Abstract
Small molecule inhibitors that target fms-like tyrosine kinase 3 (FLT3)–activating mutations have potential in the treatment of leukemias. However, certain mutations can simultaneously activate the tyrosine kinase, and confer resistance to small molecule inhibitors. We therefore tested the sensitivity of 8 FLT3 activation loop mutants to midostaurin. Each mutant conferred IL-3 factor–independent proliferation to Ba/F3 cells, and each resulted in the constitutive activation of FLT3 and its targets, signal transducer and activator of transcription 5 (STAT5) and extracellular stimuli-responsive kinase (ERK). For each mutant tested, midostaurin inhibited cell growth and phosphorylation of FLT3, STAT5, and ERK. In contrast, midostaruin did not inhibit Ba/F3 cells stably transduced with FLT3-internal tandem duplications containing a G697R mutation that confers resistance to midostaurin, demonstrating that midostaurin inhibition of FLT3 activation loop mutants was not due to off-target effects. We conclude that midostaurin is a potent inhibitor of a spectrum of FLT3 activation loop mutations, and that acute myeloid leukemia patients with such mutations are potential candidates for clinical trials involving midostaurin.
Introduction
Fms-like tyrosine kinase 3 (FLT3), a cell surface receptor tyrosine kinase, is among the most commonly mutated genes in acute myeloid leukemia (AML).1 Stimulation of FLT3 activates signal transduction pathways such as signal transducer and activator of transcription 5 (STAT5), RAS/mitogen-activated protein kinase (RAS/MAPK), phosphoinositide 3-kinase (PI3K), src homologous and collagen gene (SHC), SH2-containing inositol-5-phosphatase (SHIP), and cytoplasmic tyrosine phosphatase with 2 Src-homology 2 (SH2) domains (SHP2), which play important roles in cellular proliferation, differentiation, and survival.2,3
There are 2 types of activating mutations in FLT3 described in patients with leukemia. These include a spectrum of internal tandem duplications (ITD) occurring within the auto-inhibitory juxtamembrane domain,4–6 and activation loop mutations that include Asp835Tyr (D835Y), Asp835Val (D835V), Asp835His (D835H), Asp835Glu (D835E), Asp835Ala (D835A), Asp835Asn (D835N), Asp835 deletion and Ile836 deletion.7–10 These activating mutations result in constitutive phosphorylation and activation of FLT3, and subsequent activation of downstream targets.10,11
The importance of FLT3 mutations in the pathogenesis of leukemias has been well established and, in most studies, these have been shown to confer a poor prognosis, with decreased survival.12–14 Therefore, attention has been focused on developing small molecule inhibitors that target FLT3. Midostaurin (formerly known as PKC412) is a selective inhibitor of FLT3, as well as vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR), c-kit receptor tyrosine kinase (KIT), and fibroblast growth receptor 1 (FGFR-1).15–17 In vitro, midostaurin induces apoptosis in Ba/F3 cells that have been transformed to IL-3-independent growth by FLT3-ITD.18 Furthermore, midostaurin is effective in FLT3-ITD-induced disease in a murine bone marrow transplantation model.18
Midostaurin has been investigated in phase 1 and phase 2 studies, and is generally well tolerated. In a phase 2 study in patients with relapsed or refractory AML, single-agent midostaurin resulted in significant reductions in peripheral blood and bone marrow blasts, with a 35% response rate.19 Midostaurin also showed activity in a phase 1 trial in combination with daunorubicin and cytarabine in newly diagnosed AML, in particular those patients with FLT3-ITD mutations.20 Based on these promising findings, a randomized phase 3 Cancer and Leukemia Group B (CALGB)/Southwest Oncology Group (SWOG)/Eastern Cooperative Oncology Group (ECOG) Intergroup trial is planned to assess efficacy of induction chemotherapy with midostaurin in patients with mutant FLT3.
Studies have analyzed the sensitivity of FLT3-activating mutations to small molecule inhibitors, and have demonstrated that certain activation loop mutations simultaneously activate the kinase and confer resistance to small molecule inhibitors.21 Therefore, defining the precise spectrum of sensitivity of the range of FLT3 activation loop mutations to specific kinase inhibitors may be critical to determine patient eligibility for treatment, and evaluation of response to therapy. Here we report that midostaurin is a potent inhibitor of 8 activation loop mutations tested.
Materials and methods
DNA constructs and retroviral transduction
DNA constructs of FLT3-ITD, and FLT3 with D835A, D835E, D835H, D835N, D835V, D835 deletion, I836 deletion, and D835Y point mutations were created and cloned into the murine stem cell virus (MSCV)-neo vector as previously described.14,21 IL-3-dependent murine hematopoietic Ba/F3 cells were transduced as previously described.21,22 In addition, a DNA construct consisting of a FLT3 G697R kinase domain point mutation was cloned into the MSCV-FLT3-ITD vector and transduced into Ba/F3 cells.23 Transduced cells were grown in absence of IL-3 to confirm factor independence.
Midostaurin dosing
Midostaurin (Novartis Pharma, Basel, Switzerland) was prepared in a 10mM stock solution in DMSO and stored at −20°C. Serial dilutions were made in DMSO to obtain the final concentrations used for immunoblotting and cell growth assays.
Ba/F3 cell growth assays and dose-response curves
Each cell line (1.5 × 105 cells/mL) was grown with varying concentrations of midostaurin in DMSO for 48 hours. The number of viable cells was then determined by a colorimetric assay.21 Results are expressed as a percentage of viable cells after 48 hours in the presence of DMSO only.
Immunoblotting analysis
Immunoblotting was performed as previously described.21,24 Briefly, after incubating cells in the presence of DMSO alone or with various concentrations of midostaurin, cell lysates were prepared. Lysates were separated using SDS-PAGE and transferred onto a nitrocellulose membrane. For immunoblotting, the following primary antibodies were used: anti-phospho-FLT3 (Tyr 591; Cell Signaling, Beverly, MA), anti-FLT3/Flk-2 (S-18; Santa Cruz Biotech, Santa Cruz, CA), anti-phospho-STAT5 (Tyr 694; Cell Signaling), anti-STAT5a (L-20; Santa Cruz Biotech), anti-phospho-p44/42 MAPK (Thr202/Tyr204; Cell Signaling), anti-p44/42 MAPK (Cell Signaling). Antirabbit-immunoglobulin horseradish peroxidase (Amersham Biosciences, Piscataway, NJ) was used as a secondary antibody. Detection was performed by enhanced chemiluminescence.
Results and discussion
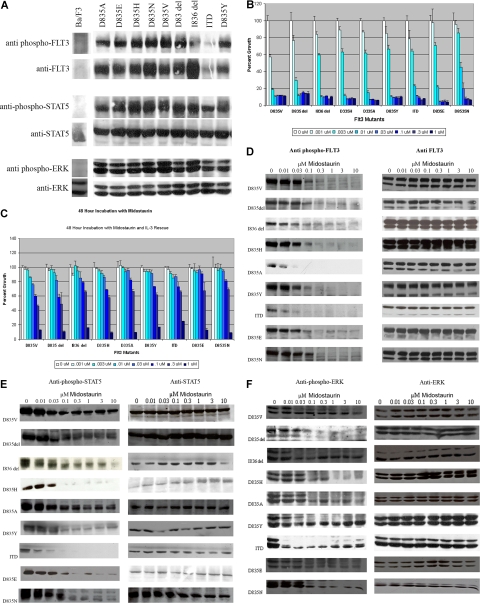
We confirmed that each of the activation loop mutations resulted in constitutive activation of FLT3 and its downstream effectors STAT5 and ERK. Figure 1A demonstrates that FLT3 was constitutively phosphorylated in all mutants. In addition, blotting with phospho-specific antibodies for STAT5 and ERK also demonstrated constitutive activation of these targets of activated FLT3.
Figure 1.
FLT3 activation loop mutants and the effects of midostaurin. (A) FLT3 activation loop mutations result in constitutive activation of FLT3, STAT5, and extracellular stimuli-responsive kinase (ERK). Ba/F3 cells expressing each of the FLT3 mutations were grown in the absence of IL-3. Cell lysates were prepared and the proteins resolved by sodium dodecyl sulfate–polyacrimide gel electrophoresis (SDS-PAGE). Immunoblotting was then performed using the indicated antibodies. (B) Midostaurin inhibits proliferation of Ba/F3 cells expressing FLT3-ITD and activation loop mutations. Ba/F3 cells expressing FLT3-ITD or FLT3 activation loop mutations were incubated with increasing concentrations of midostaurin for 48 hours. The number of viable cells was then determined using a colorimetric assay. Results are expressed as a percentage of viable cells after 48 hours of growth (± SD) in the presence of dimethyl sulfoxide (DMSO) without the drug. (C) Restoration of cellular proliferation by IL-3. The experiment was repeated in the presence of IL-3. Results are again expressed as a percentage of viable cells after 48 hours of growth (± SD) in the presence of DMSO without the drug. (D) Inhibition of FLT3 autophosphorylation by midostaurin. Ba/F3 cells expressing FLT3-ITD or FLT3 activation loop mutations were incubated with increasing concentrations of midostaurin for 20 minutes. Cell lysates were prepared, resolved by SDS-PAGE, then immunoblotted with either anti-phospho-FLT3 antibody or anti-FLT3 antibody, as indicated. (E) Inhibition of STAT5 phosphorylation by midostaurin. Whole cell lysates were prepared and resolved by SDS-PAGE as before, and immunoblotted with anti-phospho-STAT5 or anti-STAT5 antibodies, as indicated. (F) Inhibition of ERK phosphorylation by midostaurin. As before, cell lysates were prepared, resolved by SDS-PAGE, and then probed with anti-phospho–ERK or anti-ERK antibodies, as indicated.
Each of the transduced Ba/F3 cell lines was then tested for sensitivity to midostaurin. Ba/F3 cells expressing each of the activation loop mutations as well as FLT3-ITD were incubated with varying concentrations of midostaurin. At 48 hours, viable cells were quantitated using a colorimetric assay. There was inhibition of growth in all cell lines with increasing concentrations of midostaurin (Figure 1B). Inhibition of growth was abrogated by the addition of IL-3, supporting that inhibition by midostaurin is specific to the FLT3 pathway (Figure 1C). The cellular concentration that inhibited response by 50% (IC50) was less than 10 nM for all of the mutants tested, with a range of 1 nM to 10 nM (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), demonstrating that midostaurin resulted in a decrease in cellular proliferation of all of the Ba/F3 constructs at relatively low concentrations. Pharmacokinetic studies in phase 1 and 2 trials of midostaurin have shown that these are values that should be readily achieved in patients.25
To assess the effect of midostaurin on FLT3 phosphorylation, lysates were prepared from cells incubated with various concentrations of midostaurin, followed by immunoblotting with anti-phospho-FLT3 antibody. There was inhibition of FLT3 phosphorylation in all of the activation loop mutants at relatively low concentrations of midostaurin (Figure 1D), with an IC50 for inhibition of phosphorylation comparable to that observed for inhibition of cell growth. We also observed inhibition of phosphorylation of STAT5 and ERK, at comparably low concentrations of midostaurin (Figures 1E,F).
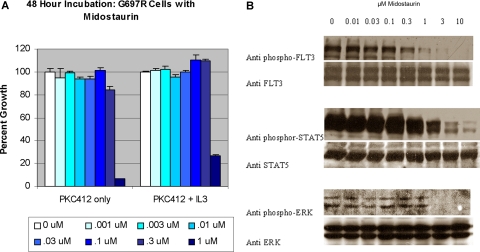
Thus, all of the activation loop mutants tested were sensitive to inhibition with midostaurin. However, midostaurin is known to inhibit kinases other than FLT3, and thus it was possible that the observed results, although consistent with FLT3 as a target of midostaurin, could be explained by simultaneous inhibition of other kinases essential for cell growth. To demonstrate that the effect of midostaurin was wholly attributable to inhibition of FLT3, and not due to off-target effects, we repeated the experiments using Ba/F3 cells stably transduced with a FLT3-ITD G697R allele (Ba/F3 FLT3-ITD-G697R cells), that we have previously reported to confer resistance to midostaurin.23 In the cell growth assay (Figure 2A), there was no inhibition of cellular proliferation at doses of midostaurin up to 300 nM. In addition, immunoblot assays (Figure 2B) demonstrated that midostaurin did not inhibit phosphorylation of FLT3, STAT5, or ERK in Ba/F3 FLT3-ITD-G697R cells. These findings indicate that inhibition of the respective mutant FLT3 activating alleles in this cellular context is due to specific inhibition of FLT3 by midostaurin.
Figure 2.
Midostaurin does not inhibit proliferation or FLT3 phosphorylation in Ba/F3 cells expressing an ITD and the G697R point mutation. (A) The proliferation of Ba/F3 cells expressing FLT3-ITD and G697R point mutation is not inhibited by midostaurin even at high doses. Ba/F3 cells expressing FLT3-ITD plus G697R were incubated with increasing concentrations of midostaurin for 48 hours. The number of viable cells was then determined using a colorimetric assay. Results are expressed as a percentage of viable cells after 48 hours of growth (± SD) in the presence of DMSO without the drug. The experiment was repeated in the presence of IL-3. (B) Midostaurin does not inhibit phosphorylation of FLT3, STAT5, or ERK in Ba/F3 ITD cells with the G697R mutation. Whole cell lysates were prepared from cells incubated with various concentrations of midostaurin for 20 minutes, resolved by SDS-PAGE as before, and immunoblotted with the antibodies indicated.
These findings have a direct bearing on phase 3 trials that are currently being planned in the Intergroup setting for treatment of newly diagnosed AML patients with FLT3-ITD mutations with midostaurin and induction chemotherapy. In contrast to other small molecule FLT3 inhibitors, midostaurin inhibits the full spectrum of FLT3 activation loop mutations. Thus, these data indicate that AML patients harboring such activation loop mutations in FLT3 should be considered as candidates for enrollment, in addition to those with FLT3-ITD alleles.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grants DK50654 and CA66996 (D.G.G.), Belgian Federation Against Cancer grant SCIE2006-34 (J.C.), and by the Leukemia and Lymphoma Society. D.G.G. is a Doris Duke Foundation Distinguished Clinical Scientist and an Investigator of the Howard Hughes Medical Institute.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.V.B. wrote the paper, J.J.C. designed the research and analyzed data, J.C. and J.R. contributed reagents, and D.G.G. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: J.R. is employed by Novartis Pharma. All other authors declare no competing financial interests.
Correspondence: Elly V. Barry, MD, Dana-Farber Cancer Institute, Department of Pediatric Oncology, 44 Binney St, SW320, Boston, MA 02115; e-mail: elly_barry@dfci.harvard.edu.
References
- 1.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Genomic structure of human FLT3: implications for mutational analysis. Br J Haematol. 2001;113:1076–1077. doi: 10.1046/j.1365-2141.2001.02821.x. [DOI] [PubMed] [Google Scholar]
- 2.Dosil M, Wang S, Lemischka IR. Mitogenic signalling and substrate specificity of the Flk2/Flt3 receptor tyrosine kinase in fibroblasts and interleukin 3-dependent hematopoietic cells. Mol Cell Biol. 1993;13:6572–6585. doi: 10.1128/mcb.13.10.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Broxmeyer HE. p85 subunit of PI3 kinase does not bind to human Flt3 receptor, but associates with SHP2, SHIP, and a tyrosine-phosphorylated 100-kDa protein in Flt3 ligand-stimulated hematopoietic cells. Biochem Biophys Res Commun. 1999;254:440–445. doi: 10.1006/bbrc.1998.9959. [DOI] [PubMed] [Google Scholar]
- 4.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 5.Horiike S, Yokota S, Nakao M, et al. Tandem duplications of the FLT3 receptor gene are associated with leukemic transformation of myelodysplasia. Leukemia. 1997;11:1442–1446. doi: 10.1038/sj.leu.2400770. [DOI] [PubMed] [Google Scholar]
- 6.Yokota S, Kiyoi H, Nakao M, et al. Internal tandem duplication of the FLT3 gene is preferentially seen in acute myeloid leukemia and myelodysplastic syndrome among various hematological malignancies. A study on a large series of patients and cell lines. Leukemia. 1997;11:1605–1609. doi: 10.1038/sj.leu.2400812. [DOI] [PubMed] [Google Scholar]
- 7.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 8.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br J Haematol. 2001;113:983–988. doi: 10.1046/j.1365-2141.2001.02850.x. [DOI] [PubMed] [Google Scholar]
- 9.Fröhling S, Schlenk RF, Breitruck J, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002;100:4372–4380. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 11.Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102:646–651. doi: 10.1182/blood-2002-11-3441. [DOI] [PubMed] [Google Scholar]
- 12.Mizuki M, Fenski R, Halfter H, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- 13.Hayakawa F, Towatari M, Kiyoi H, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 14.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 15.Fabbro D, Ruetz S, Bodis S, et al. PKC412–a protein kinase inhibitor with a broad therapeutic potential. Anticancer Drug Des. 2000;15:17–28. [PubMed] [Google Scholar]
- 16.Chen J, Deangelo DJ, Kutok JL, et al. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:14479–14484. doi: 10.1073/pnas.0404438101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotlib J, Berube C, Growney JD, et al. Activity of the tyrosine kinase inhibitor PKC412 in a patient with mast cell leukemia with the D816V KIT mutation. Blood. 2005;106:2865–2870. doi: 10.1182/blood-2005-04-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisberg E, Boulton C, Kelly LM, et al. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell. 2002;1:433–443. doi: 10.1016/s1535-6108(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 19.Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105:54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 20.Stone RM, Fischer T, Paquette R, et al. Phase IB Study of PKC412, an Oral FLT3 Kinase Inhibitor, in Sequential and Simultaneous Combinations with Daunorubicin and Cytarabine (DA) Induction and High-Dose Cytarabine Consolidation in Newly Diagnosed Adult Patients (pts) with Acute Myeloid Leukemia (AML) under Age 61 [abstract]. Blood (ASH Annual Meeting Abstracts) 2006;108:157. [Google Scholar]
- 21.Clark JJ, Cools J, Curley DP, et al. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518. Blood. 2004;104:2867–2872. doi: 10.1182/blood-2003-12-4446. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Schwaller J, Kutok J, et al. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. Embo J. 2000;19:1827–1838. doi: 10.1093/emboj/19.8.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cools J, Mentens N, Furet P, et al. Prediction of resistance to small molecule FLT3 inhibitors: implications for molecularly targeted therapy of acute leukemia. Cancer Res. 2004;64:6385–6389. doi: 10.1158/0008-5472.CAN-04-2148. [DOI] [PubMed] [Google Scholar]
- 24.Sternberg DW, Tomasson MH, Carroll M, et al. The TEL/PDGFbetaR fusion in chronic myelomonocytic leukemia signals through STAT5-dependent and STAT5-independent pathways. Blood. 2001;98:3390–3397. doi: 10.1182/blood.v98.12.3390. [DOI] [PubMed] [Google Scholar]
- 25.Propper DJ, McDonald AC, Man A, et al. Phase I and pharmacokinetic study of PKC412, an inhibitor of protein kinase C. J Clin Oncol. 2001;19:1485–1492. doi: 10.1200/JCO.2001.19.5.1485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.