Abstract
Nuclear receptor-mediated gene expression is regulated by corepressors and coactivators. In this study we demonstrate that prohibitin (PHB), a potential tumor suppressor, functions as a potent transcriptional corepressor for estrogen receptor α (ERα). Overexpression of PHB inhibits ERα transcriptional activity, whereas depletion of endogenous PHB increases the expression of ERα target genes in MCF-7 breast cancer cells. Chromatin immunoprecipitation experiments demonstrate that PHB is associated with the estrogen-regulated pS2 promoter in the absence of hormone and dissociates after estradiol treatment. We demonstrate that PHB interacts with the repressor of estrogen receptor activity (REA), a protein related to PHB, to form heteromers and enhance the protein stability of both corepressors. Interestingly, the corepressor activity of PHB is cross-squelched by the coexpression of REA (and vice versa), suggesting that PHB and REA repress transcription only when they are not paired. We further demonstrate that coiled-coil domains located in the middle of PHB and REA are responsible for their heteromerization, stabilization, and cross-squelching actions. Finally, ablation of PHB function in the mouse results in early embryonic lethality, whereas mice heterozygous for the PHB null allele exhibit a hyperproliferative mammary gland phenotype. Our results indicate that PHB functions as a transcriptional corepressor for ERα in vitro and in vivo, and that its heteromerization with REA acts as a novel mechanism to limit its corepressor activity.
LIGAND-ACTIVATED nuclear receptors (NRs) are transcription factors that regulate gene transcription through a series of molecular events centering on the dismissal of corepressors and the recruitment of coactivators (1,2,3). The steroid receptor coactivator (SRC) family is the best-characterized group of coactivators, consisting of three members, steroid receptor coactivator-1 (SRC-1), SRC-2 (TIF2, GRIP1), and SRC-3 (p/CIP/RAC3/ACTR/AIB1/TRAM-1) (4,5). The SRC/p160 family of transcriptional coactivators contains intrinsic histone acetyltransferase activity (6,7), and recruits secondary coactivators such as p300 and cAMP response element binding protein-binding protein, which possess potent intrinsic histone acetyltransferase activity (8) and coactivator-associated arginine methyltransferase 1, which contains histone methyltransferase activity (9). The physiological roles of SRC coactivators in female reproductive biology have been revealed using coactivator knockout mouse models. Mice lacking any of the SRC family coactivator genes have distinct phenotypes, such as partial hormone resistance in the SRC-1 null mice (10), severe hypofertility and infertility in SRC-2 null mice (11,12), and growth retardation in SRC-3 null mice (13,14). These data indicate that overlapping but distinct roles exist for each of these SRC family members in maintaining reproductive function, mammary gland morphogenesis and energy homeostasis.
Importantly, coactivators are opposed by NR corepressors that form a critical balance to allow an appropriate and measured response to NR hormones, such as estrogen (E) and progesterone (P). The prototypical general corepressors N-CoR (15) and SMRT (16) associate with unliganded type II NRs which bind to their target genes regardless of whether they are liganded, and repress transcription by recruitment of histone deacetylase (HDACs) (17). These corepressors also play an important role in regulating the transcriptional activities of many hormone-bound type I NRs including androgen, E, and glucocorticoid receptors (18,19,20,21).
Examples of other NR corepressors are: RIP-140 (22), SUN-CoR (23), Alien (24), Hairless (25), MTA1 (26), and LCoR (27). Corepressors such as RIP-140, LCoR, N-CoR, and SMRT directly interact with the nuclear receptor ligand-binding domain via their NR or CoRNR box (28,29).
Repressor of ER Activity (REA, also known as PHB2), a member of the prohibitin (PHB) family of corepressors, was shown to markedly repress ERα-mediated transactivation and enhance the inhibitory effectiveness of SERMs bound to ERα (30,31,32). Consistent with its role as a corepressor, mice heterozygous for the REA null allele displayed an increased response to E in both the uterus (33) and mammary gland (34). Both REA and its related protein PHB1 (referred to here as PHB) belong to a family of proteins that carry an evolutionarily conserved domain, the prohibitin homology (PHB) domain (35). PHB domain-containing proteins have been identified in both bacteria and eukaryotes and have multiple functions (35). In this study, we provide strong in vitro and in vivo evidence supporting a potent corepressor role for PHB in ERα-mediated signaling and demonstrate that its corepressor activity is controlled by heteromerization with REA.
RESULTS
Prohibitin Represses ERα-Mediated Transcription
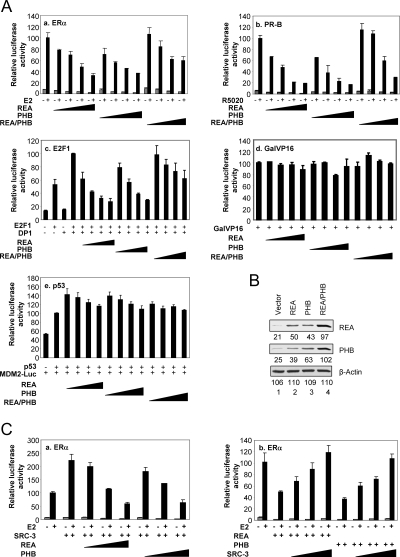
The potential tumor suppressor PHB has been reported to be a transcriptional corepressor for E2F1 (36). Its related protein REA has been reported to be a repressor for ERα (30). Because PHB and REA share high homology in their primary amino acid sequences (53% identical over 252 amino acids of PHB), we tested whether PHB also can repress the transcriptional activity of ERα, and conversely if REA can repress the transcriptional activity of E2F1. In HepG2 cells, cotransfection of a vector expressing ERα, and an ERE-luciferase reporter, ERα transcriptional activity is significantly stimulated by addition of 10 nm estradiol (E2) (Fig. 1A). Cotransfection of increasing amounts of PHB significantly reduced ERα transcriptional activity, to an extent similar to that seen for coexpression of increasing amounts of REA. Strikingly, when both PHB and REA were coexpressed with ERα and the ERE-Luc reporter, their combined ability to repress transcription consistently was less than that seen for either corepressor alone.
Figure 1.
Prohibitin Represses ERα-Mediated Transcription
A, PHB and REA are corepressors for ERα, PR-B, and E2F1, but not for Gal4-VP16 and p53. Transcriptional activities of ERα, PR-B, E2F1, Gal-VP16, and p53 were determined by cotransfection of HepG2 cells with increasing amounts of REA, PHB, or REA/PHB (50, 100, 200, and 300 ng). B, Western blot analysis to show the expression of PHB and REA protein from their respective vectors. The 293T cells were transiently transfected with empty vector (lane 1), or vector expressing REA (lane 2) or PHB (lane 3), or both expression vectors (lane 4). Expression levels were determined by using anti-REA, anti-PHB antibodies. β-Actin was analyzed as sample loading control. C, PHB and REA antagonize ERα coactivation by SRC-3. In the left panel, 300 ng of SRC-3 was cotransfected with increasing amounts of REA or PHB plasmids (100, 200, and 300 ng) in HepG2 cells. On the other hand, overexpression of SRC-3 overcomes the ERα activity repressed by PHB and REA. In right panel, 200 ng of REA or PHB were cotransfected with increasing amounts of SRC-3 (100, 200, and 300 ng).
Next, we investigated the effect of PHB and REA on the transcriptional activity of E2F1. As reported (37), when cotransfected with E2F1, DP1 (heterodimer partner for E2F1), and the CycE-Luc reporter, PHB overexpression was able to efficiently repress E2F1-mediated activity (Fig. 1A). REA repressed E2F1-mediated transcription to an extent similar to that seen with PHB. As seen above for ERα, coexpression of both PHB and REA again reduced their repressive effect on E2F1-mediated transcription. To investigate whether the corepressive effects of PHB and REA are receptor specific, we also investigated the effect of PHB and REA on the transcriptional activity of MMTV-Luc reporter mediated by the progesterone receptor B (PR-B). Similar to what we observed for ERα and E2F1, PR-B activity was repressed by both PHB and REA, and coexpression of PHB and REA reduced their repressive capabilities (Fig. 1A). The observation that REA also can repress PR-B differs from a previous report (30), where it was reported to have only a mild repressive effect on PR-B transcriptional activity in Chinese hamster ovary cells (30). Our results may be due to our use of HepG2 cells in this study where the repressive activity is more pronounced. Next, we tested whether the repressive function of PHB and REA is simply due to a general repression of RNA Pol II-dependent transcription. We assessed the effect of PHB and REA coexpression on the transcriptional activity of a Gal4-VP16 chimeric transactivator. Similar to that shown previously for REA (30), coexpression of PHB had no significant effect on the transcriptional activity of Gal4-VP16, indicating that the corepressor activity of PHB and REA does not apply to all transcriptional factors (Fig. 1A). Because it has been reported that PHB functions as a coactivator for p53 (38), we next tested whether both PHB and REA can function as coactivators for p53 on the MDM2 promoter luciferase reporter in our system. Shown in Fig. 1A-e, PHB and REA moderately enhanced p53-mediated transcription of the MDM2 promoter at the lower amounts of transfected plasmids. Transfection of high levels of PHB or REA resulted in reduced stimulation of transcription; no repression was observed at all levels tested.
To confirm the expression of PHB and REA from their mammalian expression vectors used above, we transiently transfected the 293T cells with PHB, REA, or a combination of PHB and REA. Western blot analysis (Fig. 1B) showed that PHB and REA (lanes 2 and 3) were highly expressed in comparison to endogenous PHB and REA (lane 1). Remarkably, transient transfection of REA plasmids not only increased the protein level of REA, but also that of PHB, suggesting that the overexpression of exogenous REA stabilizes the endogenous PHB. Similarly, exogenous expression of PHB increased the protein level of REA. Moreover, cotransfection of PHB and REA together produced significantly more PHB and REA protein than transfection of individual plasmids. These data indicate that PHB and REA proteins stabilize each other.
One mechanism by which REA could repress ERα activity is by functioning as a competitive inhibitor of SRC-3, which is specifically important in mammary tissue. We examined whether there is functional interplay among SRC-3, PHB, and REA. Shown in the left panel of Fig. 1C, SRC-3 increased ERα activity. Coexpression of increasing amounts of PHB or REA was able to counter SRC-3 coactivation of ERα. On the other hand, coexpression of increasing amounts of SRC-3 also was able to overcome the ERα-mediated transcriptional activity repressed by PHB or REA (right panel, Fig. 1C). These data indicate that the corepressor functions of PHB and REA competitively oppose the coactivator function of SRC-3 during ERα-mediated transcription.
Interactions among ERα, PHB, and REA
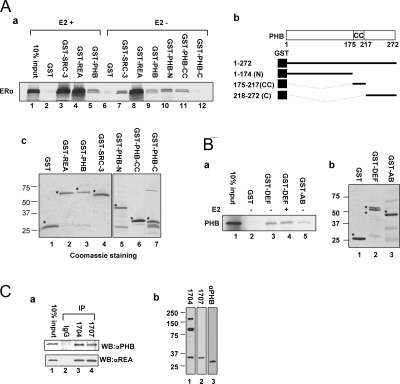
Because it has been shown that corepressors can directly interact with ERα (30), we performed a glutathione S-transferase (GST) pull-down assay to assess the interaction of ERα with PHB. Shown in Fig. 2A-a, GST-REA (lanes 4 and 8) and GST-PHB (lanes 5 and 9) fusion proteins bound well to ERα in comparison to GST alone (lanes 2 and 6). The interactions appear to be hormone independent (Fig. 2A-a, compare lane 4 with 8, and lane 5 with 9). As a control, the GST-SRC-3 showed E2-enhanced interaction with ERα in the presence of E2 (Fig. 2A-a, compare lane 3 with 7). Figure 2A-c indicates the amounts of each GST fusion protein used in these binding assays.
Figure 2.
ERα, PHB and REA Directly Interact with Each Other
A-a, GST pull-down shows a direct interaction between ERα and PHB, which is hormone independent. The interaction domains of PHB with ERα map to the NH2 terminus and the CC domain. As a control, the interaction between SRC-3 and ERα is enhanced by E2 treatment. A-b, Schematic diagram of PHB, indicating the location of the NH2 terminus [amino acids (aa) 1–174, N], CC domain (aa 175–217, CC), and carboxyl terminus (aa 218–272, C) is shown. A-c, Amounts of each GST fusion protein used in the pull-down assays. Stars indicate the expected sizes for each expressed proteins. B-a, Reciprocal GST pull-down experiment confirms a direct interaction between ERα and PHB in a hormone-independent manner. B-b, The amounts of each GST fusion protein used. AB, NH2-terminal regulatory domain that contains aa 1–180; DEF, includes hinge region, ligand binding domain, and C-terminal variable region, which contains aa 251–595. C-a, Protein extract from MCF7 cells were immunoprecipitated with two antibodies specifically against different epitopes of REA. Western blot analysis demonstrated the in vivo association of PHB with REA. As a negative control, nonspecific IgG could not precipitate PHB. C-b, In Western blot (WB) analysis, anti-REA antibodies BL1704 and BL1707 do not recognize PHB, whereas an anti-PHB antibody (rabbit polyclonal; Santa Cruz Biotechnology) can only recognize PHB.
To localize the regions of the PHB protein responsible for its interaction with ERα, GST pull-down experiments were conducted in a similar manner. The schematic presented in Fig. 2A-b illustrates the domain structure of PHB and different GST-fusion proteins used in the pull-down experiments. Shown in Fig. 2A-a, GST-PHB (1–272, full length), GST-PHB-N (1–174, NH2 terminus), and GST-PHB-CC (175–217, coiled-coil domain) interact with ERα, whereas GST-PHB-C (218–272, the carboxyl terminus of PHB) does not interact with the receptor (compare lane 9, 10, 11, and 12 with 6). The amounts of different PHB domains fused to GST used in these binding assays are shown in Fig. 2A-c. Taken together, these results indicate that the NH2 terminus and CC domains of PHB interact with ERα in vitro.
To further confirm the interaction between ERα and PHB, reciprocal GST pull-down experiments were conducted. The in vitro-translated, 35S-labeled PHB was able to interact with the GST-ERα DEF domain (Fig. 2B-a, compare lane 3 with 2) and also with the GST-ERα AB domain to a lesser extent (Fig. 2B-a, compare lane 5 with 2 and 3). The interaction appears to be E2 independent because interaction between GST-ERα-DEF and ERα is not changed by E2 treatment (Fig. 2B-a, compare lane 3 with lane 4). Figure 2B-b indicates the amounts of GST-ERα-DEF and GST-ERα-AB fusion protein used in these binding assays. Interestingly, GST-ERα-DEF migrates as two bands. The duplicated bands were also detected by in vitro-translated full-length ERα (Fig. 2A-a, input in lane 1) and by in vitro-translated ERα DEF (data not shown), suggesting that ERα DEF may be partially degraded.
It was reported that PHB and REA directly interact with each other in yeast and human fibroblast cells (39,40,41). To investigate whether these corepressors interact in the MCF-7 breast cancer cells, a coimmunoprecipitation assay was conducted with whole cell lysates of MCF-7 cells with two rabbit antibodies against REA BL1704 and BL1707. As shown in Fig. 2C-a, both antibodies efficiently precipitated PHB and REA. Importantly, in Western blot analyses, neither BL1704 nor BL1707 recognize PHB (Fig. 2C-b), indicating that there is no cross-reaction between these two REA antibodies and PHB protein. These data demonstrate that, as in fibroblast-derived cells, PHB associates with REA in a breast epithelium-derived cell line.
Depletion of Endogenous PHB Enhances Expression of ERα Target Genes
To further establish the corepressor function of PHB on ERα-mediated transcription, we performed small interfering RNA (siRNA) knock down experiments to reduce the expression of endogenous PHB (and REA as control). As indicated in Fig. 3, A-a and A-b, both 10- and 40-nm concentrations of the corresponding SMART pool siRNAs can efficiently deplete the mRNA of PHB and REA, as determined by quantitative real-time PCR. Importantly, siRNA against REA had no effect on PHB mRNA and vice versa, indicating that each siRNA SMART pool specifically depletes its intended target mRNA. Consistent with that seen for its mRNA, the PHB siRNA also efficiently reduced the level of the PHB protein (Fig. 3A-c). Similarly, REA siRNA reduced the REA protein level, as shown in Fig. 3A-c. Interestingly, REA siRNA also reduced the PHB protein level, whereas PHB siRNA reduced the REA protein level to a similar extent. This phenomenon where PHB and REA depend on each other for protein stability in MCF-7 cells is in agreement with previous observations from other studies in yeast (PHB1 and PHB2) and HeLa cells (42,43).
Figure 3.
Depletion of Endogenous PHB Increases the Expression of ERα Target Genes in MCF-7 Cells
A-a and A-b, siRNA against PHB and REA specifically depleted the mRNA of PHB and REA in MCF-7 cells, respectively, as determined by real-time PCR. A-c, The protein levels of PHB and REA were simultaneously reduced by siRNA against either PHB or REA, indicating that PHB and REA depend on each other for protein stability. B, In MCF-7 cells, expression of ERα target gene pS2 was strongly increased by depletion of REA, and moderately increased by depletion of PHB (B-a). Expression of cyclin D1 mRNA was significantly increased by depletion of PHB, but not by depletion of REA (B-b). Individual siRNA against REA mRNA nos. 2 and 3 caused a strong increase of pS2 gene expression, whereas siRNA nos. 1 and 4 only show slight effect (B-c). This is in agreement with the knock down efficiency of REA protein level with individual siRNAs (B-e). Similarly individual siRNAs against PHB mRNA nos. 2–4 caused a moderate but statistically significant increase of cyclin D1 (CCND1) gene expression, whereas siRNA no. 1 only show little effect (B-d). Shown here is a representative of three experiments (±sd). *, P < 0.05). This again is in agreement with the knock down efficiency of PHB protein level by these individual siRNAs (B-f). C, Recruitment of PHB to endogenous pS2 gene promoter. MCF-7 cells were grown in stripped media for 3 d, and treated with vehicle or 10 nm E2 for 45 min. ChIP assays were conducted with antibodies against ERα, SRC-3, and PHB. The input lanes represent for 5% of total genomic DNA used in ChIP assay. RNAi, RNA interference.
We next investigated the effect of depletion of the corepressors on expression of ERα target genes in MCF-7 cells. Shown in Fig. 3B, knock down of REA significantly increased the expression of the E-regulated pS2 gene as determined by real-time PCR, whereas knock down of PHB moderately increased the expression of pS2 mRNA. On the other hand, knock down of PHB significantly increased the expression of cyclin D1, whereas knock down of REA had little effect on cyclin D1 expression. The expression of cyclin D1 mRNA was stimulated with E2 treatment by about 1.5-fold. This moderate stimulation is consistently obtained, in agreement with previous reports that cyclin D1 promoter is a complex promoter that is regulated by numerous transcriptional factors in addition to ERα (44). Our data indicate that PHB functions as an ERα corepressor on endogenous ER target gene promoters in a promoter-dependent manner in breast epithelium-derived cells.
To rule out the possibility that the increased expression of ER target genes is due to off target effects from siRNA, we next transfected MCF-7 cells with individual siRNAs against REA or PHB and measured the pS2 and cyclin D1 mRNA expression levels. As shown in Fig. 3B-c, the level of pS2 mRNA level was significantly increased by transfection of REA siRNA nos. 2 and 3 but much less substantially affected by siRNA nos. 1 and 4. Western blot analysis (Fig. 3B-e) confirmed that REA siRNA nos. 2 and 3 more efficiently depleted REA protein, whereas nos. 1 and 4 are much less effective. Similarly we found that transfection of MCF-7 cells with PHB siRNA nos. 2–4 caused a moderate but statistically significant increase in cyclin D1 mRNA (Fig. 3B-d) (*, P < 0.05). Western blot analysis (Fig. 3B-f) confirmed that PHB siRNA nos. 2–4 are more effective in the depletion of PHB protein than siRNA no. 1.
Association of Endogenous PHB with the pS2 Gene Promoter
It has been shown that in a tetracycline inducible system, PHB is recruited to the promoter of the p53 target gene, MDM2 (38). We next wanted to see whether PHB directly interact with promoter of the ER-regulated pS2 gene. ER and coactivators have been shown to be recruited to a classical E response element located on pS2 promoter (45,46). Figure 3C shows that ERα and SRC-3 weakly associated with the pS2 promoter before E2 treatment, and that treatment with E2 significantly increased both ERα and SRC-3 recruitment. In contrast to that seen for ERα and SRC-3, endogenous PHB was detected on pS2 promoter before E2 treatment, and hormone treatment dismissed PHB from the promoter.
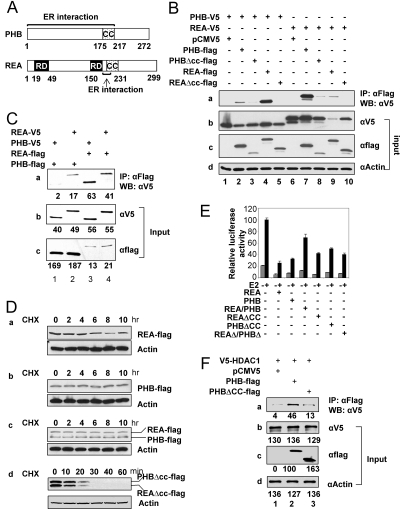
The CC Domains of PHB and REA Are Critical for Their Oligomerization, Stabilization, and Cross-Squelching Actions
Figure 4A illustrates the location of functional domains in both PHB and REA. CC domains are located in the middle of both PHB (175–217) and REA (150–231). Interestingly, the ERα-binding domains overlap with CC domains. The CC domains are responsible for protein dimerization and oligomerization. It has been shown that PHB can either form a homomer (47) or hetero-oligomerize with REA (40). As indicated from Fig. 4B, coimmunoprecipitation experiments demonstrate that in cultured 293T cells, both PHB and REA can interact with themselves (Fig. 4B-a, lanes 2 and 9); this interaction is dependent on their CC domains because PHB and REA mutants lacking CC domains barely interacted with themselves (Fig. 4B-a, lanes 3 and 10). Interestingly, heteromerization between these two corepressors appears much stronger than homomerization (Fig. 4B-a, compare lanes 2 and 9 with lanes 4 and 7). Similar to that seen above, the ability of PHB and REA to heteromerize is dependent on their CC domains as shown by lack of heteromerization between PHB and REA mutants, which lack CC domains (Fig. 4B, lanes 5 and 8). Figure 4B, b–d, indicates the amounts of protein inputs for the different tagged PHB and REA wild-type and mutants and β-actin. Interestingly, the steady-state level of V5-tagged PHB when coexpressed with flag-tagged REA (seen in Fig. 4B-b, lane 4) is significantly higher than in cases where V5-tagged PHB is cotransfected with other vectors, indicating that heteromerization with REA stabilizes the PHB protein. It is also noteworthy that the steady-state level of V5-tagged REA, when coexpressed with flag-tagged REA (seen in Fig. 4B-b, lane 9), is consistently lower than in cases where V5-tagged REA is cotransfected with other expression vectors, suggesting that V5-tagged REA became unstable when coexpressed with flag tagged REA. The above results argue that PHB and REA preferentially form heteromers instead of homomers; the heteromers are formed through interactions between the CC domains of the two proteins.
Figure 4.
The CC Domains of PHB and REA Are Required for Hetero-Oligomerization, Stabilization, and Transcriptional Cross-Squelching
A, Schematic diagram of functional domains of PHB and REA. B, Transient expression of carboxyl-terminal V5 or flag tagged PHB and REA wild-type and CC deletion mutants in 293T cells. Protein extracts were first immunoprecipitated (IP) with anti-flag antibody, and precipitated protein complexes were analyzed by Western blot (WB) analysis with an anti-V5 antibody. Bands in lanes 1–5 in B-a represent the immunoprecipitated V5-tagged PHB, whereas bands in lanes 6–10 represent the immunoprecipitated V5-tagged REA. B-b through -d, Input control of different tagged PHB and REA wild type and mutants and β-actin. C, REA-V5, PHBV5, REA-flag, and PHB-flag were overexpressed in 293T cells separately. D, Flag-tagged PHB and REA wild types or CC deletion mutants were expressed in 293T cells. The protein synthesis inhibitor cycloheximide (CHX) was added to a final concentration of 200 μg/ml and cells were harvested at the indicated time points. Protein stability was measured by Western blot analysis with anti-flag antibody (top panels of D-a through -d), and anti-β-actin antibody as an input control (bottom panels of D-a through -d). E, The CC domains are required for the cross-squelching actions between PHB and REA. The HepG2 cells were cotransfected with ERE-luc reporter vector, expression vectors of ERα (5 ng), REAΔCC (300 ng), PHBΔCC (300 ng), REAΔ/PHBΔ (150 ng of REAΔCC and 150 ng of PHBΔCC), REA (100 ng), PHB (100 ng), REA/PHB (50 ng of REA and 50 ng of PHB), REA (300 ng), PHB (300 ng), or REA/PHB (150 ng of REA and 150 ng of PHB). F, V5-HDAC1, PHB-flag, and PHBΔCC were overexpressed in 293T cells separately. Western blot analysis was used to determine the amounts of cell lysates that contain equal levels of PHB wild type and PHBΔCC. After this, the cell lysates containing either contain PHB-flag or PHBΔCC-flag was mixed with equal amounts of cell lysates containing V5-HDAC1. After overnight incubation, the cell lysates were immunoprecipitated with beads linked to anti-flag antibodies, and probed with antibodies against V5.
Because coexpression of PHB and REA results in significant stabilization of each protein, we cannot rule out the possibility that the stronger bands in Fig. 4B-a, lanes 4 and 7, are influenced by the amount of protein expressed in the cells. To further confirm whether PHB and REA preferentially form hetero-oligomers, we expressed the REA-V5, PHB-V5, REA-flag, and PHB-flag separately in 293 cells. Western blot analysis was used to determine the amounts of cell lysates that contain the same amounts of REA-V5 and PHB-V5. We then mixed equal amounts of REA-V5 and PHB-V5 with these cell lysates that contained either REA-flag or PHB-flag. After overnight incubation at 4 C, the proteins were immunoprecipitated with anti-flag antibody-linked beads. Shown in Fig. 4C-a, the band in lane 2 is much stronger than the band in lane 1, indicating that PHB/REA hetero-oligomerization is much stronger than PHB homo-oligomerization. Similarly, in Fig. 4C-a, the band in lane 3 is stronger than the band in lane 4, indicating that PHB/REA hetero-oligomerization is stronger than REA homo-oligomerization. Quantitations of input proteins are shown in Fig. 4, C-b and C-c.
Also consistent with their reported ability to stabilize each other in yeast and HeLa cells (42,43), Fig. 3A-c reveals that depletion of REA via RNA interference results in depletion of PHB to a similar extent in MCF-7 cells, and vice versa. To further test the hypothesis that PHB and REA stabilize each other by forming heteromers that are dependent on their CC domains, we measured the protein half-lives of PHB and REA (wild-type and CC deletion mutants). 293T cells were transfected with carboxyl-terminal flag tagged PHB, and/or REA wild type or mutants. The protein synthesis inhibitor cycloheximide was then added and cells were harvested at the indicated time points. Results in Fig. 4, D-a and D-b, show that when wild-type flag tagged PHB or REA was overexpressed alone, the half-life of PHB is more than 10 h, whereas the half-life of REA is about 8 h. When both flag-tagged proteins were coexpressed in the 293T cells, their half-lives exceed 10 h (Fig. 4D-c). In contrast, the half-lives of PHB and REA CC deletion mutants are about <20 min (Fig. 4D-d). These results indicate that the CC domains of both PHB and REA are essential for establishing their protein stability.
Our results in Fig. 1 indicated that there is a cross-squelching of corepressor function between PHB and REA. Because these two closely related proteins tend to form heteromers, it is possible that PHB-REA heteromers do not function as corepressors, and that the corepressor function of either PHB or REA likely arises from the unpaired forms of these two proteins. To test this hypothesis, we measured the corepressor activities of PHB and REA CC deletion mutants. Figure 4E shows that both PHB and REA ΔCC mutants repress ERα-mediated transcription, but to a lesser extent than their wild-type counterparts. Importantly, coexpression of the CC deletion mutants of PHB and REA did not result in cross-squelching of their repressive function, in contrast to the cross-squelching effect observed when wild-type PHB and REA were coexpressed. These data suggest that the cross-squelching effect between PHB and REA is dependent on the mutual interaction between these two proteins through their CC domains.
Because the CC domain of PHB was shown to directly interact with histone deacetylase HDAC1 (47), we tested whether the PHB CC deletion mutant loses its interaction with HDAC1 and is responsible for its reduction of repressive action. Because the PHB CC deletion mutant is much less stable than PHB wild type, we overexpressed PHB ΔCC, PHB wild type, and V5-tagged human HDAC1 separately in 293T cells. By Western blot analysis, we first substantiated that the amounts of cell lysate used contained the same levels of wild type and ΔCC PHB. Then we mixed the wild type and ΔCC cell lysate with equal amounts of HDAC1 cell lysate. After incubation at 4 C, the proteins were immunoprecipitated with anti-flag antibody linked agarose beads. Shown in Fig. 4F, wild-type PHB can efficiently immunoprecipitate V5-tagged HDAC1 (panel a, compare lane 2 with 1), whereas PHB ΔCC could not (panel a, compare lane 3 with 1). The amounts of input proteins were shown in Fig. 4F, b–d. Our results are in agreement with the observation that the corepressor activity of PHB ΔCC is reduced in comparison to PHB wild type (Fig. 4E). However, the fact that PHBΔCC is still able to partially repress ER activity indicates that recruitment of HDAC1 by the CC domain is not the only mechanism responsible for its ability to repress ER-mediated transcription.
Abrogation of PHB Function Results in Mouse Embryonic Lethality
To determine whether PHB exhibits a corepressor role in vivo, we created two separate lines of PHB knockout mice from two ES cells clones, in which the PHB gene was disrupted by a gene-trap vector. One ES cell clone was obtained from the Sanger Institute Gene Trap Resource (SIGTR) (cell-line ID: XT0035), whereas the other ES cell clone was obtained from Baygenomics Consortium (San Francisco, CA) (cell-line ID: BGB069). PHB knockout mice were derived from both ES clones and analyzed in parallel throughout this study. Because the results obtained from the two lines of PHB knockout mice are essentially the same, only data from ES clone XT0035 is presented here.
Information in Fig. 5A provides details of the PHB targeting mutation for ES cell clone XT0035. The gene-trap targeting vector is comprised of the En2 intron, a splice acceptor site, a β-geo fusion cassette composed of β-galactosidase and the neomycin resistance marker, and a simian virus 40 polyadenylation signal. This gene-trap targeting vector was inserted into the coding region of exon 7 of the mouse PHB gene thereby disrupting the expression of this gene through the deletion of the evolutionarily conserved carboxyl-terminal 27 amino acids of PHB. The insertion site was confirmed by PCR analysis followed by sequencing verification (data not shown). Using a neo gene fragment as a hybridizing probe, Southern analysis demonstrated that insertion into the PHB gene is the only insertion in ES cell clone XT0035 (Fig. 5C). Additionally, LacZ-positive staining indicates that the PHB promoter is active in this ES cell clone (Fig. 5B).
Figure 5.
Generation of PHB Knockout Mice
A, PHB gene trapped ES cell clone XT0035 was obtained from Sanger Institute Gene Trap Resource. The gene-trap targeting vector is comprised of the En2 intron, a splice acceptor site, a β-geo fusion cassette composed of β-galactosidase and the neomycin resistance marker, and a simian virus 40 polyadenylation signal. The targeting vector was inserted into the coding region of last exon. B, The ES cells were stained with X-gal. The blue staining indicates PHB gene promoter is active in ES cells. C, Southern blot analysis using neo probe to demonstrate the single locus insertion of the target vector. The genomic DNA purified from the ES clone XT0035 were digested with NsiI (lane 1) and SpeI (lane 2). Neither the restriction enzyme NsiI nor SpeI cuts the gene trap vector. The predicted sizes of restriction fragments resulting from NsiI and SpeI digestion are 14.6 and 16.7 kb, respectively. D, Epithelial cells were isolated from inguinal mammary glands of wild-type (WT) and PHB+/− mice, and total proteins were extracted, subject to Western blot (WB) analysis with the antibodies against PHB, REA, and β-actin. Each lane represents the protein extract from the epithelial cells isolated from two mice. E, The wild-type and PHB+/− female siblings were treated with a standard 3-wk E-P treatment regimen, which induces mammary gland ductal side branching and alveologenesis. Proteins were extracted from the whole mammary glands. Western blot analysis was conducted using different antibodies against cyclin D1 (CCND1), PHB, REA, and β-tubulin (E-a). The relative protein levels of REA, PHB, and cyclin D1 were quantitated and shown in E-b.
To study the role of PHB gene in mouse embryonic development, PHB+/− mice were crossed to obtain mice homozygous for the PHB null mutation. As shown in Table 1, live PHB−/− pups were not obtained, demonstrating that PHB in mouse, like REA (33), is essential for early embryo development.
Table 1.
Number of Pups Obtained
| No. of Pups | No. (%) of Pups
|
|||
|---|---|---|---|---|
| WT | PHB+/− | PHB−/− | ||
| Weaned | 146 | 51 (35%) | 95 (65%) | 0 (0%) |
| E8.5 | 17 | 5 (29%) | 12 (71%) | 0 (0%) |
| E3.5 | 48 | 14 (29%) | 22 (46%) | 12 (25%) |
To determine when embryonic lethality occurs during embryogenesis, we isolated E8.5 embryos from PHB+/− females after mating with PHB+/− male mice. A total of 17 embryos were genotyped, and no PHB−/− embryos were detected, indicating that embryonic lethality occurs before E8.5. We next determined whether PHB−/− embryos can survive to the blastocyst stage. PHB+/− females were superovulated, and mated with PHB+/− male mice. E3.5 blastocysts were flushed out from the uterus. PCR genotyping showed that among 48 blastocysts, 12 were PHB−/−, indicating that PHB−/− embryos can survive to the blastocyst stage. Next we determined whether PHB−/− embryos can survive to embroynic d 6.5 (E6.5). Because wild-type mouse E6.5 embryos are so small that they are barely able to be isolated from the decidual balls. We reasoned that PHB−/− embryos would be even smaller and too small to be isolated from the decidual balls even if they were able to survive to E6.5 stage. Therefore, we fixed the decidual balls, and all decidual balls were completely serially sectioned. Under a microscope, we examined the presence of embryos in each section. Surprisingly, 16 decidual balls (24% of the total 68 decidual balls isolated from PHB+/− females which mated with PHB+/− males) do not contain noticeable embryos (Table 2). Figure 6A, a and c, shows hematoxylin and eosin (H&E) staining of one representative decidual ball that contains normal E6.5 embryo. Figure 6A, b and d, shows H&E staining of one representative decidual ball that does not contain a noticeable embryo. As a control, all of the 37 decidual balls isolated from PHB+/− females who mated with wild-type males contain embryos of normal size (Table 2). Our data indicate that PHB−/− embryos die before the E6.5 stage. The presence of decidual balls that do not contain embryos suggests that PHB−/− embryos survive long enough for implantation and decidualization but are subsequently resorbed.
Table 2.
Number of Decidual Balls
| No. of E6.5 Decidual Balls Sectioned | # (%) of Decidual Balls
|
No. of Decidual Balls per Litter | ||
|---|---|---|---|---|
| Contain Embryo | No Embryo | |||
| PHB+/− × PHB+/− | 68 | 52 (76%) | 16 (24%) | 7.0 |
| WT × PHB+/− | 37 | 37 (100%) | 0 (0%) | 7.3 |
Figure 6.
H&E and Immunohistochemistry Staining of E6.5 Decidual Balls
E6.5 decidual balls isolated from PHB+/− females mated with PHB+/− male were fixed in 10% formaldehyde, sectioned, and subject to H&E and immunohistochemistry staining. A-a and A-c, The H&E staining of a representative decidual ball containing a normally developed E6.5 embryo at two different magnifications. A-b and A-d, H&E staining of a representative decidual ball which does not contain noticeable embryo. PDZ, SDZ, and EM denote the primary decidual zone, secondary decidual zone, and embryo, respectively. B-a and B-c, Immunohistochemistry staining of a representative E6.5 embryo with anti-PHB antibody. B-b and B-d, Negative control staining in which the anti-PHB primary antibody was not included. B-e, Anti-PHB staining of another section of E6.5 embryo. B-f, Anti-β-galactosidase staining of the consecutive section of E6.5 embryo used for B-e. exec, Extraembryonic ectoderm; ep, epiblast; pac, pro-amniotic cavity.
To investigate the molecular mechanism by which PHB deficiency causes embryonic lethality before E6.5 stage, we stained the E6.5 embryo sections with anti-PHB antibody. Data in Fig. 6B, a and c, indicate that PHB is highly expressed in extraembryonic ectoderm, the ectoplacental cone, and the epiblast, although at a lower level. As a negative control, Fig. 6B, b and d, did not show any positive staining when the primary antibody was not included. Because β-galactosidase expression is under control of the PHB gene promoter in our PHB+/− mouse line, we conducted immunohistochemistry staining with an anti-β-galactosidase antibody. Figure 6B-f shows strong staining in extraembryonic ectoderm, ectoplacental cone, and also epiblast for this β-galactosidase reporter. This staining pattern is similar to staining with anti-PHB antibody (6B-e), confirming the specificity of anti-PHB antibody.
PHB and REA Stabilize Each Other in Vivo
To confirm that the protein level of PHB is reduced in the mice heterozygous for the PHB null allele, and to determine whether PHB is expressed in mammary gland epithelial cells, epithelial cells were isolated from mammary glands of both wild-type and PHB+/− female virgin mice. Western blot analysis in Fig. 5D shows that PHB and REA are both expressed in wild-type mammary epithelial cells, and their levels are both reduced in PHB+/− mice. These data demonstrate that PHB and REA are both expressed in mouse mammary epithelial cells in conjunction with ERα. Also, just as seen in cell culture, PHB and REA proteins depend on each other to achieve their higher steady-state expression level in vivo.
Reduction of PHB Levels Results in Accelerated Mammary Growth in Response to Steroid Hormone Treatment
Because PHB is an ERα corepressor in cultured cells, we hypothesized that the mammary glands of PHB+/− mice should exhibit a hyperproliferative phenotype. To test this hypothesis, wild-type and PHB+/− female siblings were treated with an established 3-wk E-P treatment regimen (48), which induces mammary gland ductal side branching and alveologenesis. Data in Fig. 5E show that protein levels of PHB and REA were reduced in E-P-treated PHB+/− mouse mammary glands (Fig. 5E-a, compare lanes 1–3 with 4–6), whereas the protein level of cyclin D1 was increased in E-P-treated PHB+/− mouse mammary glands (Fig. 5E-a, compare lanes 1–3 with 4–6). The relative levels of each protein were quantitated, normalized against α-tubulin and are presented as mean ± sd. *, P < 0.05 (Fig. 5E-b).
Whole mount analysis at low-power magnification reveals a similar overall ductal patterning in the E-P-treated PHB+/− mammary gland compared with the similarly treated wild type [Fig. 7, compare panel A (wild type) to B (PHB+/−)], but examination at high-power reveals significantly more alveolar lateral budding and ductal side-branching in the hormone-treated PHB+/− mammary gland compared with wild type (Fig. 7, compare panels C and D). Increased alveolar budding was confirmed by histological analysis (Fig. 7A, compare panels E and F) in which the hormone-treated PHB+/− mammary gland shows a 1.96-fold increase in alveolar bud number per field (Fig. 7G; **, P < 0.01). 5-Bromo-2-deoxyuridine (BrdU) staining confirmed that there is increased cell proliferation in PHB+/− mammary gland in comparison to the E-P-treated wild-type mammary gland (Fig. 7, compare panels H and I). Average percentages of mammary epithelial cells (±sd) scoring positive for BrdU staining in wild type and PHB+/− mammary gland were shown in Fig. 7J. These data confirm an important in vivo role for the PHB corepressor in controlling steroid-induced mammary morphogenesis.
Figure 7.
The Mammary Glands of PHB Heterozygous Mice Showed Hyper-Proliferation after E-P Treatment
A and B, Whole mounts of inguinal mammary glands from E-P-treated wild type (WT) (A) and PHB+/− (B) mice, respectively (LN, lymph node). C and D, Higher magnifications of regions of panels A and B, respectively. E and F, H&E-stained sections of mammary glands shown in panels A and B, respectively. Compared with the E-P-treated wild-type mouse gland, note the significant increase in alveologenesis and ductal side branching (black arrowhead) in the E-P-treated PHB+/− mouse gland. G, Alveolar bud number per field (±sd). **, P < 0.01) in E-P-treated wild-type and PHB+/− mouse glands. H and I, 5-Bromo-2-deoxyuridine (BrdU) incorporation in mammary gland sections. H, Luminal epithelial cells of wild-type mammary gland, whereas panel I shows the luminal epithelial cells of PHB+/− mammary gland. J, Average percentages of mammary epithelial cells scoring positive for BrdU staining (±sd). *, P < 0.05) in E-P-treated wild-type and PHB+/− mouse glands.
DISCUSSION
In a search for tumor suppressor genes, PHB was first cloned in rat liver cells in the late 1980s (49,50). The human prohibitin gene is located on chromosome 17q12–21 in close proximity to the familial breast cancer gene BRCA1 (51). Somatic mutations in the PHB protein coding region have been identified in breast cancers (52,53), suggesting that PHB may be a tumor suppressor. PHB belongs to a family of proteins which carry an evolutionarily conserved domain, the PHB domain (35). Another member of the prohibitin family, REA, also known as PHB2, has been shown to be an ERα binding protein that represses ERα-mediated transactivation and enhances the inhibitory effectiveness of 4-hydroxytamoxifen-bound ERα (30,31,32). These observations underscored REA as a potent cellular modulator of E responsiveness. Our current study demonstrates that PHB also functions as an ERα transcriptional corepressor. In addition to HepG2 cells tested in Fig. 1A, we have also tested in CV1, HeLa, MCF-7 and T47D cells. PHB showed similar repressive activity toward ERα-mediated transcription in those cell lines (data not shown), indicating that PHB-mediated transcriptional repression is not cell line specific. We demonstrate that both PHB and REA can counteract SRC-3 enhanced ERα transcriptional activity, supporting a role for these corepressors as counterbalances to SRC-3’s influence on ERα-mediated signaling in vitro. This also is in agreement with a previous study showing that REA can counteract SRC-1 coactivator function (30,31). Although the mechanism underlying the counteraction of SRC-3 activity has yet to be delineated, the fact that PHB and REA can directly interact with ERα and recruit HDAC (54,55) suggests that these transcriptional repressors both compete with SRC-3 for ERα binding and also recruit HDACs that suppress ERα-mediated transactivation.
It has been shown that knockout of the REA gene results in a lethal phenotype during early embryo development in the mouse (33). Our current studies demonstrate that the PHB knockout exhibits a similar lethal phenotype during early embryogenesis. Together, these data indicate that PHB and REA have essential and potent in vivo roles in controlling cell growth and differentiation during murine embryogenesis, and that these corepressors are involved in cell signaling events that may be independent of ERα during prenatal development. The fact that these corepressors have been shown to interact with E2F-1 (56,57), Rb (36,55,56), p53 (38), Brg1/Brm (37,58), androgen receptor (59), and AKT (60) supports the idea that these proteins affect other biological targets in addition to ERα, which may be responsible for their embryonic functions. Apart from the nucleus, moreover, localization of some of these corepressors to the mitochondrial and plasma membranes (40,61) suggests that the phenotypes of REA- and PHB-deficient animals could result in part from their activities in these other cellular compartments.
Consistent with being an ERα corepressor, our studies show that mammary glands from adult mice heterozygous for the PHB null mutation (PHB+/−) exhibit early signs of epithelial hyperplasia, similar to that observed in REA+/− mice (34). Our previous studies also demonstrate that the REA+/− uterus displays a hyperproliferative response to E exposure (33). Collectively, these studies support an important role for the PHB family in controlling E-initiated growth responses in a subgroup of female target tissues (mammary gland and uterus) in vivo.
Our data demonstrate that PHB and REA can form both homomers and heteromers, although they strongly prefer to heteromerize. To further support this, as a part of the Nuclear Receptor Signaling Atlas (NURSA) project, the REA-associated protein complex has been purified from the HeLa cell lysates with two different antibodies against REA (J. Qin, personal communication). Both antibodies could efficiently precipitate PHB in addition to REA, although there is no cross interaction between PHB protein and the REA antibodies. Coomassie blue staining showed that PHB and REA exist in approximately 1:1 stoichiometric ratio in the REA coregulator complex, supporting that endogenous PHB and REA also preferentially form hetero-oligomers. Interestingly, our data show that there is cross-squelching of transcriptional repression between PHB and REA, indicating that heteromerization of PHB and REA inactivates their corepressor function. As shown in Fig. 8, we propose that PHB and REA repress transcription only when they are not paired, and the heteromer forms of PHB and REA are inefficient as transcriptional corepressors, and could act in mitochondria and the plasma membrane to exert other functions. This idea is in agreement with the observation that their CC domains are not only responsible for heteromerization, but also participate in interaction with ERα (31). Our observation that PHB and REA have preferential ERα target gene corepressor effects (REA more effectively corepresses the pS2 gene, whereas PHB corepresses the cyclin D1 gene) also supports the idea that PHB and REA function as coregulator homomers to selectively repress transcription when separated from each other. Another important question raised is how the equilibrium between homomer and heteromer forms of PHB and REA is regulated. The ability of CC domains to interact with each other has been shown to depend upon its phosphorylation state (62,63) and pH (64). Related to this point, we have observed that both PHB and REA are phosphorylated in MCF-7 cells (data not shown), suggesting that PHB and REA heteromerization and thereby their corepressor activities may be influenced by kinase signaling pathways.
Figure 8.
Working Model for Multifunctional Proteins PHB and REA
PHB and REA are able to interact with each other to form hetero-oligomers. There is equilibrium between homomer and heteromer forms of PHB and REA. We propose that, only when they are not paired, PHB and REA can function as transcriptional regulators for a variety of transcriptional factors, including ERα (30), E2F1 (36), PR-B, and AR (59). On the other hand, the hetero-oligomers of PHB and REA exert other functions, such as mitochondrial chaperones (40) and B-cell receptor-associated proteins (71). Moreover, it has been reported that PHB acts as a vascular marker of adipose tissue (72), inhibitor of pyruvate carboxylase (73), propigmentation effector (74), and plays critical role in Ras signaling pathway (75). It remains unknown whether REA is also involved in these biological processes as PHB partner.
In summary, our studies provide strong evidence that the potential tumor suppressor PHB functions as an ERα transcriptional corepressor and opposes the oncogenic coactivator protein SRC-3. Herein we propose a novel mechanism by which the corepressor activities of PHB and REA can be restrained by their heteromerization. We hypothesize that the balance between promotion (by SRC-3) and inhibition (by the PHB family) of the E-initiated proliferation signal in mammary epithelial cell is key to maintaining mammary gland growth homeostasis. By extension, an imbalance in activity of coactivators and corepressors (i.e. from overexpression of a coactivator or reduction in expression of one or both PHB family members) would likely lead to excessive cellular proliferation, ultimately promoting the progression of cancer or other pathologies.
MATERIALS AND METHODS
Plasmids
cDNAs for human PHB and REA were amplified by RT-PCR from HeLa cell total RNA followed by sequencing verification. Mammalian expression vectors for PHB and REA were constructed by ligation of PHB and REA PCR products between EcoRI and XbaI sites within the pCMV5 vector (Invitrogen Corp., Carlsbad, CA). Vectors expressing GST fusion proteins were constructed by PCR amplification of the corresponding regions and ligated into the pGEX vectors (Invitrogen). The V5 (peptide representing amino acid residues 95–108 (GKPIPNPLLGLDST) of RNA polymerase α subunit of simian virus 5) or flag-tagged expression vectors for PHB and REA were constructed by standard cloning techniques into the same pCMV5 vector. The deletion of CC regions were generated through a double PCR strategy as previously described (65). A mammalian expression vector for human HDAC1 was provided by Dr. Jiemin Wong (Baylor College of Medicine). This was used to construct a NH2-terminal V5-tagged form through standard PCR and cloning techniques. Mammalian expression plasmids used to express ERα, PR-B, E2 promoter binding factor (E2F1), transcription factor DP1 (TFDP1), GalVP16, SRC-3, a cyclin E promoter luciferase reporter vector (CycE-LUC), mouse mammary tumor virus luciferase reporter vector (MMTV-LUC), and an E-responsive luciferase reporter vector (pERE-E1b-LUC) have been described previously (66). A mammalian expression plasmid for p53 and a MDM2 promoter luciferase reporter vector (MDM2-LUC) were provided by Dr. Xiongbin Lu (Baylor College of Medicine).
Antibodies
For coimmunoprecipitation, affinity purified rabbit polyclonal anti-REA antibodies BL1704 and BL1707 (Bethyl Laboratories, Inc., Montgomery, TX) were used. For Western blot analysis, rabbit polyclonal anti-REA antibody was purchased from Upstate Biotechnology (Lake Placid, NY). Rabbit polyclonal anti-PHB and anti-cyclin D1 antibodies were purchased from Lab Vision Corp. (Fremont, CA). Rabbit polyclonal anti-flag antibody was purchased from Affinity BioReagents, Inc. (Golden, CO) and the mouse monoclonal anti-V5 antibody was purchased from Invitrogen. Rabbit polyclonal anti-BrdU antibody was obtained from Amersham Biosciences, Inc. (Piscataway, NJ). A rabbit polyclonal anti-β-galactosidase was purchased from Cortex Biochem (San Leandro, CA). Mouse monoclonal anti-α-tubulin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture, Transient Transfection, and Luciferase Assay
Human hepatocellular carcinoma HepG2 cells (American Type Culture Collection) were maintained in 5% CO2 at 37 C in Eagle’s MEM (Invitrogen). Cells were plated at 2 × 105 cells/well in 12-well tissue culture plates and transfected with the indicated concentrations of expression vectors and pERE-Luc reporter vector using Fugene 6 transfection reagent (Roche Diagnostics, Indianapolis, IN). Twenty-four hours after transfection, the indicated hormones, E2 or synthetic progestin (R5020), were added when appropriate. Luciferase activity was determined 24 h later using the Promega Luciferase Assay kit according to the manufacturer’s protocols (Promega Corp., Madison, WI). MCF-7 cells were maintained in DMEM supplemented with 10% fetal calf serum.
In Vitro Protein Interaction Assay
GST-PHB (1–272), GST-PHB (1–174), GST-PHB (175–217), and GST-PHB (218–272), GST-SRC-3 (581–840), GST-REA (1–299), GST-ERα-DEF (251–595), and GST-ERα-AB (1–180) were expressed from pGEX-4T-1 as GST fusion proteins. The GST fusion proteins were expressed in Escherichia coli XL1-Blue cells treated with 0.5 mm isopropyl-β-d-thiogalactopyranoside; extracted in 0.15 m NaCl, 5 mm EDTA, 10% glycerol, 100 μm phenylmethylsulfonyl fluoride, 10 mm dithiothreitol, and 50 mm Tris-HCl (pH 8.0); and incubated with glutathione-agarose beads (Amersham Biosciences) as described (65). Full-length human ERα protein was in vitro-translated in the presence of 25 μCi of [35S]methionine (PerkinElmer Life Sciences, Boston, MA) from the pCR3.1hERα vector using a TNT T7 quick-coupled transcription/translation system (Promega Corp., Madison, WI). Washed beads were boiled in sodium dodecyl sulfate (SDS)-containing buffer. Input lanes contained 10% of the binding reactions.
Immunoprecipitation and Immunoblotting
Forty-eight hours after transfection, cells were harvested and washed once with PBS. The cells were then disrupted in lysis buffer [50 mm Tris (pH 8.0), 100 mm NaCl, 0.5% Nonidet P-40, 50 mm NaF, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Roche Diagnostics)]. After incubation at 4 C for 1 h, the lysates were centrifuged, and the supernatant was incubated with 10 μl of anti-Flag-M2 Affinity Gel (Sigma Corp., St. Louis, MO) at 4 C for 4 h. After centrifugation at 1000 rpm for 3 min at 4 C, the pellet was washed three times with lysis buffer. Samples were separated on 4–15% polyacrylamide gels containing SDS. After electrophoresis, proteins were electrophoretically transferred to nitrocellulose membranes and the blots were incubated with the indicated antibodies. Immunoreactive bands were visualized using chemiluminescence (SuperSignal West Pico chemiluminescent substrate; Pierce Chemical, Rockford, IL). Chromatin immunoprecipitation (ChIP) was conducted as previously described (67). The sequences of PCR primers used to amplify the pS2 promoter have been previously described (68).
RNA Interference and Real-Time PCR
SMART pool siRNAs used to knock down PHB and REA expression were obtained from Dharmacon Research (Lafayette, CO), and transfected at the concentrations indicated. Before the siRNA transfection, MCF-7 cells were switched to phenol red-free DMEM containing 5% charcoal dextran-stripped fetal calf serum. siRNAs were introduced into cells using TransIT-TKO (Mirus Corp., Madison, WI) as a transfection agent. Twenty-four hours after transfection, the medium was replaced with fresh medium. Three days after transfection, E2 (10−8 m) or its ethanol vehicle was added. The cells were harvested and total RNA was isolated with Trizol reagent (Invitrogen). To measure the relative mRNA levels of pS2 and cyclin D1 genes, real-time RT-PCR was performed using the Taqman RT-PCR one-step master mix in conjunction with an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA). Each sample was tested in duplicate in three independent experiments.
ES Cell LacZ Staining and Southern Blot Analysis
Two ES clones, in which the PHB gene was disrupted by a gene-trap vector, were obtained from the Sanger Institute Gene Trap Resource (SIGTR, Cambridge, UK) (cell-line ID: XT0035) and Baygenomics Consortium (San Francisco, CA) (cell-line ID: BGB069). Both ES cell clones were cultured in 1× the Glasgow modification of Eagle’s MEM supplemented with 2 mm glutamine, 1 mm sodium pyruvate, 1× nonessential amino acids, 10% (vol/vol) fetal bovine serum, a 1:1,000 dilution of β-mercaptoethanol stock solution, and 1000 U/ml of leukocyte inhibitory factor. For LacZ staining, the ES cells were first placed in a fixation buffer [0.1 m phosphate buffer (pH 7.3), 5 mm EGTA, 2 mm MgCl2, and 0.2% glutaraldehyde] for 15 min at room temperature. The cells were washed twice with wash buffer [0.1 m phosphate buffer (pH 7.3), 2 mm MgCl2], then incubated in staining buffer [0.1 m phosphate buffer (pH 7.3), 2 mm MgCl2, 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, and 1 mg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside)] for overnight at 37 C. The next day, the staining buffer was aspirated and kept in the wash buffer until photomicrographs were taken.
Genomic DNA isolated from ES cells was digested with restriction enzymes overnight as previously described (48), and separated on an 0.8% agarose gel, then transferred to a nitrocellulose membrane. A neomycin resistance gene (neo) gene fragment was used as probe to demonstrate that the gene trap vector was inserted into a single insertion site.
Using standard procedures, ES cells were injected into the blastocoel of embryonic D3.5 mouse embryos to obtain PHB chimeric mice. Mice heterozygous for the PHB null mutation (F1) were obtained by breeding male PHB chimeric mice with C57 female mice.
General Mouse Manipulations, Hormone Treatments, and Histological Analysis
The mice were housed in a temperature-controlled (22 ± 2 C) room with a 12-h light, 12-h dark photocycle and fed rodent chow meal (Purina Mills, Inc., St. Louis, MO) and given fresh water ad libitum. All mice were treated humanely in accordance with institutional and Institutional Animal Care and Use Committee guidelines.
To study mammary ductal side branching and alveologenesis, 6-wk-old virgin female mice received E and P from a beeswax pellet (implanted sc in the intrascapular region) that delivered 1 μg E and 1 mg P daily for 3 wk (48). The inguinal mammary glands were processed for whole-mount staining as previously described (48).
Mammary glands were fixed, processed, embedded, and sectioned as previously described (48,69). H&E staining was performed on the mammary tissue sections as previously described (48). To BrdU incorporation, mice were injected ip with BrdU (Amersham Biosciences, Piscataway, NJ) at 0.1 ml/10 g of body weight 2 h before the mice were killed. For each tissue section, cell counting consisted of scoring the number of BrdU-stained cells in a random field of 1000 cells per section (12). The average number of BrdU-stained cells in a given tissue section was obtained by taking the average BrdU-stained cells in three separate fields of 1000 cells per section. Final counts were expressed as a percentage of epithelial cells immunopositive for BrdU. Representative sections were used in these studies, and only intensively stained nuclei were scored positive.
Collection of blastocysts was performed as previously described (70). Briefly, 4-wk-old female PHB+/− mice were superovulated by treatment with pregnant mare serum gonadotropin and human chorionic gonadotropin. The female mice were placed with male PHB+/− mice right after human chorionic gonadotropin injection. The E3.5 blastocysts were flushed out of the uterus, lysed and subjected to PCR genotyping.
The decidual balls at E6.5 were fixed with 10% formaldehyde in PBS and embedded in paraffin. The decidual balls were then completely serially sectioned, and each section was examined under microscope to determine the presence of embryos in the decidual ball.
Mammary Epithelial Cell Protein Isolation, Whole Mammary Gland Protein Isolation, and Western Blot Analysis
Mammary glands of 8-wk-old female mice were disrupted in homogenization buffer [20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EGTA, 1% Triton X-100, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail] until uniform. The samples were incubated on ice for 1 h and vortexed every 20 min. The samples were then centrifuged at 15,000 × g for 20 min, and supernatants were centrifuged for another 20 min. The supernatant protein concentration was determined using the BCA reagent (Bio-Rad, Richmond, CA), and samples were separated on 4–15% polyacrylamide SDS gels. To isolate epithelial cells from mouse mammary glands, the inguinal mammary glands were removed from host mice and lymph nodes discarded. Glands were placed in DMEM:F12 solution buffered with HEPES (pH 7.6) containing 2 mg/ml collagenase A and 100 U/ml of hyaluronidase. Glands were minced into very small pieces (<1 mm) with a razor blade, and then shaken at 37 C at 110 rpm for 2 h until the tissues were thoroughly digested. The digested material was then centrifuged at 1000 × g for 5 min. The supernatant was aspirated, and the pellet was washed 5 times with 10 ml of sterile PBS. Finally, the pellet was resuspended in lysis buffer [50 mm Tris (pH 8.0), 100 mm NaCl, 0.5% Nonidet P-40, 50 mm NaF, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail]. Western blot analyses were repeated at least three times for each protein.
Acknowledgments
We thank Drs. Austin J. Cooney, Sophia Tsai, and Ming-Jer Tsai, and Drs. Michael Lewis and Peili Gu at Baylor College of Medicine for their helpful discussions. The technical assistance of Jie Han, Jie Li, and Yan Ying is gratefully acknowledged.
Footnotes
This research was supported by the following National Institutes of Health and private grants: HD-07857 (to B.W.O.), HD-42311 (to F.J.D.), CA-07730, the Susan G. Komen Breast Research Cancer Program (to J.P.L.), and CA-18119 (to B.S.K.).
Disclosure Statement: Nothing to disclose.
First Published Online October 11, 2007
Abbreviations: BrdU, 5-Bromo-2-deoxyuridine; CC, coiled-coil domains; ChIP, chromatin immunoprecipitation; E, estrogen; E2, estradiol; E6.5, embryonic d 6.5; ER, estrogen receptor; GST, glutathione S-transferase; HDAC, histone deacetylase; H&E, hematoxylin and eosin; NR, nuclear receptor; P, progesterone; PR-B, progesterone receptor B; REA, estrogen receptor activity; PHB, prohibitin; SDS, sodium dodecyl sulfate; siRNA, small interfering RNA; SRC, steroid receptor coactivator.
References
- Evans RM 1988 The steroid and thyroid hormone receptor superfamily. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MJ, O’Malley BW 1994 Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 63:451–486 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG 2000 The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14:121–141 [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW 2006 The expanding cosmos of nuclear receptor coactivators. Cell 125:411–414 [DOI] [PubMed] [Google Scholar]
- O’Malley BW 2006 Molecular biology. Little molecules with big goals. Science 313:1749–1750 [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O’Malley BW 1997 Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194–198 [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM 1997 Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569–580 [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y 1996 The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953–959 [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR 1999 Regulation of transcription by a protein methyltransferase. Science 284:2174–2177 [DOI] [PubMed] [Google Scholar]
- Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O’Malley BW 1998 Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922–1925 [DOI] [PubMed] [Google Scholar]
- Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P 2002 The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol 22:5923–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Soyal SM, Fernandez-Valdivia R, Gehin M, Chambon P, Demayo FJ, Lydon JP, O’Malley BW 2006 Steroid receptor coactivator 2 is critical for progesterone-dependent uterine function and mammary morphogenesis in the mouse. Mol Cell Biol 26:6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O’Malley BW 2000 The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci USA 97:6379–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rose DW, Hermanson O, Liu F, Herman T, Wu W, Szeto D, Gleiberman A, Krones A, Pratt K, Rosenfeld R, Glass CK, Rosenfeld MG 2000 Regulation of somatic growth by the p160 coactivator p/CIP. Proc Natl Acad Sci USA 97:13549–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG 1995 Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397–404 [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM 1995 A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454–457 [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM 1997 Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373–380 [DOI] [PubMed] [Google Scholar]
- Berrevoets CA, Umar A, Trapman J, Brinkmann AO 2004 Differential modulation of androgen receptor transcriptional activity by the nuclear receptor co-repressor (N-CoR). Biochem J 379:731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kawate H, Ohnaka K, Nawata H, Takayanagi R 2006 Nuclear compartmentalization of N-CoR and its interactions with steroid receptors. Mol Cell Biol 26:6633–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Baek SH, Bourk EM, Ohgi KI, Garcia-Bassets I, Sanjo H, Akira S, Kotol PF, Glass CK, Rosenfeld MG, Rose DW 2006 Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell 124:615–629 [DOI] [PubMed] [Google Scholar]
- Yoon HG, Wong J 2006 The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol 20:1048–1060 [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG 1995 Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J 14:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I, Dawson J, Lavinsky RM, Glass CK, Rosenfeld MG, Lazar MA 1997 Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA 94:14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressel U, Thormeyer D, Altincicek B, Paululat A, Eggert M, Schneider S, Tenbaum SP, Renkawitz R, Baniahmad A 1999 Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol Cell Biol 19:3383–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Beaudoin 3rd GM, DeRenzo CL, Zarach JM, Chen SH, Thompson CC 2001 The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev 15:2687–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M,Vadlamudi RK, Kumar R 2001 Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol 3:30–37 [DOI] [PubMed] [Google Scholar]
- Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH 2003 Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell 11:139–150 [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA 1999 The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402:93–96 [DOI] [PubMed] [Google Scholar]
- Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, Parks DJ, Moore JT, Kliewer SA, Willson TM, Stimmel JB 2002 Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature 415:813–817 [DOI] [PubMed] [Google Scholar]
- Montano MM, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen BS 1999 An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc Natl Acad Sci USA 96:6947–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage-Mourroux R, Martini PG, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS 2000 Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J Biol Chem 275:35848–35856 [DOI] [PubMed] [Google Scholar]
- Martini PG, Katzenellenbogen BS 2003 Modulation of estrogen receptor activity by selective coregulators. J Steroid Biochem Mol Biol 85:117–122 [DOI] [PubMed] [Google Scholar]
- Park SE, Xu J, Frolova A, Liao L, O’Malley BW, Katzenellenbogen BS 2005 Genetic deletion of the repressor of estrogen receptor activity (REA) enhances the response to estrogen in target tissues in vivo. Mol Cell Biol 25:1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussi P, Liao L, Park SE, Ciana P, Maggi A, Katzenellenbogen BS, Xu J, O’Malley BW 2006 Haploinsufficiency of the corepressor of estrogen receptor activity (REA) enhances estrogen receptor function in the mammary gland. Proc Natl Acad Sci USA 103:16716–16721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow IC, Parton RG 2005 Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic 6:725–740 [DOI] [PubMed] [Google Scholar]
- Wang S, Nath N, Adlam M, Chellappan S 1999 Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 18:3501–3510 [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang B, Faller DV 2002 Prohibitin requires Brg-1 and Brm for the repression of E2F and cell growth. EMBO J 21:3019–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S 2003 Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem 278:47853–47861 [DOI] [PubMed] [Google Scholar]
- Back JW, Sanz MA, De Jong L, De Koning LJ, Nijtmans LG, De Koster CG, Grivell LA, Van Der Spek H, Muijsers AO 2002 A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci 11:2471–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA 2000 Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J 19:2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Model K, Langer T 2005 Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol Biol Cell 16:248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasashima K, Ohta E, Kagawa Y, Endo H 2006 Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J Biol Chem 281:36401–36410 [DOI] [PubMed] [Google Scholar]
- Berger KH, Yaffe MP 1998 Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol Cell Biol 18:4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Silva MD, Shang Y, Donaher JL, Brown M, Weinberg RA 2001 AIB1 enhances estrogen-dependent induction of cyclin D1 expression. Cancer Res 61:3858–3862 [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M 2000 Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F 2003 Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- Joshi B, Ko D, Ordonez-Ercan D, Chellappan SP 2003 A putative coiled-coil domain of prohibitin is sufficient to repress E2F1-mediated transcription and induce apoptosis. Biochem Biophys Res Commun 312:459–466 [DOI] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery Jr CA, Shyamala MG, Conneely OM, O’Malley BW 1995 Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev 9:2266–2278 [DOI] [PubMed] [Google Scholar]
- McClung JK, Danner DB, Stewart DA, Smith JR, Schneider FL, Lumpkin CK, Dell’Orco RT, Nuell MJ 1989 Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun 164:1316–1322 [DOI] [PubMed] [Google Scholar]
- Nuell MJ, Stewart DA, Walker L, Friedman V, Wood CM, Owens GA, Smith JR, Schneider EL, Dell’Orco R, Lumpkin CK, Danner DB, McClung JK 1991 Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol Cell Biol 11:1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen HM, Smith SA, Mazoyer S, Fujimoto E, Stevens J, Williams B, Rodriguez P, Cropp CS, Slijepcevic P, Carlson M, Robertson M, Bradley P, Lawrence E, Harrington T, Sheng ZM, Hoopes R, Sternberg N, Brothman A, Callahan R, Ponder BAJ, White R 1994 A physical map and candidate genes in the BRCA1 region on chromosome 17q12–21. Nat Genet 7:472–479 [DOI] [PubMed] [Google Scholar]
- Sato T, Saito H, Swensen J, Olifant A, Wood C, Danner D, Sakamoto T, Takita K, Kasumi F, Miki Y, Skolnick M, Nakamura Y 1992 The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res 52:1643–1646 [PubMed] [Google Scholar]
- Sato T, Sakamoto T, Takita K, Saito H, Okui K, Nakamura Y 1993 The human prohibitin (PHB) gene family and its somatic mutations in human tumors. Genomics 17:762–764 [DOI] [PubMed] [Google Scholar]
- Kurtev V, Margueron R, Kroboth K, Ogris E, Cavailles V, Seiser C 2004 Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J Biol Chem 279:24834–24843 [DOI] [PubMed] [Google Scholar]
- Wang S, Fusaro G, Padmanabhan J, Chellappan SP 2002 Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 21:8388–8396 [DOI] [PubMed] [Google Scholar]
- Wang S, Nath N, Fusaro G, Chellappan S 1999 Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol Cell Biol 19:7447–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Joshi B, Dasgupta P, Morris M, Wright K, Chellappan S 2006 Prohibitin facilitates cellular senescence by recruiting specific corepressors to inhibit E2F target genes. Mol Cell Biol 26:4161–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang B, Faller DV 2004 BRG1/BRM and prohibitin are required for growth suppression by estrogen antagonists. EMBO J 23:2293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble SC, Chotai D, Odontiadis M, Dart DA, Brooke GN, Powell SM, Reebye V, Pal P, Gkaliagkousi E, Seager M, Ferro A 2007 Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene 26:1757–1768 [DOI] [PubMed] [Google Scholar]
- Sun L, Liu L, Yang XJ, Wu Z 2004 Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J Cell Sci 117:3021–3029 [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Artal SM, Grivell LA, Coates PJ 2002 The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci 59:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilak L, Moitra J, Krylov D, Vinson C 1997 Phosphorylation destabilizes α-helices. Nat Struct Biol 4:112–114 [DOI] [PubMed] [Google Scholar]
- Szilak L, Moitra J, Vinson C 1997 Design of a leucine zipper coiled coil stabilized 1.4 kcal mol−1 by phosphorylation of a serine in the e position. Protein Sci 6:1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV 2001 Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11:82–88 [DOI] [PubMed] [Google Scholar]
- He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, Wilson EM 2004 An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J Biol Chem 279:30643–30653 [DOI] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O’Malley BW 2006 Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 26:7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai MJ, O’Malley BW 2003 Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol Cell Biol 23:3763–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wu RC, Sandquist J, Wong J, Tsai SY, Tsai MJ, Means AR, O’Malley BW 2005 Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol Cell Biol 25:9687–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon JP, Ge G, Kittrell FS, Medina D, O’Malley BW 1999 Murine mammary gland carcinogenesis is critically dependent on progesterone receptor function. Cancer Res 59:4276–4284 [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E 1994 Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Terashima M, Kim KM, Adachi T, Nielsen PJ, Reth M, Kohler G, Lamers MC 1994 The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J 13:3782–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W 2004 Reversal of obesity by targeted ablation of adipose tissue. Nat Med 10:625–632 [DOI] [PubMed] [Google Scholar]
- Vessal M, Mishra S, Moulik S, Murphy LJ 2006 Prohibitin attenuates insulin-stimulated glucose and fatty acid oxidation in adipose tissue by inhibition of pyruvate carboxylase. FEBS J 273:568–576 [DOI] [PubMed] [Google Scholar]
- Ni-Komatsu L, Orlow SJ 2007 Identification of novel pigmentation modulators by chemical genetic screening. J Invest Dermatol 127:1585–1592 [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T 2005 Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol 7:837–843 [DOI] [PubMed] [Google Scholar]