Introduction
The rapid development and application of neuroimaging over the past twenty years has led to a better understanding of brain function in health and disease. New techniques such as Magnetic Resonance Imaging (MRI) and including Diffusion Tensor Magnetic Resonance Imaging (DTI) provide powerful capabilities for non-invasive studies of the brain (1-2). Image analysis techniques also play a critical role in the extraction of meaningful information that is relevant to biomarkers for diagnosis, prognosis and following of treatment response. Moreover, the emergence of increasingly sophisticated mathematical models, image processing and visualization tools enables the comprehensive mapping of brain structure and function.
Publications in peer-reviewed journals and scientific conferences provide dissemination of new scientific principles, and deployment of the image processing algorithms, including support that enables reusability, is hampered by many factors. These include the lack of calibration and validation of the techniques required to achieve meaningful reproducibility of results. Another factor is the many sources of variability in neuroimage data. The benefits of the developed algorithms can be limited by a lack of support for finish engineering, such as providing for multi-platform support, upgrades and training materials. The major part of the development of new image analysis tools within the medical imaging community occurs at a local scale, based on focused efforts of individual research groups. As a result, valuable non-commercial software is blocked from reaching the broader scientific and clinical research communities.
Many of these obstacles have been identified by multiple groups as part of a joint effort towards the identification and characterization of the opportunities for scientific research and engineering development in biomedical imaging (3-6). To solve these challenges, the NIH Roadmap initiative has laid out a vision for a more efficient and productive system of medical research. Essential elements in the NIH Roadmap and the research pipeline are the translational steps, and global consortia such as the National Alliance for Medical Image Computing (NA-MIC) and the Center for Computational Biology (CCB), two of the seven National Centers for Biomedical Computing (NCBC), focus their efforts on the conversion of scientific advances from the biomedical imaging community into working open-source systems, so as to improve the availability and deployment of these tools on a national scale (7-9). The NA-MIC consortium involves clinical researchers in the definition of the tools that will address their hypotheses, computer scientists in the design and implementation of the corresponding algorithms, and engineers for ensuring robustness and usability of the open-source software toolset made available to the broader community. This movement in general, and the NA-MIC consortium in particular, builds upon the open-source effort such as the National Library of Medicine’s Insight Toolkit (ITK) (10), the Visualization Toolkit (VTK) (11), and the CMake build environment (12). These efforts include the integration of the best available practices in software development, including architecture design, software engineering process and automated quality assurance. Cutting-edge biomedical computing algorithms thus become accessible through end-user applications compatible with major computer platforms.
Still, such technology and knowledge-sharing systems face the challenge of translating scientific advances made by engineers and computer scientists to a broader community of clinical research scientists. It is not enough to make a new technology accessible, its deployment must be converted into new skills for these scientists and clinicians, and the most sophisticated mathematical algorithms will achieve little impact on biomedical discoveries if they do not reach the clinical researchers for whom they were developed. Potential solutions must therefore be implemented in order to bridge this gap and to ensure that clinical researchers are able to use new tools in their investigations. In this paper, we propose a perspective for translating advances in neuroimage analysis to clinical researchers.
Requirements for transferring advances in neuroimage analysis
We have identified three requirements for an ideal translation of the techniques developed by computer scientists and engineers into clinical applications: a multidisciplinary approach, a balance between theory and practice, and an immersive collaborative environment.
Multidisciplinary
The challenges inherent to posing appropriate hypotheses, designing accurate experimental paradigms to test them, developing the technologies required for image acquisition and extracting relevant image information necessary to advance our understanding of brain function are highly interdisciplinary. Any attempt at transferring new neuroimage analysis tools needs to combine clinical needs and engineering solutions.
Theory into practice
The use of advanced image analysis techniques often involves both a conceptual understanding of the underlying theory and a practical knowledge of how to use the software tool (e.g., the input parameters and processing pipeline). Scientists and clinicians interested in existing cutting-edge neuroimage analysis tools need a complementary approach of theory and practice, in order to take full advantage of the techniques and to be able to develop new skills.
Immersion in a collaborative environment
One of the difficulties in transferring medical image analysis tools developed by computer-scientists to clinicians and medical researchers is the lack of communication between the two communities. Computer-scientists often speak in terms of technology, while clinicians speak in terms of health and diseases. Bridging the communication gap requires immersion in a collaborative environment that brings together clinicians and scientists, and where the focus is on fostering interactions as well as facilitating the transfer of tacit knowledge through in-depth discussions.
For the past two years, we have been developing an initiative based on the three criteria presented for transferring advances in neuroimage analysis to clinical research applications at a national scale. In the paragraphs below, we report an example for Diffusion Tensor Imaging analysis.
Exemplar Case: Diffusion Tensor Imaging Tractography
The development of Diffusion Tensor Imaging (DTI) has led to the possibility of studying in-vivo the connectivity of white matter in the brain. The analysis of DTI data provides anisotropy maps that give insight into the structure and orientation of fiber tracts in healthy subjects, and provides clues to disturbances in disease. In particular, streamline tractography provides visualization resources to investigate major fiber bundle trajectories. Such techniques allow for quantitative comparison of white matter anatomy that reveals meaningful differences between normal and diseased patient populations. To transfer these advances to scientists and medical researchers, we developed a workshop meeting the three stated requirements for translation of advances in neuroimage analysis into clinical applications. Our workshop was focused on the clinically driven goal of studying the structure and orientation of the inferior occipito-frontal fasciculus, which is part of the temporal stem, a region known to be dysfunctional in schizophrenic individuals (13-14). This fasciculus, which connects the frontal lobe to the occipital lobe through the temporal lobe, presents key challenges for image analysis as it passes through several areas of crossing fibers that confound the tractography solutions.
Our workshop was organized in four hands-on sessions that guided the participants through a logical progression, from data loading and visualization to tractography seeding from user-defined regions of interest. We analyzed selected exemplar datasets with the open-source 3DSlicer package developed by the NA-MIC consortium (15). The sessions provided the audience with theoretical background on the specificity of tensor data, and practical experience regarding DTI analysis and visualization. At the end of the workshop, all the participants could successfully compute and visualize the inferior occipito-frontal fasciculus bundle trajectory from the dataset provided. Figure 1 demonstrates the results of this fiber tractography analysis.
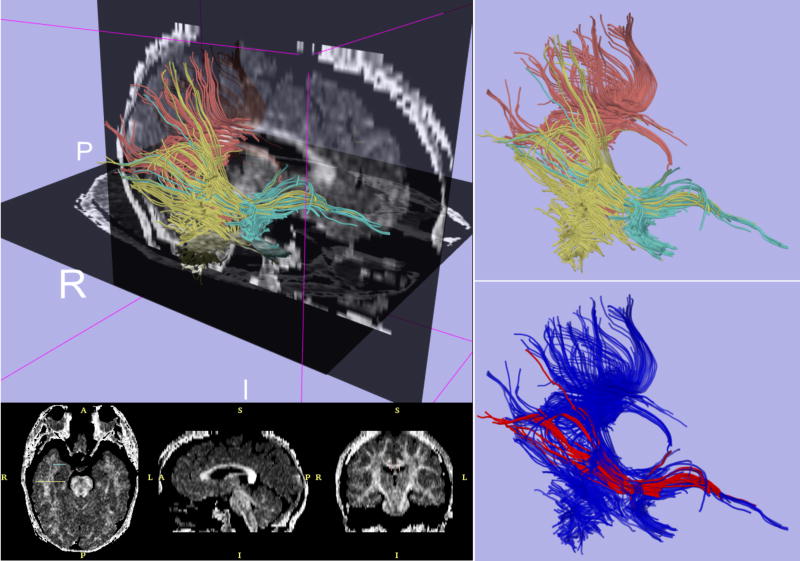
Figure 1. Exemplar case for Diffusion Tensor Imaging Tractography.
Upper left panel displays fiber tractography combined with sagittal and axial slices from a fractional anisotropy map computed with 3DSlicer. Upper-right panel shows fiber tracts generated from regions of interest in the posterior-temporal lobe (yellow), splenium of the corpus callosum (pink), and temporal stem (green). Lower-right figure shows 3D visualization of the computed trajectory of the inferior occipito-frontal fasciculus bundle (red).
We delivered this 8-hour workshop multiple times, including once at the National Library of Medicine, as part of the dissemination events presented at the NIH Roadmap National Centers for Biomedical Computing (NCBC) All-Hands Meeting (16).
In two years, we delivered 10 hands-on workshops to 370 clinicians and scientists following the model presented above. The geographical distribution of attendees showed that our workshops gathered a broad audience representing more than 50 different universities from the medical and scientific communities. The areas of expertise of the participants were equally distributed among medical imaging (23%), computer science (24%), and neuroscience (25%). 43% of the participants had a doctorate level, with a proportion of two thirds of PhDs and one third of MDs. We diversified our workshops in two main categories: general introduction to advanced neuroimaging analysis and specialized programs for expert research groups. To ensure compatibility with the different operating systems used in hospital and research laboratories, all analyses were designed and tested on major computer platforms. Our syllabus includes topics such as statistical analysis of functional Magnetic Resonance Imaging (fMRI) data and automatic cortical segmentation in structural MRI data. We made all of the courses, exemplar datasets and software tools of our compendium publicly available to the scientific community (17). The number of clinical researchers that attended our sessions, and the 12,000 hits on our compendium webpage are promising outcomes. We collected feedback from a sample of 64 voluntary respondents. Preliminary results showed that the majority of participants estimated both the content of the courses and the speed of the sessions to be appropriate. Participants’ ratings of the quality of the workshops were uniformly good to excellent. Future work includes the validation of the impact of our courses on the participants’ learning, as well as the development of an evaluation procedure for the long-term benefit of our initiative on their research.
Conclusion and perspectives
The evolution of brain imaging and neuroimage analysis has seen tremendous advances. However, translating the latest image processing algorithms developed by computer-scientists to clinical research applications remains a challenge. The long term risk of conducting parallel activities is the delayed benefit for patients: engineers and scientists developing a technology with a theoretical interest to the computer science community, but in search of a clinical use, and clinicians and medical researchers in search of a solution that might already exist. Cutting-edge studies that cross disciplines would benefit from complementary activities towards a clinically driven goal. Pragmatic steps need to be taken to maximize the long-term positive impact of consortia gathering the two communities.
In this report, we highlighted three criteria for facilitating the transfer of advances in neuroimaging techniques: a multidisciplinary approach, the combination of theory and practice, and an immersive collaborative environment. We presented a concrete implementation of our initiative for Diffusion Tensor Imaging analysis. This preliminary work focused on image processing techniques that are critical to brain studies based on neuroimage analysis findings. The life cycle of such studies also involve steps prior to the analysis, such as defining the hypothesis, screening for subjects who meet the inclusion/exclusion criteria, doing clinical and diagnostic measures and bringing the subjects to the magnet. The approach presented in this article could be extended to the transmission of such knowledge from the clinical researchers’ community to the computer scientists’ community.
Sharing the advances of neuroimaging and translating the latest analysis tools into new skills of clinical researchers could speed the discovery of new scientific principles. Our initiative addressed the vision expressed by the biomedical imaging community for fostering interdisciplinary collaborations: clinical researchers involved in developing the tools needed to address their hypotheses and computer scientists interested in developing algorithms related to clinical applications.
This paradigm could be deployed in other domains of biomedical research with similar obstacles to the successful translation of engineering solutions to clinical researchers, and ultimately to patient care.
Acknowledgments
The authors wish to acknowledge Professor Martha Shenton, Director of the Psychiatry Neuroimaging Laboratory, Harvard Medical School, for her thoughtful comments on the manuscript.
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sonia Pujol, Surgical Planning Laboratory, Harvard Medical School, Department of Radiology, L1-050, Brigham and Women’s Hospital, 75 Francis Street, Boston MA 02115
Ron Kikinis, Surgical Planning Laboratory, Harvard Medical School, Department of Radiology, L1-050, Brigham and Women’s Hospital, 75 Francis Street, Boston MA 02115
Randy Gollub, Athinoula A. Martinos Center for Biomedical Imaging, Harvard Medical School, Department of Psychiatry, Massachusetts General Hospital, Building 149 - 13th Street - Room 2660, Charlestown, MA 02129-2000
References
- 1.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66(1):259–67. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44(4):625–32. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 3.Carson PL, Giger M, Welch MJ, et al. Biomedical Imaging Research Opportunities Workshop: report and recommendations. Radiology. 2003;229(2):328–39. doi: 10.1148/radiol.2292030807. [DOI] [PubMed] [Google Scholar]
- 4.Hendee WR. Biomedical Imaging Research Opportunities Workshop II: a summary of findings and recommendations. Med Phys. 2005;32(8):2484–6. doi: 10.1118/1.1947196. [DOI] [PubMed] [Google Scholar]
- 5.Hendee WR. Biomedical Imaging Research Opportunities Workshop III: summary of findings and recommendations. Radiology. 2006;238(2):402–4. doi: 10.1148/radiol.2382051462. [DOI] [PubMed] [Google Scholar]
- 6.Hendee WR. Special report: biomedical imaging research opportunities workshop IV--a summary of findings and recommendations. Radiology. 2007;242(2):338–41. doi: 10.1148/radiol.2422060733. [DOI] [PubMed] [Google Scholar]
- 7.http://www.na-mic.org
- 8.http://www.loni.ucla.edu/CCB/
- 9.http://www.bisti.nih.gov/ncbc/
- 10.Ibanez L, Schroeder W, Ng L, Cates J. The ITK software guide. Clifton Park, NY: Kitware Inc.; 2003. [Google Scholar]
- 11.Schroeder W, Martin K, Lorensen W. The Visualization Toolkit. Third. Clifton Park, NY: Kitware Inc.; 2004. [Google Scholar]
- 12.Martin K, Hoffman B. Mastering Cmake. Second. Clifton Park, NY: Kitware Inc.; 2006. [Google Scholar]
- 13.Kubicki M, Park H, Westin CF, et al. DTI and MTR abnormalities in schizophrenia: analysis of white matter integrity. Neuroimage. 2005;26(4):1109–18. doi: 10.1016/j.neuroimage.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns J, Job D, Bastin ME, Whalley H, Macgillivray T, Johnstone EC, Lawrie SM. Structural disconnectivity in schizophrenia: a diffusion tensor magnetic resonance imaging study. Br J Psychiatry. 2003;182:439–43. [PubMed] [Google Scholar]
- 15.http://www.slicer.org
- 16.http://www.bisti.nih.gov/ahm2006/index.htm
- 17.http://www.na-mic.org/Wiki/index.php/Slicer:Workshops:User_Training_101