Abstract
A serpin was identified in normal mammary gland by differential cDNA sequencing. In situ hybridization has detected this serpin exclusively in the myoepithelial cells on the normal and noninvasive mammary epithelial side of the basement membrane and thus was named myoepithelium-derived serine proteinase inhibitor (MEPI). No MEPI expression was detected in the malignant breast carcinomas. MEPI encodes a 405-aa precursor, including an 18-residue secretion signal with a calculated molecular mass of 46 kDa. The predicted sequence of the new protein shares 33% sequence identity and 58% sequence similarity to plasminogen activator inhibitor (PAI)-1 and PAI-2. To determine whether MEPI can modulate the in vivo growth and progression of human breast cancers, we transfected a full-length MEPI cDNA into human breast cancer cells and studied the orthotopic growth of MEPI-transfected vs. control clones in the mammary fat pad of athymic nude mice. Overexpression of MEPI inhibited the invasion of the cells in the in vitro invasion assay. When injected orthotopically into nude mice, the primary tumor volumes, axillary lymph node metastasis, and lung metastasis were significantly inhibited in MEPI-transfected clones as compared with controls. The expression of MEPI in myoepithelial cells may prevent breast cancer malignant progression leading to metastasis.
The serine protease plasminogen/plasmin activator(PA) system contributes significantly to extracellular proteolysis in a wide variety of physiological processes of normal development and pathological processes in the etiology of diseases such as tumor invasion and metastasis (1–2). Compelling experimental evidence has suggested an important and apparently causal role for the tumor-associated urokinase type PA (uPA) and the receptor uPAR in cancer invasion and metastasis (1–3). Consistent with its role in cancer metastasis, overexpression and unrestrained activity of u-PA has been shown clinically to be a prognostic marker in many different types of human cancer (4–10). The down-regulation of uPA may occur at the level of transcriptional regulation of the genes and through interaction with specific endogenous inhibitor serpin such as plasminogen activator inhibitor (PAI). The best characterized serpins are PAI-1 and PAI-2. Both PAI-1 and PAI-2 have been shown to inhibit extracellular matrix degradation in vitro (11–12). These results suggest that the inhibitory activity of PAIs may be important in inhibiting tumor metastatic progression leading to metastasis. In fact, administration of a recombinant PAI-2 to mice decreases tumor growth (13), whereas overexpression of either PAI-1 or PAI-2 inhibits tumor metastasis (14–15).
It has been demonstrated extensively that high tumor levels of uPA and PAI-1 are statistically independent and poor prognostic factors for disease-free and overall survival in breast cancer (6, 16–19). In contrast to PAI-1, high levels of PAI-2 expression may be a favorable prognostic marker in breast cancer (2–3). In breast carcinomas with high uPA values, PAI-2 was associated with a prolonged relapse-free survival, metastasis-free survival, and overall survival (20–21). These results indicate that PAI-2 may play a critical role in inhibition of extracellular matrix degradation mediated by PA during tumor cell invasion and metastasis.
We describe here a serpin MEPI from normal mammary gland and discuss its biological relevance in inhibiting human breast cancer progression. In contrast to PAI-1 and PAI-2, whose expressions are elevated in breast carcinomas as compared with normal breasts, the expression of MEPI was exclusively detected in myoepithelial cells adjacent to the basement membrane of normal and noninvasive benign mammary gland, and no MEPI expression was detected in malignant breast carcinomas.
MATERIALS AND METHODS
Reagents.
Restriction enzymes, T7 polymerase, random primer DNA labeling kit, and digoxigenin-labeled nucleotides were obtained from Boehringer Mannhem. [32P]dATP was purchased from Amersham Pharmacia.
Molecular Cloning of MEPI Full Length cDNA Sequence.
As we previously described (24–25), we have used expressed sequence tag (EST)-based differential cDNA sequence analysis to search for new genes differentially expressed in breast cancer vs. noncancerous breast cells. Among several differential expressed EST groups (24), we revealed one EST group from a noncancerous breast and pancreas libraries having translated sequences >40% homologous to PAI-1. One clone, HPASD50, encoding an intact N-terminal signal peptide was identified in the pancreas library and selected for further investigation. It was fully sequenced on both strands, and its homology was confirmed. It was named MEPI.
In Situ Hybridization.
The colorimetric in situ hybridization was carried out as described (24–25). A 1,176-bp digoxigenin-labeled nucleotide was used as antisense probe for MEPI. The probe was generated by EcoRI digestion of MEPI cDNA plasmid and followed by transcription of digoxigenin-labeled antisense probe with T7 RNA polymerase.
Northern Blot and Reverse Transcription–PCR Analysis.
Detection of MEPI mRNA expression was analyzed by using Northern blot analysis as described (24–25). Reverse transcription–PCR analysis was performed by using a standard reversed transcription–PCR with the primers corresponding to the 5′ and 3′ sequence of the cDNA (5′ primer, GGAAGTCAAGCCTCAAGATGCTCA; 3′ primer, GGGATTTGTCACTCTTCCCATAAA).
Transfection.
The full-length MEPI cDNA was inserted into a pCI-neo mammalian expression vector. The resulting vector was transfected into MDA-MB-435 cells followed by G418 selection and cloning as described (25–26).
In Vitro Assay for Cell Growth.
Exponentially growing cultures of different MDA-MB-435 clones were detached with trypsin, and the trypsin was neutralized with DMEM/10% serum. Cells were counted, diluted, and seeded in triplicate at 3,000 cells per well (24-well plate) in 1 ml of DMEM/5% serum. Cell growth was measured by using CellTiter 96 AQueous nonradioactive cell proliferation assay kit (Promega).
Detection of PA Activity.
All of the clones were maintained in subconfluent monolayers with 10% fetal calf serum. The medium was discarded, and the monolayers were washed twice with PBS. The monolayers were cultured in the absence of serum, in DMEM supplemented with transferrin (1 mg/liter), fibronectin (1 mg/liter), and trace elements (Biofluids, Rockville, MD). After 24 hours, the serum-free medium was discarded, and the cells were replenished with fresh serum-free medium. The conditioned media (CM) were collected 40 hours later. Media were then centrifuged at 1,200 × g, and supernatants were saved and concentrated approximately 20-fold by using an Amicon hollow fiber concentrator with a 10,000-molecular weight cutoff at 4°C. The protein concentrations of CM were determined and normalized. The PA activity was subsequently analyzed as follows: 140 μl of reaction mixture was added to the 96-well ELISA plate. The reaction mixture was composed of 1 mg per 100 ml plasminogen and 0.5 mM plasmin substrate d-Val-Phe-Lys p-nitroanilide (both from Sigma) dissolved in reaction buffer (50 mM Tris⋅HCl, pH 7.5/20 ng/ml leupeptin/20 ng/ml pepstatin A/0.5 mM o-phenanthroline/1 mg/ml fibrin). Ten microliters of concentrated CM was added into the mixture and incubated at 37°C. A405 was measured after 1 hour.
In Vitro Invasion Assay.
The modified Boyden chamber invasion assay was performed as described (26).
Tumor Growth and Lymph Node and Lung Metastasis in Athymic Nude Mice.
A nude mouse orthotopic tumor growth and metastasis was performed as we previously described (25–26).
Statistical Analysis.
Values were expressed as means ± standard errors (SEs). Comparisons were made by using the two-tailed Student’s t test. Where appropriate, the χ2 test was used to compare proportions.
RESULTS
Molecular Cloning of MEPI cDNA.
We generated cDNA libraries from a breast cancer biopsy specimen and a normal breast and analyzed these libraries by EST-based differential cDNA sequencing approach (23–24). As previously demonstrated (23), we identified three classes of EST groups that were differentially expressed in normal breast vs. breast cancer: (1) genes that are more abundant in breast cancer than in normal breast; (2) genes that are more abundant in normal breast than in breast cancer; and (3) genes that are selectively expressed in breast relative to other tissue types. Within the second class, the automated screening revealed a group of ESTs encoding a novel gene with homology to human PAI-1 and PAI-2 (greater than 40% homology). Among this EST group, most of the distinctive PAI-related EST clones were derived from the normal pancreas library and the normal breast library. No ESTs were derived from the breast cancer libraries. After sequencing of these cDNA fragments, one clone encoding an intact N-terminal signal peptide was identified in a pancreas library and selected for further characterization. A start codon (ATG) and an ORF were identified in this clone.
The ORF encodes a protein of 405 amino acids. A hydrophobic leader sequence at the amino terminus conforms to a consensus signal peptide with a predicted cleavage site following an alanine residue located at position 18 in the precursor. Removal of the signal sequence results in a mature protein of 387 amino acids having a calculated molecular weight of 44 kDa, which is in close agreement with the molecular mass range of the PAI family. Like PAI-1, MEPI is a secreted protein. Comparison of the predicted amino acid sequence with the sequences of human PAI-related proteins is shown in Fig. 1. After optimal alignment, the putative protein shows 33% sequence identity and 58% similarity to PAI-1 and PAI-2.
Figure 1.
Comparison of the predicted amino acid sequence of MEPI with PAI-1 and PAI-2. The available amino acid sequence of PAI-1 and PAI-2 were obtained from the SwissProt database and aligned with the MEPI deduced sequence by using the clustal method of the megalign program from the dnastar software package. Conserved amino acids are shaded. The putative 18 hydrophobic signal peptide is located between two arrows.
Expression of MEPI in Human Breast Myoepithelial Cells.
We speculate that the expression of MEPI is lost or decreased during the breast cancer progression because the differential cDNA sequencing revealed MEPI expression in the normal breast library but not in the breast cancer library. In an attempt to evaluate the potential biological significance of MEPI on human breast cancer development and progression, we first studied MEPI gene expression in human breast cancer cells as well as normal human mammary epithelial cells. Northern blot analysis failed to detect MEPI expression in MCF-7, T47D, MDA-MB-231, MDA-MB-435, MDA-MB-436, and Hs578T human breast cancer cells and HME4144 and HME4244 (Clonetics) normal human mammary epithelial cells (data not shown). We also did RT-PCR analysis of MEPI expression in MCF-7, T47D, MDA-MB-231, and MDA-MB-435 cells and no MEPI signals were detected (data not shown). The inability to pick up the MEPI mRNA in both normal and malignant breast epithelial cells suggest that MEPI may be expressed in nonepithelial stromal or myoepithelial cells.
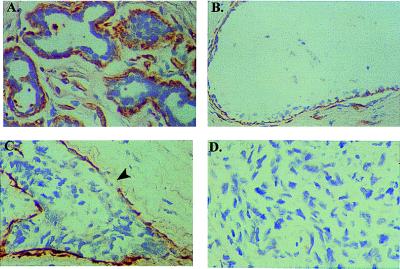
To localize the cellular source of MEPI expression and to assess the biological relevance of MEPI expression in breast cancer progression, we next performed in situ hybridization on the fixed sections from a variety of different human breast specimens. In these experiments, we examined two aspects of MEPI expression, including the cellular localization and the potential correlation of the loss of MEPI expression and breast cancer malignant phenotype. We found a strongly positive MEPI hybridization in the myoepithelial cells, which lie on the epithelial side of the basement membrane of the normal lobular (Fig. 2A), and normal duct (Fig. 2B). The expression of MEPI mRNA was detectable in the myoepithelial cells in all 5 normal reduction mammoplasty specimens and in 5 benign hyperplasia lesions. In contrast, expression of MEPI was absent in 5 of 5 cases of infiltrating breast carcinomas. The loss of MEPI expression in the malignant breast carcinomas may be because of the loss of putative MEPI-producing myoepithelial cells during the malignant progression. A representative negative stained metastatic breast carcinoma was shown in Fig. 2D. No MEPI expression can be detected in both normal and malignant mammary epithelial cells. The stromal fibroblasts were also negative for MEPI expression. In some cases we found a strong MEPI transcript in the endothelial cells of small vessels (data not shown). These in situ hybridization results are consistent with (a) our differential cDNA sequencing cloning strategy, which suggests a down-regulation or loss of MEPI in breast cancer progression; and (b) the Northern blot analysis which showed no MEPI expression in both normal and malignant breast epithelial cells.
Figure 2.
In situ hybridization analysis of MEPI expression in human breast. Cells stained brown indicate MEPI gene expression. All sctions were counterstained lightly with hematoxylin for viewing negatively stained epithelial and stromal cells. (A) Myoepithelial cells surrounding lobules from a normal breast-reduction mammoplasty specimen showed strong MEPI expression. (B) A strong positive staining of MEPI in myoepithelial cells surrounding a normal duct. (C) A DCIS showed a partial MEPI expression; arrow indicates the loss of MEPI expression. (D) Negative staining of MEPI in an infiltrating breast cancer. A total of 30 clinical breast specimens were analyzed. Five of five normal breast-reduction mammoplasty samples and five of five benign hyperplasias showed strong expression of MEPI in myoepithelial cells as a continuing layer. Nine of 15 DCIS expressed MEPI in the myoepithelial cells as a continuing layer and the the other 6 DCIS showed partial expression. Five of five infiltrating breast carcinomas were negative. The normal breast section was also hybridized with the sense probe, and no detectable background staining was observed at the same conditions for the antisense probe. All of the sections presented in the figure were derived from the same experiment.
It is interesting to note that although MEPI was detected in the myoepithelial cells as a continuing layer lying on the epithelial side of the basement membrane of normal mammary gland, 6 of 15 ductal carcinomas in situ (DCIS) showed a partial MEPI expression. As shown in Fig. 2C, MEPI mRNA was detected in most part of the myoepithelial layer surrounding the carcinoma, but its expression was missing in some areas (indicated by an arrow). It is well recognized that an intact myoepithelial layer, like an intact basement membrane, can help distinguish benign epithelial proliferation and in situ carcinomas from invasive disease. The loss of MEPI expression in the in situ carcinomas may indicate an initiation of invasion. Our results, which demonstrated a stage-specific MEPI expression from the expression in the complete myoepithelial layer surrounding the normal breasts to the partial loss in some in situ carcinomas and to the complete loss in the infiltrating malignant breast carcinomas, suggest an association of loss of MEPI expression with breast cancer progression.
Tissue-Specific Expression of MEPI.
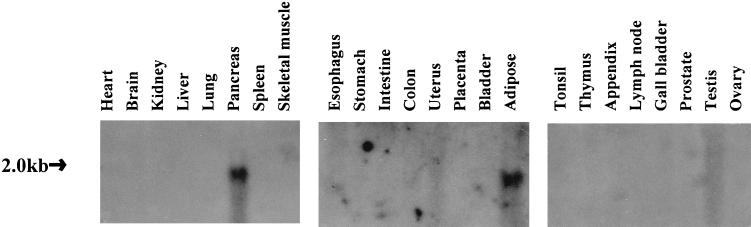
We investigated the expression of MEPI gene in a variety of human tissues by Northern blot analysis (Fig. 3). As expected, the Northern blot showed maximal MEPI transcript levels in pancreas, the tissue the gene was cloned from. Adipose also demonstrated a high abundant MEPI expression. No bands were present in other organs analyzed. This unique expression pattern suggests that MEPI may function in a tissue-specific fashion as part of proteolysis.
Figure 3.
The expression of MEPI gene in a variety of normal human adult tissues. Three blots containing approximately 20 μg of total RNA per line for the above tissues were purchased from Invitrogen. By using a full-length cDNA hybridization probe, a high abundance of 2-kb transcripts was detected in pancreas and adipose tissue.
Transfection and Selection of MEPI Positive Clones.
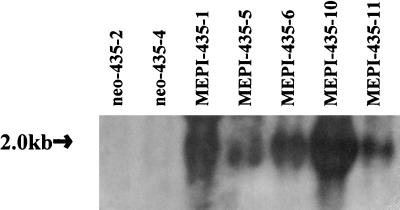
Because we demonstrated a loss of MEPI expression in breast cancer, we reason that the exclusive expression of MEPI in the myoepithelial cells may contribute the roles of myoepithelial layer as a paracrine cellular suppression of invasion. We therefore asked whether we could suppress breast cancer growth and metastasis by overexpression of MEPI gene in breast cancer cells. We selected MDA-MB-435 cell line as recipient for MEPI mediated gene transfection because: 1) it lacks detectable MEPI transcript; and 2) it is relatively highly tumorigenic and metastatic in nude mice. The full-length MEPI cDNA was inserted into pCI-neo mammalian expression vector. The resulting vector was transfected into MDA-MB-435 cells. The same cells were also transfected with the vector containing no insert as a control. MDA-MB-435 subclones transfected with MEPI cDNA were designated MEPI-435, and MDA-MB-435 subclones transfected with pCI-neo were designated neo-435. Fig. 4 shows the Northern blot analysis of MEPI expression in selected clones. All five selected MEPI-435 clones expressed MEPI mRNA transcripts. In contrast, none of the neo-435 clones produced any detectable MEPI transcripts. No changes in morphology were observed in these clones. We selected MEPI-435–1, MEPI-435–10, neo-435–2, and neo-435–4 clones for the subsequent studies.
Figure 4.
Northern blot analysis of MEPI transfection of MDA-MB-435 cells. Total RNAs were isolated from two control pCI-neo-transfected clone and five MEPI-transfected clones. Strong MEPI transcripts were detected in MEPI-positive clones. In contrast, no endogenous MEPI transcripts were detected in control clones. The integrity of the RNAs and loading control were ascertained by visualization of the 18S rRNA bands in stained gel (data not shown).
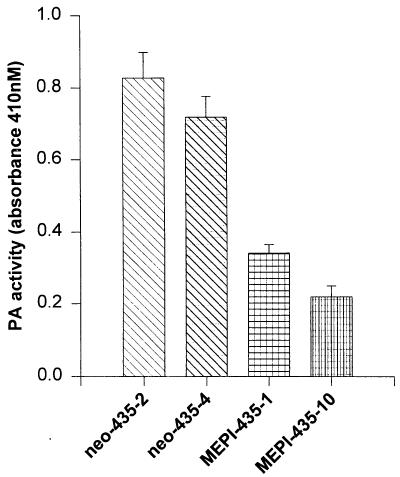
Decreased PA Activity in MEPI-435 Clones.
As an initial attempt to evaluate the potential PAI activity, we analyzed the total net PA activity, which reflects a balance between PA and PAI including transfected MEPI activity. In this regard, the CM from clones of MEPI-435–1, MEPI-435–10, neo-435–2, and neo-435–4 cells were collected, concentrated, and analyzed for PA activity (Fig. 5). A significant reduction in PA activity was noted in MEPI-435 clones. The PA activity from MEPI-435–10 cells was only 26% of that in neo-435–2 cells, and 31% of that in neo-435–4 cells. The PA activity of MEPI-435–1 cells was also significantly reduced, with 42% and 48% of that as compared with neo-435–2 and neo-435–4 cells, respectively.
Figure 5.
The activities of the CM from MEPI-435 and neo-435 clones on plasminogen activation. CM were collected, concentrated, normalized, and subjected to plasminogen activation analysis as described in Materials and Methods. The numbers represent the mean ± SE of three measurements. Statistical comparisons for pooled MEPI-435 clones relative to pooled neo-435 clones indicated P < 0.001 for the PA activity.
In Vitro Growth of MEPI-435 Cells.
To determine whether MEPI expression affects the growth of MDA-MB-435 cells, the growth rates of MEPI-435–1 and MEPI-435–10 cells were compared with that of neo-435–2 and neo-435–4 cells in the monolayer culture. No significant differences in growth rate were observed among MEPI-positive and MEPI-negative cells (data not shown).
Inhibition of in Vitro Invasion.
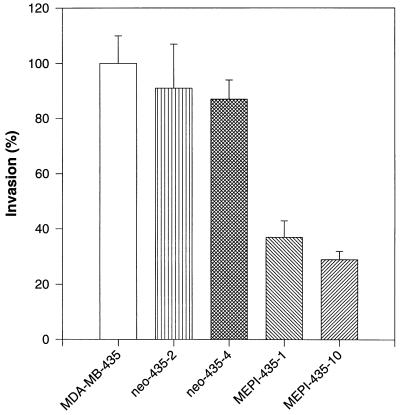
We used an in vitro-reconstituted basement membrane invasion assay to determine whether MEPI expression affects breast cancer cell invasion. Both parental MDA-MB-435 cells and neo-435 cells were moderately invasive. At the end of a 48-h incubation, about 5.1% of MDA-MB-435 cells, 4.7% of neo-435–2 cells, and 4.5% of neo-435–4 cells had crossed the Matrigel barrier. A significant reduction in invasive potential was noted in two MEPI-expressing clones with percentages of invasion for MEPI-435–1 and MEPI-435–10 being 1.9% and 1.5%, respectively. To facilitate the comparison of the relative invasiveness between controls and MEPI-transfected clones in this study, all values were normalized to the percent invasion of parental MDA-MB-435 cells, which was taken as 100% (Fig. 6).
Figure 6.
Inhibition of cell invasion by MEPI. Comparison of invasion potentials of MEPI-positive and MEPI-negative cells. The invasion of MDA-MB-435 cells was used as control and was taken as 100%. The invasion potentials of all of the other clones were expressed as a percentage of the control. The numbers represent the mean ± SE of three cultures. Statistical comparisons for pooled MEPI-positive MEPI-435–1 and MEPI-435–10 clones relative to pooled MEPI negative MDA-MB-435, neo-435–2, and neo-435–4 clones indicated P < 0.001 for the invasion.
Effect of MEPI Transfection on Tumorigenicity.
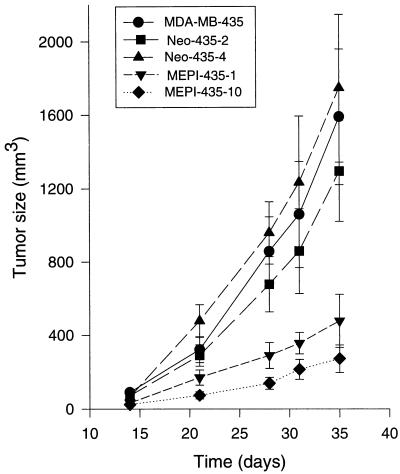
An orthotopic mammary fat pad nude mouse model was used to study the effects of MEPI on tumor growth and metastasis (Table 1). After a lag phase of 7–10 days, 46 of 48 (96%) injections in the mice given implants of MEPI-negative MDA-MB-435, neo-435–2, and neo-435–4 cells developed tumors. In contrast, only 17 of 32 (53%) injections in the mice given implants of MEPI-positive MEPI-435–10 and MEPI-435–1 cells developed tumors. The tumor growths in MEPI-435 clones were significantly inhibited. At 35 days after tumor-cell injection, the size of MEPI-435–10 tumors, which expressed relative high level of MEPI mRNA, was only 17% of that in parental MDA-MB-435 tumors, 21% of that in neo-435–2 tumors, and 15% of that in neo-435–4 tumors. In addition, the tumor incidence also was greatly decreased. With 16 injections, only 4 implants developed tumors. The tumor growth of MEPI-435–1 cells also was significantly reduced, with 30%, 37%, and 27% of tumor size observed as compared with MDA-MB-435, neo-435–2, and neo-435–4 tumors, respectively. Fig. 7 shows growth kinetics of parental MDA-MB-435, neo-435–2, neo-435–4, MEPI-435–1, and MEPI-435–10 tumors. After a slow growth phase of 14 days, tumors from both parental MDA-MB-435 cells and two neo-MDA-435 clones increased in volume at an exponential rate. In contrast, the growth of MEPI-435–1 and MEPI-435–10 cells was dramatically inhibited. Thus, the tumorigenicity of the human breast cancer cells was significantly inhibited by the expression of MEPI.
Table 1.
Effects of MEPI expression on tumor size, tumor incidence, lymph node status, and lung metastasis of MDA-MB-435 cells
| Treatment group | Tumor vol of primary size, mm3 | Tumor incidence tumor/total (%) | Lymph node positive/total (%) | Lung metastasis positive/total (%) |
|---|---|---|---|---|
| MDA-MB-435 | 1589 ± 368 | 15/16 (100) | 7/16 (44) | 2/8 (25) |
| Neo-435-2 | 1295 ± 275 | 15/16 (94) | 9/16 (56) | 3/8 (38) |
| Neo-435-4 | 1745 ± 401 | 16/16 (100) | 8/15 (53) | 4/8 (50) |
| MEPI-435-1 | 478 ± 145 | 13/16 (81) | 3/15 (20) | 1/8 (13) |
| MEPI-435-10 | 271 ± 75 | 4/16 (25) | 1/16 (6) | 0/8 (0) |
On day 1, 300,000 cells were injected with MEPI into the mammary fat pads, and tumor volumes, lymph node and lung metastasis were determined as described in Materials and Methods. Volumes are expressed as mean ± SE (number of tumors assayed). There were 16 total injections for eight mice in each group, and each mouse received 2 injections. The mice were sacrificed 35 days after injection. Statistical comparisons for pooled MEPI-positive clones relative to pooled MEPI-negative clones indicated P < 0.001 for the mean tumor sizes and P < 0.01 for the lymph node and lung metastasis. Statistical comparison for primary tumors was analyzed by Student’s t test. A χ2 test was used for statistical analysis of lymph node and lung metastasis.
Figure 7.
In vivo tumor growth of MDA-MB-435, neo-435–2, neo-435–4, MEPI-435–1, and MEPI-435–10 cells in nude mice. Each of the eight mice in each group received two injections (one on each side) in the mammary fat pads between the first and second nipples. Tumor size was determined at intervals by three-dimensional measurements (mm) by using a caliper. Only measurable tumors were used to calculate the mean tumor volume for each clone at each time point. Each point represents the mean of tumors ± SE (bars).
Regional and Metastatic Tumor Dissemination.
To study tumor dissemination, hematoxylin/eosin-stained paraffin sections of axillary lymph nodes and lungs were examined for morphologic evidence of tumor cells by light microscopy. MEPI-positive MEPI-435–10 and MEPI-435–1 clones showed an average lower proportion of lymph node positivity of 6% and 20% as compared with the average of 51% from MEPI-negative MDA-MB-435, neo-435–2, and neo-435–4 cells (Table 1). MEPI-positive clones also yielded significantly less lung micrometastasis than MEPI-negative clones, with a combined 6% of lungs positive for MEPI-435–10 and MEPI-435–1 tumors as compared with a combined 38% for MDA-MB-435, neo-435–2, and neo-435–4 tumors.
DISCUSSION
Serpins are a highly diverse group of proteins, most of which are inhibitors of serine proteinases. We have cloned an unnamed serpin, on which we confer the name MEPI because it was exclusively expressed in the myoepithelial cells of mammary gland. The consensus sequence of FRADHPFLFVI in the reactive center loop near the C terminus has been proposed to serve a diagnostic hallmark of the serpin superfamily (27). A similar sequence of FIANHPFLFI is conserved in the MEPI. A hinge region with a sequence of EDGSEAA is present in MEPI. Because this consensus sequence is proposed to be important for the inhibitory serpins (28), MEPI is probably one of the serpins that inhibit serine proteinases.
It is interesting to note that MEPI expression is seen exclusively in the myoepithelial cells adjacent to the basement membrane of normal and benign mammary gland. No MEPI was detected in the metastatic breast carcinomas. Myoepithelial cells, which are lost during the cancer malignant progression and normally surround ducts of glandular organs such as breast, contribute to the synthesis of a surrounding basement membrane and exert important paracrine effects on epithelial mitogenesis and morphogenesis (29). In normal or noninvasive benign mammary cells, cell–stroma contact is mediated by myoepithelial cells, which secrete relatively low levels of matrix-degrading proteinases but relatively high levels of maspin and various other antiinvasive proteinase inhibitors (30). Myoepithelial cells can also induce differentiation of breast cancer cells (31) and inhibit tumor cell invasion (29). The exclusive expression of MEPI in myoepithelial cells of normal or benign mammary gland may represent one of the major antiinvasive and antimetastatic phenotype mediated by the host defensive system. In this regard, the expression of MEPI in myoepithelial cells would create a microenvironment in the epithelial–stromal interface where the inhibitory effect of MEPI prevents the excessive proteolytic actions and preserve the epithelial–stromal structure integrity. It will be interesting to investigate whether the loss of MEPI expression in DCIS may indicate a malignant progression leading to invasion and metastasis. There is cause for concern about the large number of DCIS cases—which are developing as a consequence of screening mammography—most of which are treated by some form of surgery. In addition, the proportion of cases treated by mastectomy may be inappropriately high (32). If the loss of MEPI expression can provide some prognostic information on distinguishing the DCIS unlikely to become invasive from the DCIS most likely to become invasive, this will help to direct the treatment strategies and to reduce some inappropriate or unnecessary mastectomies.
Our data of the loss of MEPI expression in malignant breast cancer are different from the previous studies that have linked excessive PAI-1 and PAI-2 expression to breast cancers as compared with normal breast (3, 6, 16–20). Both in situ hybridization and immunohistochemical staining have demonstrated a strong PAI-1 and PAI-2 expression in the stroma surrounding breast carcinomas or at tumor margins (19, 3). It is not easy to understand why the elevated tumor tissue content of PAI-1 indicates a poor prognosis for the breast cancer patients. One explanation is that the increased expression of PAI-1 may be reciprocally related to the increased expression of uPA and other proteinases during the tumor-mediated degradation of extracellular matrix. Therefore, these elevated levels of PAI-1 in the stroma adjunct to the invasive breast carcinomas may represent one of the subsequent acute host responses to the remodeling stimuli and attempts to balance the local tissue degradation. In this regard, the invasive tumor cells may also need some PAI-1 to protect them in the proteolysis-rich microenvironment. In fact, it has been recently demonstrated that the absence of host PAI-1 prevents cancer invasion (33). These data suggest that PAI-1 produced by host cells promotes cancer invasion presumably by protecting tumor cells from proteolysis. Alternatively, the high-level expression of PAI-1 in the breast cancer may favor the proposed coexpression model that uPA and PAI-1 have to be present in the tumor to achieve focused and optimal uPAR-mediated proteolysis and invasiveness (34). Nevertheless, in contrast to the PAI-1 and PAI-2, the loss of MEPI expression in the malignant breast carcinomas favors its role as a metastasis-suppressing gene.
Acquisition of invasive/metastatic potential is a key event in tumor progression, implicating the plasminogen system. Transfection of the MDA-MB-435 cells with an MEPI cDNA leads to increased expression of MEPI transcript and reduced PA activity when compared with control cells. The reduced in vitro invasiveness of MEPI-435 clones compared with control cells suggests that the production of MEPI altered the invasive potential of breast cancer cells. These results are consistent with the previous reports on the inhibition of the invasion by PAI-1 (35) and PAI-2 (36–37). In the nude mouse model of mammary tumor, overexpression of MEPI resulted in several phenotypic changes, including (i) a significant reduction in incidence and size of primary tumors; (ii) a reduction in number of microscopic metastatic lesions in the lung and lymph node. Although a similar growth rate of MEPI-positive clones vs. MEPI-negative clones was observed in vitro, the orthotopic tumor growth of MEPI-435 clones was significantly suppressed in vivo. We reason that the slower in vivo growth of MEPI-435 tumors may be explained, in part, by MEPI-mediated inhibition of tumor angiogenesis. It has been reported that uPA stimulates components of angiogenesis including chemotaxis, proteolytic matrix degradation, and the release of basic fibroblast growth factor from its storage in the basement membrane (38–41). Furthermore, both PAI-1 (35) and PAI-2 (42) have been demonstrated to have an antiangiogenic activity, presumably through inhibition of uPA expressed by endothelial cells in newly forming capillary sprouts. Our data indicate that despite the lack of growth inhibition of MEPI on breast cancer cells in vitro, MEPI significantly inhibits tumor growth and metastasis, presumably because of its anti-PA activity and antiangiogenic activity.
The magnitude of the tumor-suppressing activity of MEPI on human breast cancer is comparable to that observed for tumor suppressor Rb and p53 (43). The exclusive expression of MEPI in the myoepithelial cells of normal and benign mammary glands and the inhibition of breast tumor growth and metastasis by MEPI expression suggest that MEPI is one of the local myoepithelium-related paracrine factors that preserve the normal epithelial–stromal integrity and prevent the malignant progression from benign or in situ lesion to the metastatic phenotype. The potential application of MEPI as a cytostatic agent for the gene therapy treatment of cancer warrants further investigation.
Acknowledgments
We thank Dr. Shijie Sheng for discussion. This work was supported in part by Grant CA68064-01 from National Institute of Health, Grant DAMD17-94-J-4149 from U.S. Department of Army Breast Cancer Research Program, and Helen and Irving Schneider.
ABBREVIATIONS
- DCIS
ductal carcinoma in situ
- PA
plasminogen activator
- PAI
plasminogen activator inhibitor
- MEPI
myoepithelium-derived serine proteinase inhibitor
- uPA
urokinase-type PA
- uPAR
uPA receptor
- CM
conditioned media
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF130470).
References
- 1.Andreasen P A, Kjoller L, Duffy M J. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 2.Schmitt M, Harbeck N, Thomssen C, Wilhelm O, Magdolen V, Reuning U, Ulm K, Hofler H, Janike F, Graeff H. Thromb Haemostasis. 1997;78:285–296. [PubMed] [Google Scholar]
- 3.Duggan C, Kennedy S, Kramer M-D, Barnes C, Elvin P, McDermott E, O’Higgins N, Duffy M J. Br J Cancer. 1997;76:622–627. doi: 10.1038/bjc.1997.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt, M., Janicke, F. & Graeff, F. (1992) Fibrinolysis6, Suppl. 4, 3–26.
- 5.Schmitt M, Wilhelm O, Janicke F, Magdolen V, Reuning U, Ohi H, Moniwa N, Kobayashi H, Weidle U, Graeff H. J Obstet Gynaecol Br Emp. 1995;21:151–165. doi: 10.1111/j.1447-0756.1995.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 6.Brunner N, Pyke C, Hansen C H, Romer J, Grondahl-Hansen J, Dano K. Cancer Treat Rev. 1994;71:299–309. doi: 10.1007/978-1-4615-2592-9_16. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn W, Pache L, Schmalfeldt B, Dettmar P, Schmitt M, Janicke F, Graeff H. Gynecol Oncol. 1994;55:401–409. doi: 10.1006/gyno.1994.1313. [DOI] [PubMed] [Google Scholar]
- 8.Ganesh S, Sier C F, Griffioen G, Vloedgraven J H, Verheijen J H, Lamers C B, Verspaget H W. Cancer Res. 1994;54:4065–4071. [PubMed] [Google Scholar]
- 9.Nekarda H, Schmitt M, Ulm K, Wenninger A, Vogelsang H, Becker K, Roder J D, Fink U, Siewert J R. Cancer Res. 1994;54:2900–2907. [PubMed] [Google Scholar]
- 10.Duffy M J. J Clin Cancer Res. 1996;2:613–618. [PubMed] [Google Scholar]
- 11.Cajot J F, Bamat J, Bergonzelli G E, Kruithof E, Medcalf R L, Testuz J, Gordat B. Proc Natl Acad Sci USA. 1990;87:6939–6943. doi: 10.1073/pnas.87.18.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker M S, Bleakley P, Woodrow G C, Doe W F. Cancer Res. 1990;50:4676–4684. [PubMed] [Google Scholar]
- 13.Astedt B, Billstrom A, Lecander I. Fibrinolysis. 1995;9:175–177. [Google Scholar]
- 14.Muller B, Yu Y B, Laug W. Proc Natl Acad Sci USA. 1995;92:205–209. doi: 10.1073/pnas.92.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soff G A, Sanderowitz J, Gately S, Verrusio E, Weiss I, Berm S, Kwann H C. J Clin Invest. 1995;96:2593–2600. doi: 10.1172/JCI118323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffy M, O’Grady P, Devaney D, O’Siorain L, Fennelly J J, Lijnen H J. Cancer. 1988;62:531–533. doi: 10.1002/1097-0142(19880801)62:3<531::aid-cncr2820620315>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 17.Duggan C, Maguire T, McDermott E, O’Higgins N, Fennelly J J, Duffy M J. Int J Cancer. 1995;61:597–600. doi: 10.1002/ijc.2910610502. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt M, Thomssen C, Ulm K, Seiderer A, Harbeck N, Hofler H, Janicke F, Graeff H. Br J Cancer. 1997;76:306–311. doi: 10.1038/bjc.1997.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianchi E, Cohen R L, Dai A, Thor A T, Schuman M A, Smith H S. Int J Cancer. 1995;60:597–603. doi: 10.1002/ijc.2910600505. [DOI] [PubMed] [Google Scholar]
- 20.Bouchet C, Spyratos F, Martin P M, Hacene K, Gentile A, Oglobine J. Br J Cancer. 1994;69:398–405. doi: 10.1038/bjc.1994.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sumiyoshi K, Serizawa K, Urano T, Takada Y, Takada A, Baba S. Int J Cancer. 1992;50:345–348. doi: 10.1002/ijc.2910500303. [DOI] [PubMed] [Google Scholar]
- 22.Adams M A, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F, et al. Science. 1991;252:1651–1655. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Hong J, Liu Y E, Jia T, Wang M, Liu J, Xiao G, Joseph B K, Rosen C, Shi Y E. Cancer Res. 1997;57:759–764. [PubMed] [Google Scholar]
- 25.Shi Y E, Ni J, Xiao G, Liu Y E, Fuchs A, Yu G, Su J, Cosgrove J M, Xing L, Zhang M, et al. Cancer Res. 1997;57(15):3084–3091. [PubMed] [Google Scholar]
- 26.Wang M, Liu Y E, Greene J, Sheng S, Fuchs A, Rosen E M, Shi Y E. Oncogene. 1997;14(23):2767–2774. doi: 10.1038/sj.onc.1201245. [DOI] [PubMed] [Google Scholar]
- 27.Tsujimoto M, Tsuruoka N, Ishida N, Kurihara T, Iwasa F, Yamashiro K, Rogi T, Kodama S, Katsuragi N, Adachi M, et al. J Biol Chem. 1997;272(24):15373–15380. doi: 10.1074/jbc.272.24.15373. [DOI] [PubMed] [Google Scholar]
- 28.Hopkins P C R, Whisstock J. Science. 1994;265:1893–1894. [PubMed] [Google Scholar]
- 29.Sternlicht M D, Barsky S H. Med Hypotheses. 1997;48:37–46. doi: 10.1016/s0306-9877(97)90022-0. [DOI] [PubMed] [Google Scholar]
- 30.Sternlicht M-D, Safarians S, Rivera S-P, Barsky S-H. Lab Invest. 1996;74(4):781–796. [PubMed] [Google Scholar]
- 31.Bani D, Riva A, Bigazzi M, Bani Sacchi T. Br J Cancer. 1994;70:900–904. doi: 10.1038/bjc.1994.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernster V L, Barclay J, Kerlikowske K, Grady D, Henderson C. J Am Med Assoc. 1996;275:913–918. [PubMed] [Google Scholar]
- 33.Bajou K, Noel A, Brunner N, Holst-Hansen C, Fusenig N, Carmeliet P, Collen D, Foidart J M. Proc Am Assoc Cancer Res. 1998;39:83. doi: 10.1038/nm0898-923. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Shuman M A, Cohen R L. Int J Cancer. 1995;60:501–506. doi: 10.1002/ijc.2910600413. [DOI] [PubMed] [Google Scholar]
- 35.Soff G-A, Sanderowitz J, Gately S, Verrusio E, Weiss I, Berm S, Kwaan H C. J Clin Invest. 1995;96:2593–2600. doi: 10.1172/JCI118323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller B M, Tu Y B, Lang W E. Proc Natl Acad Sci USA. 1995;92:205–209. doi: 10.1073/pnas.92.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang W E, Cao X R, Yu Y B, Shimada H, Kruithof E K. Cancer Res. 1993;53:6051–6057. [PubMed] [Google Scholar]
- 38.Yasunaga C, Nakashima Y, Sueishi K. Lab Invest. 1989;61:689–704. [PubMed] [Google Scholar]
- 39.Mignati P, Tsuboi R, Robbins E, Rifkin D B. J Cell Biol. 1989;108:671–682. doi: 10.1083/jcb.108.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saksela O, Rifkin D B. J Cell Biol. 1990;110:767–775. doi: 10.1083/jcb.110.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flaumenhaft R, Moscatelli D, Saksela O, Rifkin D B. J Cell Physiol. 1989;140:75–81. doi: 10.1002/jcp.1041400110. [DOI] [PubMed] [Google Scholar]
- 42.Holmgren L, O’Reilly M S, Folkman J. Nat Med. 1995;1:149. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 43.Wang N P, To H, Lee W H, Lee E Y. Oncogene. 1993;8:279–288. [PubMed] [Google Scholar]