Abstract
CREB-binding proteins (CBP) and p300 are essential transcriptional coactivators for a large number of regulated DNA-binding transcription factors, including CREB, nuclear receptors, and STATs. CBP and p300 function in part by mediating the assembly of multiprotein complexes that contain additional cofactors such as p300/CBP interacting protein (p/CIP), a member of the p160/SRC family of coactivators, and the p300/CBP associated factor p/CAF. In addition to serving as molecular scaffolds, CBP and p300 each possess intrinsic acetyltransferase activities that are required for their function as coactivators. Here we report that the adenovirus E1A protein inhibits the acetyltransferase activity of CBP on binding to the C/H3 domain, whereas binding of CREB, or a CREB/E1A fusion protein to the KIX domain, fails to inhibit CBP acetyltransferase activity. Surprisingly, p/CIP can either inhibit or stimulate CBP acetyltransferase activity depending on the specific substrate evaluated and the functional domains present in the p/CIP protein. While the CBP interaction domain of p/CIP inhibits acetylation of histones H3, H4, or high mobility group by CBP, it enhances acetylation of other substrates, such as Pit-1. These observations suggest that the acetyltransferase activities of CBP/p300 and p/CAF can be differentially modulated by factors binding to distinct regions of CBP/p300. Because these interactions are likely to result in differential effects on the coactivator functions of CBP/p300 for different classes of transcription factors, regulation of CBP/p300 acetyltransferase activity may represent a mechanism for integration of diverse signaling pathways.
Keywords: p300, CBP interacting protein, p300, CBP associated factor, E1A
The regulation of gene transcription by DNA-binding transcription factors has been linked to the recruitment of CBP/p300, the p300/CBP associated factor (p/CAF) complex, as well as other cofactors, such as the p160/SRC/TIF2/p/CIP (1–5) family of factors (6). The CREB-binding protein (CBP) (7, 8) and the p300 adenoviral protein E1A interacting protein (9) have been implicated in the actions of a large number of regulated transcription factors (10), based on experiments using neutralizing antibodies against CBP/p300, in vivo gene deletion, and specific ribozymes (3, 6, 11, 12). The discovery that GCN5 (13), CBP/p300 (14, 15), and p/CAF (16) harbor intrinsic acetyltransferase activities for histones and for other proteins (17, 18), has led to a model of the role of these factors in the regulation of chromatinized DNA templates (19, 20). Indeed, in both biochemical and cell-based assays the acetyltransferase functions of CBP and/or p/CAF have proved critical for transcriptional function (7, 21). CBP/p300 and p/CAF are large proteins that contain conserved domains, each of which appear to interact with large numbers of distinct DNA-binding transcription factors, which include nuclear receptors (6), CREB (8), and STAT proteins, (22–24). In addition, CBP/p300 interact with other classes of modulating proteins such as RNA helicase A, the p160/SRC-1/p/CIP family of factors, and S6 kinase (25, 26). This property may underlie the putative ability of CBP/p300 to serve as nuclear integrators of transcriptional responses (6, 27).
In addition to potential roles of CBP/p300 in the modification of chromatin structure, a number of additional substrates have been identified that include DNA-binding transcription factors, such as p53 (28), GATA-1 (29), T cell factor-1 (17), and high mobility group (HMG) I/Y (18). Acetylation of these proteins can increase DNA binding (28, 29), decrease binding (18), or inhibit protein–protein interaction (17).
The adenoviral immediate early gene product, E1A, is well characterized as an inhibitor of many classes of CBP-dependent transcription factors (19, 20). Although E1A was initially found to bind to the C/H3 domain in CBP/p300 (16) there appear to be additional interaction domains that are of differential functional importance for different classes of transcription factors. Because E1A binds to critical control regions of CBP, models predicting competition between E1A and functional CBP interacting proteins, including p/CAF, RNA helicases, and p/CIP have been suggested (25, 26). Thus, a series of DNA-binding transcription factors that directly interact with the C/H3 domain of CBP/p300 might directly compete for access to this cofactor.
The cAMP-dependent transcription factor CREB interacts strongly with CBP, in response to CREB phosphorylation at Ser-133. CBP is required for CREB function (27), dependent on its acetyltransferase activity (3, 27). Similarly, the IFN-γ-dependent transcription factor STAT1 also binds to CBP and requires the CBP-HAT (histone acetyltransferase) activity for function (21). In contrast, nuclear receptors require additional factors, including p160/SRC/p/CIP (6), to recruit CBP complexes. Intriguingly, the retinoic acid receptor appears to require the acetyltransferase function of p/CAF, rather than that of CBP (21), raising questions concerning the potential regulation of CBP/p300 HAT activities by these and other coregulatory molecules.
In this paper, we report that the interaction of E1A with the C/H3 region is capable of strongly inhibiting CBP HAT function on a variety of substrates, including histones H4 and H3, HMG I/Y, HMG 14/17 and, to a limited extent, on CBP itself. In contrast, binding of CREB, or even of a CREB/E1A fusion protein, to the KIX domain does not have this effect. Surprisingly, the CBP interaction domain of p/CIP (3) also strongly inhibits CBP acetyltransferase function, whereas on a number of substrates the presence of additional p/CIP domains overcomes the inhibition of CBP HAT function. These data suggest that p/CIP is an allosteric regulator of the acetyltransferase activity of CBP, and that this effect may be modified further by interacting factors and/or covalent modifications. Together we speculate that a component of the CBP integration function reflects substrate-specific regulation of its acetyltransferase functions. Similar events may modulate p/CAF acetyltransferase function.
MATERIALS AND METHODS
Single Cell Microinjection Assay.
Quiescent insulin-responsive Rat-1 fibroblasts were seeded on acid-washed glass coverslips at subconfluent density and grown in MNE/F12 medium, supplemented with 10% FBS, gentacin, and methotrexate. Expression plasmids were injected into the nuclei of cells at 100 μg ml−1 by using an Eppendorf semiautomated microinjection system mounted on an inverted Zeiss microscope. About 1 hr after injection cells were stimulated, where indicated, with the appropriate ligand. In rescue experiments, cells were stimulated with ligand 6 hr after injection to allow protein expression. After overnight incubation, cells were fixed and stained to detect injected IgG and β-galactosidase expression (6).
Protein Preparation and Acetylation Assays.
For bacterial expression all vectors were constructed as glutathione S-transferase or HisG fusion, and purified from cultures grown to OD = 0.6–0.8, and induced 90–120 min at 37°C with 1 mM isopropyl β-d-thiogalactoside. Bacteria were lysed and purified in the presence of a phenylmethylsulfonyl fluoride/Boehringer–Mannheim protease inhibitor cocktail by using glutathione agarose or nickel columns (1–2 hr, 4°C). Flag-tagged CBP was expressed in a baculoviral expression vector and purified by using an anti-Flag IgG column. For CREB and CREB (1–198)/E1A (29–242) fusion protein interactions with CBP, recombinant proteins were incubated with the protein kinase A catalytic domain (Sigma) in the presence of 0.2 mM ATP (30 min, 4°C), stopped, and the phosphorylated proteins purified on affinity beads. All proteins were checked for purification to apparent homogeneity by SDS/PAGE. Acetylation reactions were performed in 20 μl of 10 mM sodium butyrate, 50 mM Tris⋅HCl (pH 7.6), 0.5 mM DTT, ∼2 μg of each protein, and 50 μM [14C]acetyl-CoA (50 mCi/mmol), and were incubated at 30°C for 30 min (CBP) or 1 hr (p/CAF) before electrophoresis on SDS/polyacrylamide gels.
Mutations of CBP and E1A were generated with the PCR or the Quick-Change Mutagenesis kit (Stratagene), confirmed by sequence analysis and substitution of a region containing the mutation into the wild-type vector backbone.
RESULTS
E1A Actions on CBP HAT Function.
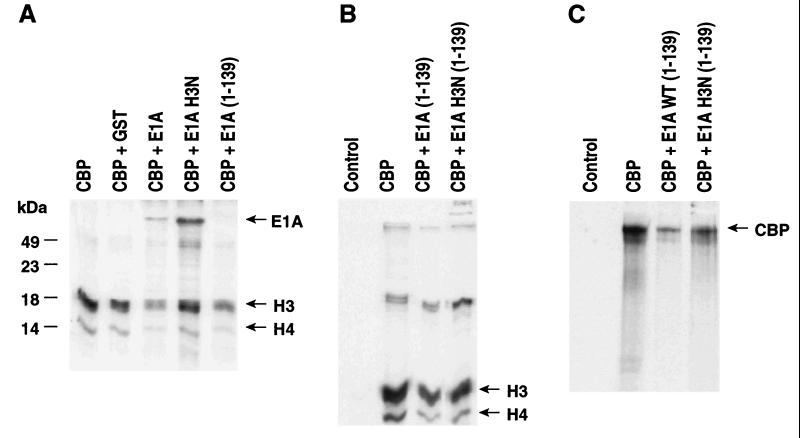
The effects of E1A on the HAT activity of CBP were initially carried out by using Flag-tagged CBP expressed in baculovirus-infected SF-9 cells. The addition of bacterially expressed E1A caused a marked inhibition of both histone H4 and histone H3 acetylation (Fig. 1). Deletion of exon 2 information at amino acid 139 [E1A(139)] reduced but did not abolish the ability of E1A to inhibit histone acetylation activity (Fig. 1 A and B). However, truncation to amino acid 82, excluding CR2 and CR3, showed a marked loss in inhibitory activity and truncation to amino acid 36 abolished activity (data not shown).
Figure 1.
Effects of E1A on CBP acetyltransferase function. (A) Effects of CBP-dependent histone H3 and histone H4 acetylation are shown using 12S E1A holoprotein (1–242) or mutant (H3N) E1A(1–242) or E1A(1–139). The H3N mutation causes loss of binding to the C/H3 domain of CBP. (B) Actions of E1A(1–139) or a H3N mutant of E1A(1–139). (C) Comparison of CBP autoacetylation in the presence of CBP(1–139) or the H3N mutant E1A(1–139). We note that the C terminus of E1A (amino acids 139–242) represents a substrate for CBP-dependent acetylation.
To examine the substrate specificity of these effects, the acetyltransferase activity of CBP was assayed by using HMG I/Y, HMG 14, and HMG 17 as substrates. E1A(1–139) was an effective inhibitor of acetylation of all of these substrates (Figs. 1 and 2). Even CBP autoacetylation was somewhat impaired by E1A(1–139) (Fig. 1C). Based on the extensive study of E1A, mutants that impair specific functions have been identified. An H3N mutation at the N terminus inhibits binding of E1A to the C/H3 domain, but only modestly impairs binding of E1A to the N terminus and p/CIP interaction domains of CBP (12). This mutant E1A protein is nearly as effective as wild-type E1A in inhibiting retinoic acid receptor function, but is much less active as an inhibitor of STAT function (12). The H3N E1A holoprotein or H3N E1A(1–139) was a much weaker inhibitor of CBP acetyltransferase function (Fig. 1 A and B). In contrast to E1A wild-type protein, this mutant E1A does not effectively inhibit CBP HAT function on either histone H3, H4, or HMG I/Y (Figs. 1 and 2A). Inhibitory effects on HMG14 or HMG17 were less marked (Fig. 2B). Therefore, effects of E1A may correlate, to some extent, with the region of CBP with which it interacts, and the interaction with the C/H3 domain appears to be particularly important for E1A inhibitory effects on CBP acetyltransferase function. Interestingly, E1A also inhibited p/CAF HAT activity, and H3N E1A was almost equivalent to the wild-type protein with respect to this inhibition (Fig. 3A).
Figure 2.
Effects of E1A on CBP-dependent acetyltransferase of HMG I/Y, HMG 14, or HMG 17. (A) Effects of the addition of E1A or H3N E1A on CBP-dependent acetylation of HMG (I/Y). (B) Effects of E1A on HMG 14 and HMG 17 acetylation with less severe inhibition than with HMG I/Y.
Figure 3.
HAT specificity and factor specificity of enzyme inhibitory actions. (A) Effects of E1A on p/CAF-dependent acetylation of histone H3. (B) Effect of CREB or CREB/E1A fusion protein or HAT activity of CBP on histone H3/H4. Neither CREB nor the CREB fusion protein inhibited CBP-dependent acetylation. Similar results were obtained with CBP/H3N E1A(1–242) fusion protein.
CREB Binding Does Not Inhibit CBP HAT Activity.
Based on the actions of E1A, it became of interest to determine whether other factors, such as CREB, affect HAT function on interaction with CBP. CREB exhibits high-affinity binding to the KIX domain after activation by protein kinase A phosphorylation of Ser-133 (8, 30). Microinjection experiments have previously indicated that CREB requires CBP HAT function to activate CRE-dependent transcription. Therefore, bacterially expressed, purified CREB was phosphorylated by using protein kinase A and assessed for effects on CBP HAT function. As shown in Fig. 3B, addition of phosphorylated CREB did not inhibit CBP acetyltransferase activity by using either histone H3 and H4 or HMG I/Y as substrates (18), and there was actually a slight increase of CBP autoacetylation (Fig. 3B). Because C-terminal E1A information was required for effective inhibition of CBP acetyltransferase activity, we inquired as to whether fusing N-terminally truncated E1A to the CREB C terminus would alter its effects on CBP acetyltransferase function on substrates tested. Here, protein kinase A-phosphorylated CREB or a CREB/E1A fusion protein was used. Interestingly, in contrast to wild-type E1A, the CREB/E1A fusion protein exhibited no detectable ability to inhibit CBP HAT function. Consistent with a requirement for CBP HAT function for actions of CREB, interactions dependent on the KIX domain did not negatively control CBP acetyltransferase activity. One possible interpretation is that the specific interaction domain on CBP is a critical component in determining the ability of specific protein sequences to inhibit acetyltransferase activity.
p/CIP/CBP Functional Interactions.
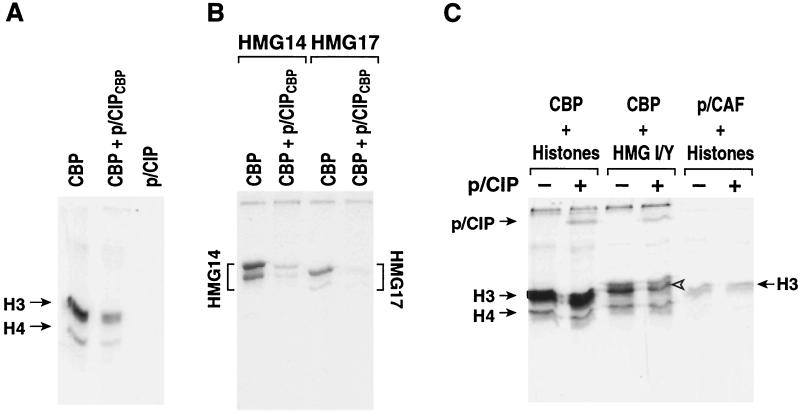
The SRC/NCoA family of p160 factors, including p/CIP, have been shown to exhibit ligand-dependent interactions with a number of nuclear receptors and to be capable of enhancing transcription on specific promoters. Indeed, nuclear receptor interactions with these factors, dependent at least in part on LXXLL-containing helical motifs (31), are more robust than with CBP, and it is postulated that CBP is recruited to liganded nuclear receptors by means of the p160 factors (6, 32). This recruitment involves a 100-aa domain that is highly conserved among the three known members of this family, SRC/NCoA (1–3), GRIP/TIF2/NCoA2 (1, 2), or p/CIP/ACTR/AIB (3–5). This interaction appears to require a domain in CBP C terminal to the C/H3 domain (3, 31). Therefore, we assessed the effects of the CBP interaction domain of p/CIP (947–1084) on CBP acetyltransferase function. Unexpectedly, this region of p/CIP (p/CIPCBP) was capable of marked inhibition of CBP acetyltransferase activity when histone H3, histone H4, HMG 14, or HMG 17 were used as substrates (Fig. 4 A and B). This result was particularly surprising because p/CIP often synergizes with CBP in receptor activation (3). Therefore, to further examine this issue, we expressed and purified a p/CIP protein that encompassed all known regulatory domains, including the nuclear receptor and CBP interaction domains, and a C-terminal domain that has been reported to have weak transactivation properties (3) and weak HAT activity (4, 33). However, when we evaluated the purified p/CIP protein, expressing this putative HAT domain, which migrated as a single band of the expected molecular weight, for its ability to alone acetylate histones, no detectable activity was observed (Fig. 4C). When this longer version of p/CIP was bound to CBP and CBP acetyltransferase activity was assessed, there was no detectable inhibition of CBP acetyltransferase activity on any substrate tested. Therefore, the inhibitory activity apparently present in the CBP interaction domain is overcome by the presence of the adjacent domains of p/CIP.
Figure 4.
Role of p/CIP on CBP protein acetyltransferase activity. (A) Inhibiting effects of the p/CIP CBP interaction domain (aa 947–1089), on histone acetylation. (B) Inhibiting effects of the p/CIP CBP interaction domain on HMG 14 and HMG 17 acetylation. (C) Inhibitory effects of p/CIP encompassing known functional domains (aa 636–1304) on CBP-dependent histone, HMG I/Y acetylation, or p/CAF-dependent histone H3 or histone H4 acetylation. While P/CIPCBP is not acetylated, the longer form is a CBP substrate.
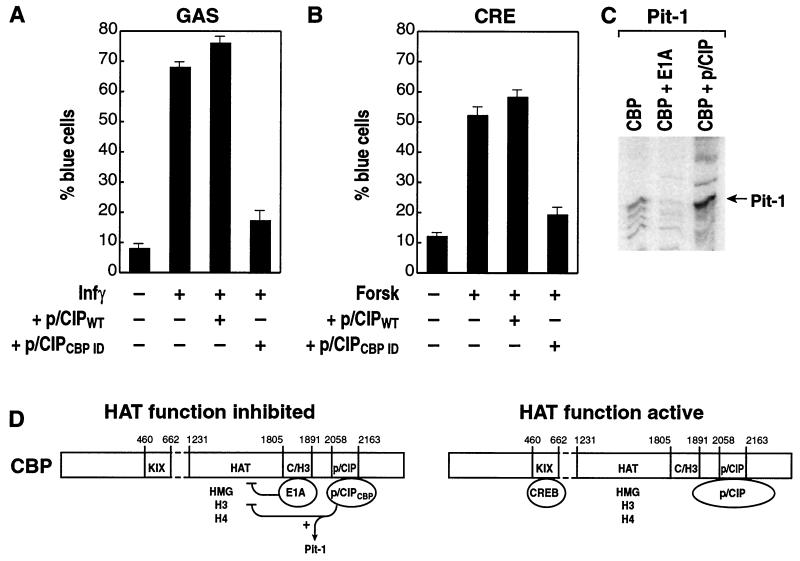
Consistent with these differential actions of the p/CIP CBP interaction domain alone versus the extended molecule on CBP acetyltransferase activation we found that the presence of the p/CIP CBP interaction domain, in contrast to the p/CIP holoprotein, inhibited both CREB-dependent and IFN-γ-dependent transactivation events (Fig. 5 A and B) by using the single-cell microinjection assay. When the two p/CIP LXXLL-related helical motifs in the CBP interaction domain were mutated, which abolished interaction with CBP (31), it now failed to inhibit CREB or IFN-γ-dependent transcription (data not shown). Although there are many potential explanations for these data, they are consistent with the observation that HAT activity of CBP, required for CREB-dependent gene activation in response of CBP activation (19), is inhibited in a manner that depends on the specific domains of p/CIP that are expressed.
Figure 5.
Correlation of p/CIP effects on function and CBP HAT activity. (A) Effects of p/CIPCBP or p/CIP holoprotein on interferon-dependent gene activation by using the nuclear microinjection assay and a gal4 activation sequence (GAS)-dependent reporter as described (3). (B) Effects of p/CIP holoprotein and p/CIP ligand binding interaction domain on forskolin-induced CREB-dependent activation as described (3). (C) Effects of E1A and the p/CIP CBP interaction domain on CBP HAT function, with Pit-1 as a substrate. (D) Factor interactions with CBP can result in inhibition of acetyltransferase functions or not interfere with the CBP acetyltransferase function. The differential effect of the p/CIP CBP interaction domain suggests combinatorial, allosteric regulation of CBP acetyltransferase function.
Finally, we wanted to determine whether there might be substrate specificity associated with the observed inhibitory effects of p/CIP. Therefore, we examined Pit-1 as a potential substrate, because the transcriptional actions of Pit-1 have been linked to CBP (34). Pit-1 holoprotein proved a poor substrate for activation by CBP HAT, but this acetylation was inhibited by E1A. Unexpectedly, the addition of the p/CIP CBP interaction domain, in contrast to actions of histone H3 or H4, or HMG I/Y acetylation, actually enhanced Pit-1 acetylation (Fig. 5C).
DISCUSSION
The discovery that CBP/p300 and p/CAF possess intrinsic HAT activities that are critical to their ability to activate transcription has intriguing implications for multifactorial regulation of gene expression. In this paper we report initial biochemical and cell culture experiments indicating that CBP and p/CAF HAT activities can be regulated, in some cases, in a substrate-specific or selective fashion.
The ability of E1A to inhibit multiple transcription factors can be correlated, for many cases, with the ability of E1A to bind to CBP/p300 (12). Because we find that the H3N E1A mutant protein is less effective than wild-type E1A at inhibiting CBP HAT activity and has selectively lost the ability to interact with the C/H3, but not with other domains of CBP, we suggest that the domain of CBP to which E1A binds is a critical component of its ability to exert inhibitory actions on HAT activity. Conversely, the requirement for C-terminal sequences of E1A for effective inhibition suggests that both specific interaction regions within CBP and distinct domains within E1A appear to be necessary for effective inhibition.
In contrast, consistent with the requirement of CBP HAT function for its activity, CREB fails to inhibit CBP HAT function, and this is true also of a CREB/E1A fusion protein that harbors the region of E1A required for its inhibitory function. Therefore, when bound to the KIX domain, these sequences fail to inhibit HAT function, supporting further the idea that both the domain of interaction and the information in the interacting protein are determinants of the ability to perturbate CBP HAT activity.
Investigation of the interactions between the coactivators p/CIP and CBP has led to a second, potentially important aspect of regulation of CBP HAT activity. Unexpectedly, the minimal CBP interaction domain of p/CIP harbors information that, on binding to a specific C-terminal domain of CBP, has proven to inhibit CBP HAT function by using histones and HMG proteins as substrates, but to stimulate acetyltransferase activity when Pit-1 was used as a substrate. Furthermore, with the inclusion of additional functional domains, p/CIP failed to inhibit histone acetylation. In contrast to previous reports, we have been unable to document significant acetyltransferase activity of p/CIP itself, and such an activity does not account for the effects of p/CIP on Pit-1 acetylation. In concert, these observations raise the possibility that p/CIP may regulate CBP function in vivo by either stimulating or inhibiting its acetyltransferase. Such effects may in turn be regulated by interactions of p/CIP with other cellular factors or by posttranslational modifications. Our data are most consistent with an allosteric model for the effects of CBP interacting factors, because of the observation that the p/CIP interaction domain, which inhibited acetylation of histones H3 and H4, enhanced acetylation of Pit-1. Distinct regions of CBP may therefore act in a potentially combinatorial fashion to modulate its acetyltransferase function, providing an additional mechanism for integration of nuclear signaling events (see Fig. 5D).
Acknowledgments
We thank A. Aggarwal for advice, P. Meyer for help in preparing the figures, and M. Fisher for help in preparing the manuscript. V.P. is supported by the Consiglio Nazionale delle Ricerche and E.K. is supported by a U.S. Army Medical Research Program Award. M.G.R. is an Investigator with the Howard Hughes Medical Institute and C.K.G. is an Established Investigator of the American Heart Association. These studies were supported by an Award from CapCURE to M.G.R. and grants from the National Institutes of Health to C.K.G. and M.G.R.
ABBREVIATIONS
- CBP
CREB-binding protein
- E1A
adenoviral oncoprotein E1A
- p/CAF
p300/CBP associated factor
- HAT
histone acetyltransferase
- p/CIP
p300/CBP interacting protein
- HMG
high mobility group
References
- 1.Hong H, Kohli K, Garabedian M J, Stallcup M R. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 4.Chen H W, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 5.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 6.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 7.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 8.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 9.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 10.Torchia J, Glass C K, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarti D, Lamorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 12.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 13.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 14.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 15.Ogryzko V V, Shiltz O L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 16.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 17.Lucas W, Mariann B. Nature (London) 1998;395:521–525. [Google Scholar]
- 18.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 19.Whyte P, Williamson N M, Harlow E. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 20.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 21.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Sun Z W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 24.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfeld M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 27.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch’ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Cell. 1998;93:351–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Roeder R G. Cell. 1997;99:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 29.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Nature (London) 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 30.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 31.McInerney E M, Rose D W, Flynn S W, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, et al. Genes Dev. 1998;21:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Nature (London) 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 33.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J X, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, Omalley B W. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 34.Xu Lan R M, Lavinsky J S, Dasen S E, Flynn E M, McInerney T-M, Mullen T, Heinzel D, Szeto E, Korzus R, et al. Nature (London) 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]