5 (d, C-8), 116.4 (s, C-12), 120.1 (d, C-22), 129.0 (s, C-21), 138.6 (s, C-7 and C-14), 141.6 (s, C-23), 156.3 (s, C-15), 160.2 (s, C-19), 161.5 (s, C-11), 161.8 (s, C-9), 163.3 (s, C-1), 164.7 (s, C-5), 169.9 (s, C-3), 179.9 (s, C-20), 190.6 (s, C-17), 194.5 (s, C-13).
TW93f (6).
HRFABMS m/z 465.0833 (calculated for C24H17O10, 1.2 mmu error); 1H NMR δ 2.00 (s, H-24, 3H, C-18, C-22, C-23), 4.35 (s, H-16, 2H, C-6, C-14, C-15, C-17), 5.34 (d, J = 1.9, H-2, 1H, C-1, C-3, C-4), 6.07 (d, J = 1.9, H-22, 1H, C-18, C-20, C-21, C-24), 6.08 (s, H-10, 1H, C-8, C-9, C-11, C-12), 6.13 (d, J = 1.8, H-4, 1H, C-2, C-3, C-5, C-6), 6.18 (d, J = 2.4, H-20, 1H, C-17, C-18, C-19, C-21, C-22), 7.43 (s, H-14, 1H, C-5, C-6, C-8, C-9, C-11, C-12, C-15, C-16, C-17), 9.70 (s, C-21 hydroxyl, 1H, C-20, C-21, C-22), 10.07 (s, C-19 hydroxyl, 1H, C-18, C-19, C-20), 10.11 (s, 1H), 11.87 (br s, 1H), 13.15 (br s, 1H); 13C NMR δ 19.7 (q, C-24), 48.0 (t, C-16), 89.6 (d, C-2), 99.7 (d, C20), 104.5 (d, C-4), 108.8 (d, C-22), 109.6 (d, C-10), 112.4 (s, C-8), 118.5 (s, C-18), 120.2 (d, C-14), 127.7 (s, C-6), 130.5 (s, C-13), 138.2 (s, C-23), 142.3 (s, C-15), 155.6 (s, C-5), 157.1 (s, C-19), 157.9 (s, C-11), 159.1 (s, C-21), 162.6 (s, C-7), 163.0 (s, C-1), 169.1 (s, C-3), 180.3 (s, C-12), 189.9 (s, C-9), 200.4 (s, C-17).
TW93g (7).
HRFABMS m/z 469.1116 (calculated for C24H21O10, −1.9 mmu error); 1H NMR δ 2.06 (s, H-24, 3H, C-18, C-22, C-23), 2.48 (d, J = 14.2, H-6a, 1H, C-4, C-5, C-7, C-8, C-16), 2.58 (d, J = 17.9, H-8a, 1H, C-6, C-7, C-9, C-10, C-16), 2.62 (d, J = 14.2, H-6b, 1H, C-4, C-5, C-7, C-8, C-16), 2.98 (d, J = 17.9, H-8b, 1H, C-6, C-7, C-9, C-16), 5.03 (s, H-16, 1H, C-6, C-7, C-8, C-10, C-14, C-15, C-17, C-18), 5.17 (d, J = 1.9, H-2, 1H, C-1, C-3, C-4), 5.87 (d, J = 1.9, H-4, 1H, C-2, C-3, C-5, C-6), 6.09 (d, J = 1.9, H-22, 1H, C-18, C-20, C-21, C-23, C-24), 6.15 (d, J = 2.5, H-14, 1H, C-10, C-11, C-13, C-14), 6.22 (d, J = 1.9, H-20, 1H, C-18, C-19, C21, C-22), 6.51 (d, J = 2.5, H-14, 1H, C-10, C-12, C-13, C-15, C-16), 12.77 (s, C-21 hydroxyl, 1H, C-9, C-10, C-11, C-12); 13C NMR δ 19.9 (q, C-24), 44.0 (t, C-6), 44.8 (t, C-8), 59.1 (d, C-16), 72.8 (d, C-7), 88.3 (d, C-2), 99.7 (d, C-20), 100.6 (d, C-12), 102.9 (d, C-4), 108.6 (d, C-14), 109.3 (d, C-22), 109.4 (s, C-10), 120.5 (s, C-18), 139.5 (s, C-23), 143.6 (s, C-15), 157.0 (s, C-19), 159.0 (s, C-21), 161.0 (s, C-5), 163.1 (s, C-1), 163.9 (s, C-11), 164.2 (s, C-13), 169.4 (s, C-3), 200.4 (s, C-9), 202.2 (s, C-17).
TW93h (8).
HRFABMS m/z 487.1234 (calculated for C24H23O11, −0.6 mmu error); 1H NMR δ 1.20 (s, H-24, 3H, C-21, C-22, C-23), 1.51 (dd, J = 11.7, 1.2, H-10eq, 1H, C-8, C-9, C-11, C-12, C-20), 1.68 (br d, J = 12.4, H-22eq, 1H, C-8, C-20, C-21, C-23), 1.76 (d, J = 12.4, H-22ax, C-7, C-8, C-20, C-21, C-22), 2.19 (d, J = 12.1, H-10ax, 1H, C-8, C-9, C-11), 2.87 (d, J = 16.7, H-12a, 1H, C-10, C-11, C-13, C-14, C-18, C-20, C-21), 3.12 (d, J = 16.7, H-12b, 1H, C-11, C-13, C-14, C-18), 3.18 (br d, J = 1.2, H-8, 1H, C-6, C-7, C-9, C-10, C-20, C-21, C-22), 3.54 (s, H-20, 1H, C-8, C-11, C-19, C-21, C-22), 3.80 (d, J = 17.6, H-6a, 1H, C-4, C-5, C-7), 3.87 (d, J = 17.6, H-6b, 1H, C-4, C-5, C-7), 5.21 (d, J = 1.9, H-2, 1H, C-1, C-3, C-4), 5.67 (s, C-21 hydroxyl, 1H, C-8, C-9, C-20, C-21), 5.97 (d, J = 1.9, H-4, 1H, C-2, C-3, C-5, C-6, C-7), 6.09 (d, J = 1.9, H-16, 1H, C-14, C-15, C-17, C-18, C-19), 6.20 (br s, H-14, 1H, C-12, C-16, C-17, C-18), 7.22 (br s, C-9 hydroxyl, 1H); 13C NMR δ 25.8 (q, C-24), 39.2 (t, C-12), 39.4 (t, C-10), 44.7 (t, C-22), 49.0 (d, C-20), 51.0 (t, C-6), 62.2 (d, C-8), 72.2 (s, C-21), 75.3 (s, C-11), 88.3 (d, C-2), 94.6 (s, C-9), 99.0 (s, C-23), 100.1 (d, C-16), 102.7 (d, C-4), 108.0 (d, C-14), 109.2 (s, C-18), 143.6 (s, C-13), 158.0 (s, C-5), 163.4 (s, C-1), 164.6 (s, C-15), 165.7 (s, C-17), 170.1 (s, C-3), 203.1 (s, C-19), 204.6 (s, C-7).
LC-ESMS Analysis of the YU105/pIJ4293 Crude Extract.
The YU105/pIJ4293 crude extract was prepared as described above, dissolved in MeOH (25 mg/ml), and analyzed (10 μl). A Shimadzu LC-10AD pump was linked to a Quattro II mass spectrometer operating in the positive ion-electrospray mode. The same HPLC column as described above was used at a flow rate of 2 ml/min and eluted with increasing amounts of CH3CN in 1% AcOH (20–50%) over a period of 30 min followed by a 100% CH3CN wash.
RESULTS
LC-ESMS Analysis of the Minimal whiE PKS-Encoded Polyketides.
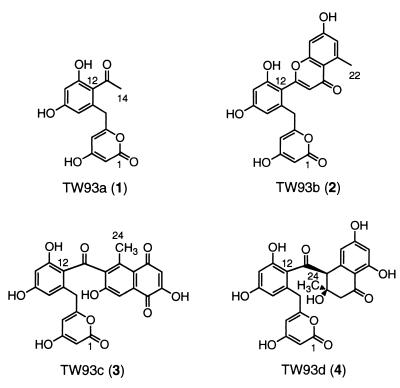
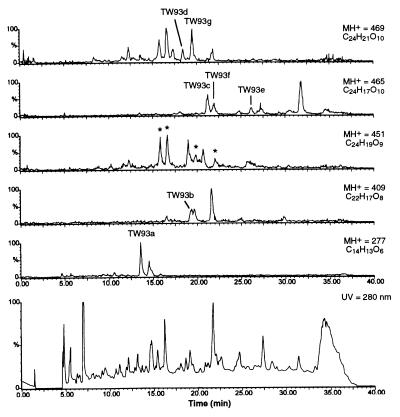
We recently reported that the expression of the minimal whiE PKS by S. coelicolor YU105/pIJ4293 resulted in the production of a mixture of pigmented polyketides, including the heptaketide TW93a (1), the undecaketide TW93b (2), and the dodecaketides TW93c (3) and TW93d (4) (Fig. 1) (13). These four polyketides share an identical structural motif from carbons 1 through 12, characteristic of a common C7/C12 first cyclization. Further analysis of the YU105/pIJ4293 polyketides by LC-ESMS demonstrated the presence of a complex series of polyketides numbering well more than 30 metabolites (Fig. 2), several of which depart from this structural motif.
Figure 1.
Structures of TW93a–d (1–4).
Figure 2.
LC-ESMS analysis of minimal whiE PKS polyketides from YU105/pIJ4293. HPLC was monitored at 280 nm. Ion chromatograms for the pseudomolecular ion MH+ of hepta-, undeca-, and dodecaketides are displayed. Note that several of the 451 ions are fragments from MH+ 469 resulting from the loss of H2O and are indicated with an asterisk.
Dodecaketides were the most numerous class of polyketides, with several isomers of mass 464 (C24H16O10) and 468 (C24H20O10) (Fig. 2). In addition to the previously reported 3 and 4, several new dodecaketides were identified, four of which were isolated and are characterized below as TW93e–h. Each of the identified dodecaketides has a distinct carbon skeleton, thus indicating that the common poly-β-ketoacyl thioester intermediate probably cyclizes to the corresponding dodecaketide end products spontaneously. On the basis of this lack of enzymatic control, we were surprised that the dodecaketide TW95a (MH+ 451), the chief product of the minimal whiE PKS plus the whiE-ORFVI cyclase (YU105/pIJ4293) (13), was not detected in this mixture. Two new dodecaketides of mass 450 (C24H18O9) were, however, identified (Fig. 2).
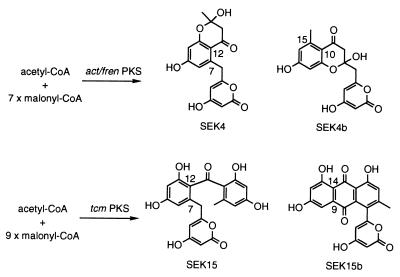
Because the whiE PKS probably synthesizes a dodecaketide spore pigment (13), it came as no surprise that dodecaketides dominated among the minimal PKS products. However, shorter polyketides were present as well. In addition to the previously reported heptaketide 1 and undecaketide 2, additional heptaketides and undecaketides were evident (Fig. 2). Octaketides, nonaketides, and decaketides possibly were present as well based on the LC-ESMS data. However, the previously reported products from other artificially expressed minimal PKSs were not detected. These included the octaketides SEK4 (18) and SEK4b (19), products of the minimal PKS for the benzoisochromanequinone actinorhodin (act from S. coelicolor) and frenolicin/nanaomycin (fren from S. roseofulvus), and the decaketides SEK15 (18) and SEK15b (12), products of the tetracenomycin F2 (tcm from S. glaucescens) minimal PKS (Fig. 3). The lack of these polyketides among the minimal whiE PKS products does not necessarily mean that they were not formed, just that they were not observed by ESMS.
Figure 3.
Structures of minimal PKS products from the act/fren (SEK4 and SEK4b) and the tcm (SEK15 and SEK15b) pathways.
Structures of the Dodecaketides TW93e–h.
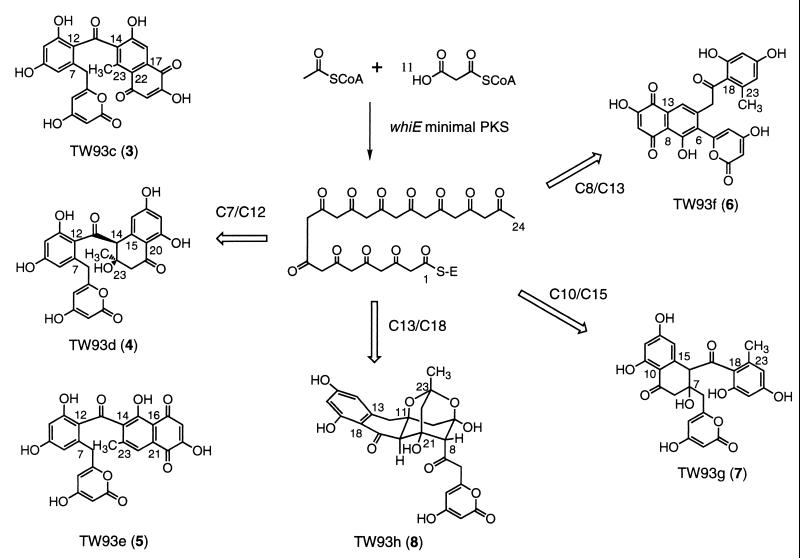
Four new dodecaketides, TW93e–h (Fig. 4), were isolated from the minimal whiE PKS recombinant S. coelicolor YU105/pIJ4193 by reversed-phase chromatography and characterized by (13C,1H) gradient-enhanced HMQC (16) and HMBC (17) spectroscopy. TW93e (5), which had the molecular formula C24H16O10 on the basis of HRFABMS, was structurally identical with 3 and 4 from carbons 1 through 13 but differed in the cyclization pattern for the remaining 11-carbon fragment (C14-C24). The NMR spectral data indicated that, in addition to 4-hydroxy-2-pyrone and o,p-dihydroxyphenyl ketone residues common in 3 and 4, 5 contained a 6-acyl-2-hydroxy-7-methyljuglone moiety in which the polyketide chain has cyclized C16/C21 and C14/C23. The quinone unit in 5, which is structurally related to that in 3 except for the exchange of the C15 phenol and the C23 methyl groups, was readily identified by means of key HMBCs, including that from H22 to C23.
Figure 4.
Structures of the minimal whiE PKS dodecaketides TW93c–h (3–8) and of their common poly-β-ketoacyl thioester biosynthetic precursor. Carbons are labeled according to their number in the polyketide backbone.
Although TW93f (6) analyzed for the same molecular formula as 5, it was very apparent from the NMR analysis that 6 has a completely different carbon skeleton. The 4-hydroxy-2-pyrone unit in 6 is attached directly, without a methylene bridge as in 1–5, to a 2,6,7-trisubstituted juglone residue that is similar to that in 5. This attachment of the pyrone directly to the juglone was assigned on the basis of a key HMBC from the H4 pyrone doublet (δ 6.13) to C6 (δ 127.7) of the juglone. This structural feature is also present in the octaketide RM77 (20) and shares similar chemical shift values with it. A methylene group (C-16: δC 48.0, δH 4.35) was deduced on the basis of HMBCs (C16 → H14; H16 → C6, C14, C15, C17) to bridge the juglone to a 2,4-dihydroxy-6-methylphenyl ketone unit, identical to that in TW95a and TW95b (13).
TW93g (7) was assigned the molecular composition C24H20O10 by HRFABMS, indicating that 7 had two degrees of unsaturation fewer than 5 and 6. The presence of 4-hydroxy-2-pyrone and 2,4-dihydroxy-6-methylphenyl ketone functionalites was assigned readily by NMR spectral analysis. An α-tetralone unit was also evident by NMR analysis and was deduced to be similar to that in 4 (13). The H6 AB quartet (δ 2.48 and 2.62) was shifted 1 ppm up-field relative to that in 4, indicating that the C6 methylene group is directly attached to an sp3 hybridized carbon. Attachments of the pyrone residue through the C6 methylene group to the δ-position (C7) and of the phenyl ketone unit to the γ-position (C16) of the α-tetralone were established readily by HMBCs.
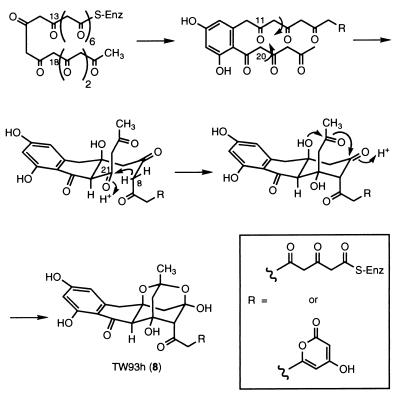
TW93h (8), which analyzed for C24H22O11 by HRFABMS, was the most unusual dodecaketide characterized from YU105/pIJ4193. The NMR spectral data indicated that although 8 contained the common 4-hydroxy-2-pyrone and 2,4-dihydroxy-6-alkylphenyl ketone structural units present in the majority of the TW93 polyketides, 8 possessed a completely novel skeleton. Except for an additional ketone carbonyl, all of the remaining 11 carbons were sp3-hybridized, including four isolated, nonequivalent methylene groups and two acetals. Altogether, these functional groups account for 10 of the required 13 sites of unsaturation, indicating that 8 must contain three additional rings. The HMBC data allowed for the assembly of a 2,4-dioxaadamantane moiety, a unique acetal (C23: δ 99.0) with both oxygens axial on tetrahydropyran rings (21). Important correlations for assigning the tricyclic ring system included those from protons H8 and H10 to the C9 hemiacetal (δ 94.6) and the C20 methine (δ 49.0), allowing for the formation of the cyclohexane ring, and from the H22 methylene to C8, C20, C21, C23, and C24. Attachment of the phenyl ketone unit to the dioxaadamantane residue was established by correlations from the C19 ketone carbonyl to H20 and from the aromatic C13 to H11 and H12. The attachment of the pyrone through an acetyl linkage to C8 of the dioxaadamantane was determined through correlations from the methylene C6 to H4 and H8.
The relative stereochemistry of 8 needed to be assigned only at C8, because the remaining five chiral centers are fixed in the rigid 2,4-dioxaadamantane unit, and was established by using difference-NOE experiments. Observation of an NOE signal between H8 and H22eq, together with the 1.2-Hz W-coupling between H8 and H10eq, indicated that the H8 hydrogen is equatorial and the acetylpyrone fragment is axial. This assignment was supported by the enhancement of H10ax upon irradiation of H20, indicating that only these hydrogens are in axial positions.
Testing is currently underway with the pure isolates 1–8 and the crude YU105/pIJ4293 polyketide extract for antimicrobial and antiviral activities.
DISCUSSION
We have demonstrated that a single recombinant expressing the minimal whiE PKS is capable of generating a large array of well more than 30 polyketides of various sizes and shapes. Many of these polyketides, so far, are unique to this recombinant and are present in comparable concentrations, each between 2 and 10 mg/liter of culture. This combinatorial biosynthetic library is, by far, the largest and most complex of its kind described to date and indicates that the minimal PKS, at least in the case of the whiE system, alone does not dictate chain length or the first cyclization event. Rather, the minimal PKS enzyme complex must rely on the stabilizing effects of additional subunits. We recently reported that the minimal whiE PKS requires the cyclase whiE-ORFVI to stabilize the complex between the long nascent polyketide chain and the minimal PKS to ensure that the chain reaches the full 24 carbons and correctly cyclizes at C9/C14 (13).
The growing polyketide chain apparently can dissociate prematurely from the minimal whiE PKS in the absence of the cyclase. Similar expressions of artificial minimal PKS gene sets from the act, fren, and tcm clusters have resulted in much simpler polyketide profiles (4). In each case, just two major products of the same chain length but of different cyclization patterns were reported. This discrepancy among minimal PKSs raises the question of the uniqueness of the whiE system. Does the capacity of the whiE PKS to generate larger polyketides [dodecaketides vs. octaketides (act and fren) and decaketides (tcm)] inherently make it a more fragile and, hence, “sloppier” system when the minimal PKS is expressed without the stabilizing effects of the cyclase? If so, will artificially expressed minimal PKSs from other aromatic PKS gene clusters that encode the biosynthesis of larger aromatic polyketides, such as fredericamycin A (22) and simaomicin α (23), yield even larger polyketide arrays?
In regard to the regiospecificity of the first cyclization involving an aldol condensation, four different regiochemistries are represented among the six characterized dodecaketides, including C7/C12 (TW93c–e), C8/C13 (TW93f), C10/C15 (TW93 g), and C13/C18 (TW93h) (Fig. 4). Other first-ring regiochemistries, such as C9/C14, C11/C16, and C12/C17, are potentially represented among the many remaining and uncharacterized dodecaketides (Fig. 2). Interestingly, none of the characterized TW93 polyketides bears the natural C9/C14 regiospecificity of the initial cyclization proposed for the whiE-encoded spore pigment (13). This dramatic loss of control implies that the growing polyketide chain does not remain enzyme-bound, resulting in the spontaneous cyclization of the methyl terminus. Alternatively, because synthesis takes place in vivo, transient interactions with unrelated enzymes in the host possibly could influence folding, cyclization, and chain length.
This incomplete control by the minimal PKS in catalyzing the initial cyclization has been observed to a much lesser extent in the act, fren, and tcm PKS systems (Fig. 3). In the case of the minimal act and fren PKSs, only the octaketides SEK4 and SEK4b were reported (10, 18, 19). The regiospecificity of the initial cyclization in SEK4 (C7/C12) is the same as that observed for the end products actinorhodin (act) and nanaomycin (fren) and thus is considered to be enzymatically controlled by the minimal PKS. On the other hand, the predominant minimal act PKS product SEK4b is not regiospecifically cyclized by the minimal PKS because it “unnaturally” cyclizes C10/C15. Rather, the minimal PKS has been proposed to influence the cyclization process by progressively releasing the growing chain (19). Similarly, the minimal tcm PKS products, the decaketides SEK15 and SEK15b, differ in the initial cyclization event (12, 18). SEK15b contains the natural tetracenomycin F2 C9/C14 cyclization pattern, whereas SEK15 is a product of an “unnatural” C7/C12 first cyclization.
The reoccurring incidence of minimal PKS products with a C7/C12 first cyclization [SEK4 (act/fren), SEK15 (tcm), and TW93a–e (whiE)] indicates that the regiochemistry of this cyclization probably is favored over the many other potential combinations for the initial cyclization. This cyclization event may, in part, be controlled indirectly by the minimal PKS, which tethers the carboxyl terminus to the ACP, thus limiting the conformational mobility of the linear poly-β-ketoacyl thioester at the carboxyl end. This, in turn, may favor a spontaneous aldol condensation between C7/C12 by decreasing the change in entropy toward the transition state. The minimal act/fren PKS product, SEK4, which harbors the “natural” C7/C12 initial cyclization, consequently may be a spontaneous and, hence, fortuitous product.
The addition of TW93e–h to the growing list of minimal whiE PKS products represents an important lesson exemplifying the caution necessary in deducing genetic functions based on the isolation of one or even a few engineered products. On the basis of the structures of TW93a–d, we were compelled to propose originally that the initial cyclization event is catalyzed by the minimal whiE PKS (13). However, the addition of several new TW93 dodecaketides exhibiting different cyclization patterns points to the modified conclusion that the minimal PKS does not catalyze the initial cyclization event, at least when expressed alone.
The dioxaadamantane TW93h (8) represents a new structural class of polyketides with no related structures isolated from natural or engineered organisms, thus supporting the claim that engineered biosynthesis is capable of producing novel chemotypes. TW93h possesses a unique 2,4-dioxaadamantane ring system, which before has been known only through synthesis (24) and in two unusual marine natural products. The potent neurotoxin tetrodotoxin (25) contains a hemilactal 2,4-dioxaadamantane, and the polypropionate muamvatin (26) is a unique 2,4,6-trioxaadamantane. TW93h is a polyketide with minimal aromatic character resulting from the expression of an aromatic PKS, indicating that truly novel compounds that bear little or no resemblance to their parent metabolite are attainable by this method. The proposed pathway of formation to TW93h is summarized in Fig. 5 and probably involves the spontaneous attack of the axial C11 hydroxyl on the axial C23 carbonyl to form an intermediary hemiacetal, which, in turn, participates in the subsequent formation of a hemiacetal with the C9 ketone to complete the 2,4-dioxaadamantane ring system.
Figure 5.
Proposed formation of TW93h (8).
Finally, the results described here are relevant to the growing use of combinatorial biosynthesis in the search for novel molecules with applications as pharmaceuticals (1) or as platforms for (combinatorial) synthesis. The generation of diverse sets of metabolites from a single recombinant is pertinent to one of the goals in the emerging field of combinatorial biosynthesis, which is to extend natural product structural diversity. With the results presented here, we have demonstrated that engineered aromatic PKSs are capable of generating polyketide libraries of different molecular sizes and shapes. We have also shown recently that modular PKSs are susceptible as well. In the case of the assembly of the polyketide backbone of rifamycin B (27), inactivation of the rifF amide synthase gene, which releases the completed undecaketide as its macrocyclic lactam, results in the accumulation of a series of linear polyketides ranging in size from tetra- to decaketides (T.-W.Y., Y.S., B.S.M., and H.G.F., unpublished data). If biologically active or chemically interesting polyketides are generated in such mixtures, then a subsequent rational engineering of their biosynthesis may be feasible.
Acknowledgments
This research was supported in part by the Washington Sea Grant Program (National Oceanic and Atmospheric Administration NA76RG0119, project no. R/B-28, to B.S.M.), the National Institutes of Health (AI 20264 to H.G.F.), the John Innes Foundation (D.A.H.), and the U.K. Biotechnology and Biological Sciences Research Council (D.A.H.).
ABBREVIATIONS
- PKS
polyketide synthase
- LC-ESMS
liquid chromatography–electrospray MS
- HMBC
heteronuclear multiple bond correlation
- HMQC
heteronuclear multiple quantum correlation
- NOE
nuclear Overhauser effect
- HRFABMS
high-resolution fast atom bombardment MS
Footnotes
A Commentary on this article begins on page 3336.
References
- 1.Fu H, Khosla C. Mol Diversity. 1995;1:121–124. doi: 10.1007/BF01721327. [DOI] [PubMed] [Google Scholar]
- 2.Tsoi C J, Khosla C. Chem Biol. 1995;2:355–362. doi: 10.1016/1074-5521(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 3.O’Hagan D. The Polyketide Metabolites. Chichester, U.K.: Ellis Horwood; 1991. [Google Scholar]
- 4.Hopwood D A. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 5.Cortes J, Haydock S F, Roberts G A, Bevitt D J, Leadlay P F. Nature (London) 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 6.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Moreno M A, Martínez E, Bibb M J, Kieser H M, Bibb M J, Hopwood D A. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 8.Bibb M J, Biró S, Motamedi H, Collins J F, Hutchinson C R. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Nature (London) 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 10.Kramer P J, Zawada R J X, McDaniel R, Hutchinson C R, Hopwood D A, Khosla C. J Am Chem Soc. 1997;119:635–639. [Google Scholar]
- 11.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 12.McDaniel R, Ebert-Khosla S, Fu H, Hopwood D A, Khosla C. Proc Natl Acad Sci USA. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu T-W, Shen Y, McDaniel R, Floss H G, Khosla C, Hopwood D A, Moore B S. J Am Chem Soc. 1998;120:7749–7759. [Google Scholar]
- 14.Davis N K, Chater K F. Mol Microbiol. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 15.Yu T-W, Hopwood D A. Microbiology. 1995;141:2779–2791. doi: 10.1099/13500872-141-11-2779. [DOI] [PubMed] [Google Scholar]
- 16.Hurd R E, John B K. J Magn Reson. 1990;91:648–653. [Google Scholar]
- 17.Ruiz-Cabello J, Vuister G W, Moonen C T, van Gelderen P, Cohen J S, Van Zijl P C M. J Magn Reson. 1992;100:282–302. doi: 10.1016/j.jmr.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Fu H, Ebert-Khosla S, Hopwood D A, Khosla C. J Am Chem Soc. 1994;116:4166–4170. [Google Scholar]
- 19.Fu H, Hopwood D A, Khosla C. Chem Biol. 1994;1:205–210. doi: 10.1016/1074-5521(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 20.McDaniel R, Hutchinson C R, Khosla C. J Am Chem Soc. 1995;117:6805–6810. [Google Scholar]
- 21.Briggs, A. J. & Kirby, A. J. (1988) Synthesis 66–67.
- 22.Byrne K M, Hilton B D, White R J, Misra R, Pandey R C. Biochemistry. 1985;24:478–486. doi: 10.1021/bi00323a035. [DOI] [PubMed] [Google Scholar]
- 23.Carter G T, Goodman J J, Torrey M J, Borders D B. J Org Chem. 1989;54:4321–4323. [Google Scholar]
- 24.Burgey C S, Vollerthum R, Fraser-Reid B. Tetrahedron Lett. 1994;35:2637–2640. [Google Scholar]
- 25.Kao Y, Lovinson S R. Tetrodotoxin, Saxitoxin and Molecular Biology of the Sodium Channel. New York: New York Academy of Sciences; 1986. [PubMed] [Google Scholar]
- 26.Roll D M, Biskupiak J E, Mayne C L, Ireland C M. J Am Chem Soc. 1986;108:6680–6682. [Google Scholar]
- 27.August P R, Tang L, Yoon Y J, Ning S, Müller R, Yu T-W, Taylor M, Hoffman D, Kim C-G, Zhang X, et al. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]