Abstract
G1/S and G2/M cell cycle checkpoints maintain genomic stability in eukaryotes in response to genotoxic stress. We report here both genetic and functional evidence of a Gadd45-mediated G2/M checkpoint in human and murine cells. Increased expression of Gadd45 via microinjection of an expression vector into primary human fibroblasts arrests the cells at the G2/M boundary with a phenotype of MPM2 immunopositivity, 4n DNA content and, in 15% of the cells, centrosome separation. The Gadd45-mediated G2/M arrest depends on wild-type p53, because no arrest was observed either in p53-null Li–Fraumeni fibroblasts or in normal fibroblasts coexpressed with p53 mutants. Increased expression of cyclin B1 and Cdc25C inhibited the Gadd45-mediated G2/M arrest in human fibroblasts, indicating that the mechanism of Gadd45-mediated G2/M checkpoint is at least in part through modulation of the activity of the G2-specific kinase, cyclin B1/p34cdc2. Genetic and physiological evidence of a Gadd45-mediated G2/M checkpoint was obtained by using GADD45-deficient human or murine cells. Human cells with endogenous Gadd45 expression reduced by antisense GADD45 expression have an impaired G2/M checkpoint after exposure to either ultraviolet radiation or methyl methanesulfonate but are still able to undergo G2 arrest after ionizing radiation. Lymphocytes from gadd45-knockout mice (gadd45 −/−) also retained a G2/M checkpoint initiated by ionizing radiation and failed to arrest at G2/M after exposure to ultraviolet radiation. Therefore, the mammalian genome is protected by a multiplicity of G2/M checkpoints in response to specific types of DNA damage.
Mammalian cells have evolved an intricate defense network to maintain genomic integrity by preventing the fixation of permanent damage from endogenous and exogenous mutagens. Cell-cycle checkpoints, a major genomic surveillance mechanism, exist at the G1/S and G2/M transitions that are regulated in response to DNA damage (1). Defects in these steps may result in a mutator phenotype that is associated with tumorigenesis.
Tumor suppressor gene product p53 is implicated to be one of the essential components of cell-cycle checkpoints (2–5). p53 is a transcription factor that up-regulates a number of important cell cycle-modulating genes, including p21WAF1/CIP1/SDI1 (p21) (6–8) and GADD45 (9). p21 is a cyclin-dependent kinase inhibitor that is capable of inhibiting the activity of every member of the cyclin/CDK family in vitro (10) and inhibits cell growth when overexpressed (6). The ability of DNA-damaging agents to trigger the G1/S checkpoint is caused at least in part by p53-dependent p21 activation (11). Inactivation of wild-type p53, a common event during human carcinogenesis (12, 13), results in loss of the G1/S checkpoint after DNA damage (9). Cells lacking p21 function, an infrequent event in human tumors (14), also are deficient in the G1/S checkpoint (11, 15). However, unlike p53-deficient mice, p21-deficient mice undergo normal development and normal apoptotic response, and do not have an increased frequency of spontaneous malignancies (11, 16). These data indicate that factors other than p21 in the G1/S checkpoint pathway may be required for p53-mediated tumor suppression. Alternatively, other p53-mediated checkpoints may play more important roles in the prevention of tumorigenesis.
In contrast to the G1/S checkpoint, the mammalian G2/M checkpoint is poorly understood. This checkpoint prevents the improper segregation of chromosomes, which is likely to be important in human tumorigenesis (17, 18). Genetic studies of Saccharomyces cerevisiae and Saccharomyces pombe have identified genes that are responsible for the G2/M checkpoint. Some of those genes are conserved between yeast and mammalian cells. For example, MEC1 and TEL1 in S. cerevisiae and RAD3 in S. pombe (19–22) share significant homology with the human ATM gene, which is mutated in individuals with ataxia telangiectasia (23). Mammalian cells lacking functional ATM display abnormal cell-cycle checkpoints after ionizing radiation (IR) (9, 24), and a reduction or delay in radiation-induced p53, p21, and GADD45 induction (25, 26). The human homologues of yeast Chk1, members of the14-3-3 family, and Cdc25 have been recently identified to be responsible for the IR-induced G2/M checkpoint (27–29). p53 also modulates the G2/M transition (30–34). Recent studies indicate that p53 can up-regulate 14-3-3σ, which can inhibit G2/M progression (35). A requirement of p53 and p21 to sustain G2 arrest after DNA damage has also been reported (36). In addition, p53 has been implicated in both a mitotic-spindle checkpoint and centrosome duplication, which are required for proper chromosome segregation (31, 37).
GADD45 was identified on the basis of its rapid transcriptional induction after UV irradiation (38). Induction of GADD45 also is observed after several types of pathological stimuli including various environmental stresses, hypoxia, IR, genotoxic drug exposure, and growth-factor withdrawal (25). Gadd45 binds to the proliferating cell nuclear antigen and p21 proteins (39–42) that are important for DNA replication and cell growth (6, 43). In addition, increased expression of Gadd45 causes growth inhibition of cells in culture (44), suggesting that Gadd45 may be involved in the cell-cycle checkpoints.
By using a genetic approach, we report here that Gadd45 is necessary for a G2/M checkpoint control. Interestingly, Gadd45 is only responsible for either UV or methyl methanesulfonate (MMS)-induced, but not IR-induced G2 arrest. These data indicate a multiplicity of G2/M checkpoints in mammalian cells.
MATERIALS AND METHODS
Plasmids.
p53–143ala and p53–249ser encode missense mutants of human p53, Val143Ala and Arg249Ser, respectively (45). pCMV45 encodes a human GADD45 cDNA (44). pcDNA-cycB1 encodes a 1.6 kb fragment of the human cyclin B1 cDNA. pcDNA-cdc25A, -cdc25B, and -cdc25C encode the human cdc25A, cdc25B, and cdc25C cDNA, respectively. These genes, subcloned by Koichi Hagiwara (National Cancer Institute), are under the control of the cytomegalovirus (CMV) early promoter. pactβgal encodes a β-galactosidase (β-gal) gene under the control of chicken β-actin promoter (46). pGreen-Lantern (green fluorescent protein) was obtained from GIBCO/BRL.
Cell Culture and Microinjection.
Primary human fibroblasts (GM07532, GM00038, and GM03395) were obtained from Coriell Cell Repositories (Camden, NJ). A spontaneously immortal LFS041 fibroblast cell line with homozygous deletion of wild-type p53 was derived from a Li-Fraumeni patient as described (47). These cells were grown in Ham’s F-10 medium supplemented with 15% FBS. Only early passaged primary human cells (before passage 9) were used in experiments. A colon carcinoma cell line HCT-116 contains a functional p53 and p21, and HCT-116(−/−) cells have a homozygous deletion of the p21 gene (15). These cells were grown in McCoy’s 5A medium with 10% FBS. The HCT-116-E6 cell line was derived from HCT-116 transfected with HPV-16 E6 gene, and this cell line has undetectable p53 protein (data not shown). A colon carcinoma cell line RKO and its derivative RKO-E6 cells were described previously (48). All cells were maintained in the appropriate media at 37°C in a humidified atmosphere containing 5% CO2. The microinjection procedure was described previously (45). Plasmid cDNA at a concentration of 100–200 μg/ml was used for microinjection, which results in an average of 10–20 molecules per cell. Because plasmids with broken ends were shown to be sufficient to induce p53 accumulation (49), all of the microinjected plasmids were purified by the double CsCl centrifugation protocol. At least 50 positive cells were analyzed for each experiment. Data were obtained from at least three independent microinjection experiments. Control plasmids include pactβgal and pGreen-Lantern (green fluorescent protein).
Antibodies and Immunocytochemistry Analysis.
Cells were fixed and stained for appropriate antibodies as described previously in detail (45). Anti-Gadd45 4T-27 mAb or anti-Gadd45 H-165 rabbit polyclonal antibody (Santa Cruz Biotechnology) was used for Gadd45 staining; a mitosis-specific MPM-2 mAb (Upstate Biotechnology) was used as a mitosis marker; anti-β-gal antibody was used for β-gal staining. The corresponding secondary anti-rabbit IgG or anti-mouse IgG antibodies conjugated to fluorescein isothiocyanate (FITC) or Texas Red (Vector Laboratories) were used. The centrosome-specific antiserum (SPJ) was used to visualize centrosome. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
DNA Ploidy Analysis.
Cells were fixed in acetone at −20°C and quantitatively stained for DNA content by the Feulgen reaction by using manufacturer’s protocols (Becton Dickinson). Quantitative DNA analysis was performed by the CAS-200 image analyzer (Becton Dickinson). Calibration was performed by using the rat hepatocyte. For a quiescent population, fibroblasts were incubated in F-10 medium containing 0.1% FBS for 72 hr. Cells with the Gadd45-induced distinct morphology were located and marked under phase-contrast microscope before CAS image analysis.
Cell Cycle Analysis.
For flow cytometry analysis, cells were harvested with 3.5 mM EDTA–PBS buffer, fixed with 70% ethanol for at least 1 hr at 4°C, treated with 20 μg/ml RNase A for 30 min, stained with 60 μg/ml propidium iodide for DNA content, and analyzed for cell cycle status with a FACScan cell sorter (Becton Dickinson). At least 10,000 cells were collected. The cell cycle profiles were calculated by using the cellquest and modfit lt software. Experiments were repeated at least twice.
Mitotic Index Assay.
Splenic lymphocytes were isolated from B16/129 mice at 4–15 weeks of age. For each experiment, lymphocytes from spleens of two or three animals that were littermates were pooled either from gadd45−/− animals or age-matched wild-type (gadd45+/+) animals. The genotypes of these cells were verified by using Southern blot and PCR-based methods (data not shown). The cells were grown in RPMI 1640 medium (GIBCO/BRL) supplemented with 10% heat-inactivated fetal calf serum, glutamine, and antibiotics. Immediately after harvest, 45 ng/ml 12-O-tetradecanoyl phorbol-13-acetate (PMA) (Sigma) and 0.75 μM ionomycin were added; on the following day cells were diluted 1:2 with fresh medium that did not contain additional PMA or ionomycin. Two days after harvest, the suspension cells were centrifuged, transferred to a fresh flask, and suspended in medium containing 40 units/ml IL-2 (Tecin, Roche) and 50 μM 2-mercaptoethanol (Sigma). Cells were diluted daily with medium without cytokines to maintain a density of approximately 106 per ml and used 1 or 2 days after the addition of IL-2, when they were growing maximally. Under these conditions, the doubling time was ≈15 hr and the G2/M fraction was approximately 16% by fluorescence-activated cell sorting analysis; the mitotic index of unirradiated cells was 2–3%, and it was estimated that the duration of G2 was 2 hr or less. For UV irradiation, lymphocytes were irradiated with 254-nm germicidal bulbs in 150 mm dishes containing 15 ml of medium. Cells were then incubated at 37°C for 1 or 2 hr. At each time point, cells were harvested and treated with 0.075 M KCl for 12 min at room temperature, fixed in methanol/acetic acid (3:1), spread on a glass microscope slide, air dried, and stained with 5% Giemsa. At least 3,000 cells were counted in each preparation. Data were obtained from three determents of the pooled lymphocytes from each collection and were expressed as the mean ± SD.
RESULTS
Gadd45 Arrests Normal Human Fibroblasts at the G2/M Boundary.
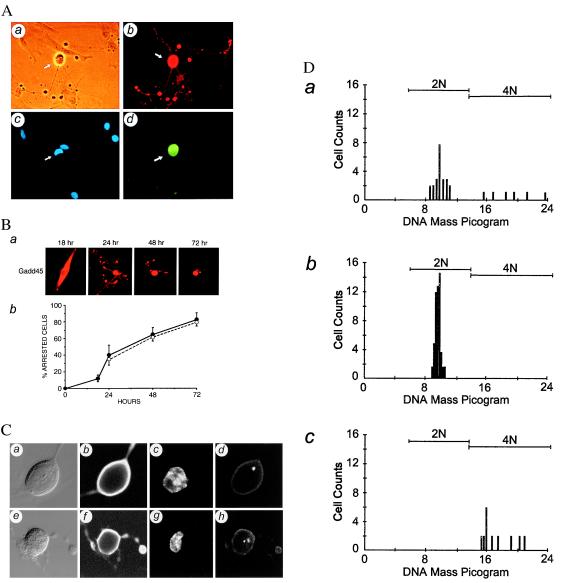
p53 is a cell-cycle checkpoint gene (6, 9) and also a potent inducer of apoptosis (5, 50–52). We recently demonstrated that increased expression of p53 by microinjection of an expression vector in either primary normal human fibroblasts or mammary epithelial cells predominantly led to apoptosis (45). However, analysis of normal primary human fibroblasts microinjected with a GADD45 expression vector revealed that Gadd45 predominantly induces a G2/M arrest (Fig. 1). Increased expression was detected as early as 3 hr after microinjection of the GADD45 expression vector (data not shown). Twenty-four hours after microinjection, 44 ± 9% (mean ± SD) of the Gadd45-overexpressing cells displayed morphologies that resemble cells arrested in an early phase of mitosis (Table 1). These include the cells with a completely rounded shape and with positive staining for MPM2 (Fig. 1A, d), a mAb that specifically recognizes certain phosphorylated proteins only present in mitosis (40). These cells also showed extensive incompletely retracted cytoplasm (Fig. 1A, a) and a partially condensed chromatin (Fig. 1 A, c and C, c and g) with an intact nuclear membrane. Because the centrosome replicates during the S phase and separates into two functional centrosomes during mitosis, we determined the status of centrosomes in the Gadd45-arrested cells by a centrosome-specific antibody SPJ (53). Approximately 85% of the Gadd45-arrested cells contained nonseparated centrosomes as analyzed by immunocytochemistry and confocal microscopy, and the remaining 15% of the arrested cells showed two separate centrosomes (Fig. 1C). These data indicate that the cells were arrested in early prophase. Similar results also were obtained by using normal primary human fibroblasts from a different donor (GM00038) (data not shown). However, the Gadd45-associated morphological changes were not observed in cells microinjected with either β-gal expression vector or green fluorescent protein expression vector (pGreen-Lantern) (data not shown). Gadd45 induction of these morphological changes was not altered by the amount of GADD45 DNA microinjected, which ranged from an average of 2 molecules per cell to 40 molecules per cell (data not shown).
Figure 1.
Gadd45-mediated G2/M cell cycle arrest in normal primary human fibroblasts. (A) At 24 hr, Gadd45-positive cells were identified by double-immunostaining with anti-Gadd45 antibody (b) and a mitosis-specific mAb MPM2 (d). A phase-contrast photomicrograph (a) and a DAPI stain (c) of the same field as b and d. (B) Time course study of Gadd45-mediated G2/M cell cycle arrest was analyzed at 18 hr, 24 hr, 48 hr, or 72 hr. Gadd45-positive cells were counted, and cells displaying a distinct mitosis-like morphology with extensive cytoplasmic extensions were scored as arrested cells. Representative Gadd45-positive cells stained with anti-Gadd45 antibody at the indicated time point (a) and the quantitative data (●) obtained from three independent time course experiments, expressed as mean +/− SD (b), are shown. ○ represent a percentage of the BrdUrd-labeled cells from the same population as an indicator of the ability of these primary cultured human fibroblasts to enter into cell cycle. (C) Gadd45-arrested cells display partially condensed nuclei and contain separated or nonseparated centrosomes, identified by double-immunostaining with anti-Gadd45 antibody (b and f) and a centrosome-specific antiserum (d and h) analyzed by confocal microscopy. The phase-contrast image (a and e) and DAPI-stained nucleus (c and g) of the same cell also are shown. (D) The DNA ploidy of a logarithmically growing fibroblast population (a), cells that were serum-starved for 3 days (b), or cells with Gadd45-induced distinct morphology (c) also were analyzed by Feulgen staining and CAS image analysis.
Table 1.
G2/M cell cycle arrest induced by increased expression of Gadd45 in different types of human cells
| Cells | Genotypes
|
Gadd45 expressing-cells exhibiting G2/M arrest, % | n | P* | ||
|---|---|---|---|---|---|---|
| p53 | p21 | ATM | ||||
| GM07532 | wt | wt | wt | 44 ± 9 | 15 | |
| LFS041 | −/− | wt | wt | 0 | 7 | <0.001 |
| GM03395 | wt | wt | −/− | 52 | 2 | 0.32 |
| HCT116 | wt | wt | wt | 46 ± 21 | 4 | |
| 80S4 | wt | −/− | wt | 25 | 2 | 0.25 |
| 80S14 | wt | −/− | wt | 59 ± 22 | 5 | 0.41 |
GM07532, primary normal human fibroblasts; LFS041, an immortalized fibroblast cell line derived from a Li-Fraumeni patient 041; GM03395, primary fibroblasts from an ataxia telangiectasia patient; HCT116, a human colon carcinoma cell line; 80S4 and 80S14, two clones derived from HCT116 cells where the p21 gene is homozygously deleted. n, Number of independent microinjection experiments conducted. Data, expressed as mean ± SD, were obtained at 24 hr after microinjection. wt, wild-type.
Student’s t-test was used to compare mutant cells with corresponding wild-type cells (GM07532 or HCT116) of the same cell type.
Time-course study indicates that the fraction of arrested cells increases with the incubation time and reaches 83% at 72 hr (Fig. 1B, b). Also, all of the cells entering into the cell cycle appear to be arrested by Gadd45 because the numbers of the bromodexoyuridine (BrdUrd)-labeled cells (Fig. 1B, b) at the same passages coincide with the numbers of the Gadd45-arrested cells. Of interest, the cytoplasmic retraction associated with Gadd45-arrested cells eventually went to completion (Fig. 1B, a), as confirmed by the time-lapse video analysis (data not shown). To further demonstrate that the Gadd45-arrested cells had passed through S phase, we performed DNA ploidy analysis with a CAS imaging system. Unsynchronized normal primary human fibroblasts displayed a normal logarithmic-phase distribution (79% in 2n and 21% in 4n), whereas virtually all of the cells were 2n after being serum-starved for three days (Fig. 1D). However, the Gadd45-arrested cells displayed 4n DNA contents. Taken together, these data indicate that increased expression of GADD45 in the normal primary human fibroblasts results mainly in a cell-cycle arrest at the G2/M transition. Unlike p53 overexpression, increased expression of GADD45 did not lead to the typical morphological changes associated with either p53 or ICE-induced apoptosis (45). Gadd45-overexpressing cells also were negative by terminal deoxynucleotidyltransferase-mediated UTP end labeling assay (data not shown) (54). Expression of Gadd45 also causes a G2/M arrest only in a colon carcinoma cell line HCT116 with wild-type p53, as analyzed by fluorescence-activated cell sorting analysis (data not shown).
Gadd45-Induced Cell Cycle Arrest Is p53-Dependent.
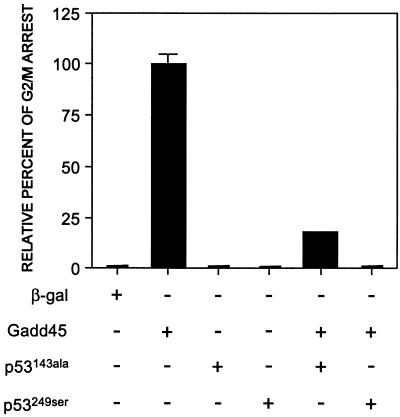
Although Gadd45 induced a G2/M arrest in normal human fibroblasts, it did not induce a G2/M arrest in a fibroblast cell line derived from a Li-Fraumeni patient (LFS041) that does not contain p53 protein (47) (Table 1). Furthermore, the dominant-negative p53 mutants can also abrogate a Gadd45-induced arrest in normal fibroblasts when coexpressing together, presumably by neutralizing the endogenous wild-type p53 (Fig. 2). A protective effect was not observed when GADD45 was coexpressed with either a β-gal expression vector or a green fluorescent protein expression vector (data not shown). Thus, Gadd45-induced G2/M arrest is p53-dependent.
Figure 2.
Modulation of Gadd45-induced G2/M arrest by p53 mutants in normal primary human fibroblasts. Cells were either microinjected alone with β-gal, GADD45, and two p53 mutants or coinjected with GADD45 and the p53 mutants. Gadd45-positive cells displaying an arrested morphology were scored at 24 hr. Anti-p53 CM-1 polyclonal antibody was used to screen for p53-positive cells. At least three independent microinjection experiments were averaged, and data are expressed as the mean ± SD.
Gadd45-Deficient Cells Are Defective in Either UV or MMS-Induced G2/M Arrest.
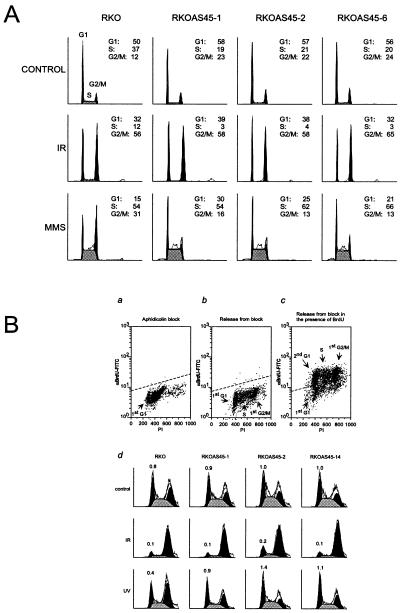
A genetic approach was then used to test the hypothesis that Gadd45 is required for the DNA damage-induced G2/M checkpoint. By using flow cytometry analysis, we first compared the cell-cycle distributions of unsynchronized cell populations from the parental human RKO colon carcinoma cells and three isogenic RKO-derived clones stably expressing antisense GADD45 mRNA established previously (55, 56). These cells have a diminished constitutive Gadd45 protein level and are more sensitive to UV radiation but not to γ-radiation in a clonogenic survival assay (55, 56) (data not shown). Cells were treated with either 6.3 Gy of γ-radiation, 5 J/m2 of UV, or 25 μg/ml MMS. We selected these doses for MMS and UV radiation because they were not toxic to these cells in a short-term growth assay and because higher doses primarily caused a delay in S phase, which may be caused by a high frequency of DNA lesions blocking replication or initiating an S phase checkpoint (data not shown). Fig. 3A depicts a representative flow cytometric analysis profile of these cells. The three untreated antisense GADD45 clones had similar G1 fractions, slightly lower S fractions, and slightly increased G2/M fractions compared with parental RKO cells (Fig. 3A). IR caused a significant accumulation in G2/M with concomitant reductions in G1 and S in all four cell lines. After MMS exposure, the parental RKO cells clearly accumulated in S and G2/M. However, the three GADD45 antisense clones failed to arrest in G2/M after MMS treatment, although the S phase accumulation still occurred (Fig. 3A). Similar results also were obtained with cells synchronized by either a uridine biosynthesis inhibitor (phosphoacetyl)-l-aspartate (PALA) or aphidicolin (data not shown). RKO-E6 cell line also did not accumulate in G2/M after UV or MMS exposure, consistent with the model that p53 may be required for these types of damage-induced G2/M checkpoint (data not shown).
Figure 3.
Defect in a UV-induced G2/M checkpoint in Gadd45-deficient cells. (A) Flow cytometric analysis of the human colon carcinoma cell line RKO and its GADD45 antisense overexpressing clones in response to IR or MMS treatment. Unsynchronized cells were treated with either 6.3 Gy IR or 25 μg/ml MMS and analyzed by FACScan after a 24-hr incubation. The G1, S, and G2/M fractions are indicated, as calculated by modfit analysis. (B) Increased expression of antisense GADD45 allows RKO cells to escape the G2/M checkpoint arrest induced by UV-induced damage. Parental RKO cells and three antisense GADD45-expressing clones were synchronized with aphidicolin for 24 hr (>95% at G1) and release from blocking in the presence of BrdUrd. (a–c) Representative profiles of RKO cells subjected to synchronization. The intensities of fluorescein isothiocyanate (anti-BrdUrd signal) and PI (DNA contents) are indicated. Three antisense GADD45 clones show similar synchronization profiles as their parental RKO cells (data not shown). On release from late G1, cells were treated with 6.3 Gy IR or 5 J/m2 UV and incubated for an additional 20 hr in the presence of BrdUrd. The BrdUrd-positive cells were subjected to FACScan and modfit analysis (d). The first solid peaks, the gray area, and the second solid peaks represent the second G1 (2nd G1), first S (S) and first G2/M (1st G2) fractions, respectively. No second S phase should be observed in this condition. The numbers above the second G1 peaks are the ratio of G1/G2 (percent of G1 fraction escaped from G2/M arrest).
A BrdUrd labeling protocol was used to further examine whether the lack of a G2/M arrest is caused by failure in controlling the G2 cell cycle exit. The RKO parental cells and three antisense Gadd45-expressing clones were synchronized at the G1/S transition by treatment with 1 μg/ml aphidicolin for 24 hr. Greater than 95% of cells had a G1 DNA content following synchronization (Fig. 3B). On release from the aphidicolin block, cells were treated with various DNA-damaging agents and incubated for an additional 20 hr in the presence of BrdUrd. Cells were harvested and the BrdUrd-positive cell populations were subjected to FACScan analysis. Untreated parental RKO cells and three antisense clones (RKOAS45–1, RKOAS45–2, and RKOAS45–14) displayed similar cell cycle distributions (28, 30, 29, and 31% in second G1; 35, 39, 40, and 39% in first S; 37, 31, 31, and 30% in first G2/M, respectively) (Fig. 3B). The G1/G2 ratios from parental and RKOAS45–1, RKOAS45–2, and RKOAS45–14 were 0.8, 0.9, 1.0, and 1.0, respectively. After treatment with 6.3 Gy IR, most of the cells in the parental and antisense clones (>74%) were accumulated in the first G2/M, indicating a functional γ-radiation-induced arrest (Fig. 3B). After exposure to a 5 J/m2 UV, many of the parental RKO cells (51%) were arrested at the first G2/M, with a small population (22%) in the second G1. However, the three antisense Gadd45-expressing clones (RKOAS45–1, RKOAS45–2, and RKOAS45–14) did not accumulate at the first G2/M and continued into the second G1 of the cell cycle (Fig. 3B). Similar results were obtained when these cells were treated with 25 μg/ml MMS (data not shown). Collectively, the above data indicate that the parental RKO cells have a physiologically normal G2/M checkpoint in response to UV, MMS, and IR treatment and that increased expression of either antisense GADD45 mRNA or E6 allows the RKO cells to escape the G2/M arrest induced by either UV or by MMS treatment but not by IR.
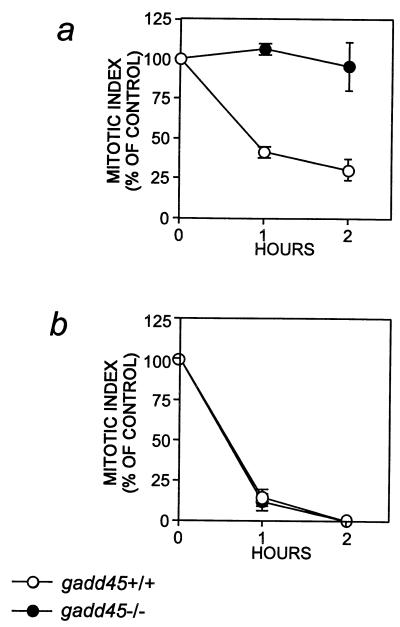
To extend the above results to murine cells, we compared the mitotic index of lymphocytes derived from normal and gadd45 knockout (−/−) mice in response to UV or IR irradiation. These cells were exposed to either UV radiation at 7.5 J/m2 or to IR at 5 Gy. A significant reduction in the percentage of the mitotic cells was observed at both time points in gadd45 +/+ lymphocytes after 7.5 J/m2 of UV treatment (Fig. 4), indicating a functional G2/M checkpoint in these cells. However, no alteration of mitotic index in the gadd45 −/− lymphocytes exposed to UV radiation was observed. Similar results were observed in lymphocytes exposed to UV radiation at either 7.5 or 20 J/m2 from two individual gadd45 mice (data not shown). However, lymphocytes from gadd45 +/+ and gadd45 −/− mice displayed normal arrest when irradiated with 5 Gy IR (Fig. 4). Taken together, we conclude that cells lacking Gadd45 are defective in a G2/M checkpoint induced by either UV or MMS.
Figure 4.
Failure of the UV-induced mitotic delay of murine gadd45 (−/−) lymphocytes. Proliferating lymphocytes from gadd45 (+/+) mice (○) or gadd45 (−/−) mice (●) were treated with UV radiation at 7.5 J/m2 (A) or with IR at 5 Gy (B). Mitotic indices were determined at the indicated times after irradiation and are shown as a percentage of the appropriate unirradiated control cells. For A, pooled lymphocytes from two gadd45 (−/−) male mice at 11 weeks of age and two gadd45 (+/+) male mice at 15 weeks of age were used. For B, pooled lymphocytes from three gadd45 (−/−) male mice at 4 weeks of age and three gadd45 (+/+) male mice at 6 weeks of age were used. Similar results were observed from three separate cultures that also included female mice (not shown).
Cyclin B1 and Cdc25C Attenuate the Gadd45-Induced G2/M Arrest.
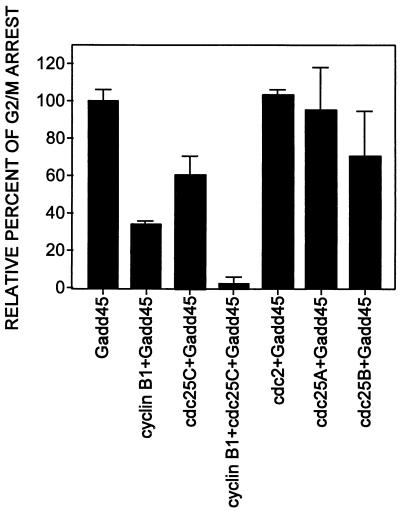
The mechanism(s) of Gadd45-mediated G2/M arrest was explored by investigating the effects of G1-to-S phase or G2-to-M phase cell cycle-modulating genes. Various cell cycle-related cDNA expression vectors were coinjected with the GADD45 expression vector into normal primary human fibroblasts. Increased expression of either human cyclin B1 or Cdc25C, proteins required for the G2/M transition (57), partially blocked the Gadd45-induced G2/M arrest (P < 0.05) (Fig. 5). Coexpression of cyclin B1 and Cdc25C almost completely abolishes Gadd45-mediated G2/M arrest (Fig. 5). However, increased expression of either the human G1/S phase-specific protein phosphatases cdc25A or cdc25B, or the human G2-specific cyclin-dependent kinase p34cdc2 had little effect on the Gadd45-induced G2/M arrest. Although these data do not provide conclusive evidence, they are consistent with the plausible model that Gadd45 arrests cells through inactivation of these factors.
Figure 5.
Effect of various cell cycle-modulating genes on the Gadd45-induced G2/M arrest. Normal primary human fibroblasts were either microinjected alone with GADD45 or coinjected with cyclin B1, cdc25A, cdc25B, cdc25C, or p34cdc2 (cdc2). Cells were fixed at 24 hr, and Gadd45-positive cells displaying an arrested morphology were scored. At least three independent microinjection experiments were averaged, and data are expressed as mean ± SD. By using Student’s t test, a statistically significant P value (<0.05) was obtained between GADD45 alone and coinjection with cyclin B1 and/or cdc25C.
Because p21 binds to Gadd45 in vivo (40, 42), we then determined whether p21 is required for the Gadd45-mediated G2/M arrest. GADD45 expression vector was microinjected into either HCT116 cells or two clonal derivatives in which the p21 gene was homozygously deleted (15). GADD45 was capable of inducing G2/M arrest in all three cell lines (Table 1). We also tested whether fibroblasts from patients with ataxia telangiectasia (AT) display an abnormal G2/M arrest by Gadd45, because a deficiency in G2/M checkpoint in response to IR treatment was associated with cells from AT patients (9, 24) suggesting that ATM is required for the G2/M checkpoint, similar to RAD3 in S. pombe and MEC1 and TEL1 in S. cerevisiae. Increased expression of Gadd45 efficiently led to a G2/M arrest in fibroblasts from an AT patient (GM03395), which is indistinguishable from the normal fibroblasts (Table 1). Thus, we conclude that both p21 and ATM are dispensable for the Gadd45-mediated G2/M arrest in normal human fibroblasts.
DISCUSSION
Our findings demonstrate that Gadd45 can mediate a G2/M cell-cycle checkpoint in both human and murine cells. Genetic evidence of the importance of Gadd45 in a G2/M checkpoint is provided by a defective UV but not IR-induced G2/M arrest in gadd45 −/− murine lymphocytes and in antisense GADD45-expressing human colon carcinoma cells. Increased expression of Gadd45 in normal primary human fibroblasts arrests the cells in G2/M. The data also agree with the previous reports that antisense GADD45-expressing RKO cells displayed a decreased clonogenic survival to UV radiation but not IR radiation (39, 56). Increased expression of Gadd45 in normal primary human fibroblasts also arrests the cells at the G2/M transition. The arrested cells show an early mitotic morphology and mitotic markers, with 4n DNA content, a partially condensed nucleus with an intact nuclear membrane, a delay in cytoplasmic retraction, and with 15% of the arrested cells having two separated centrosomes. Taken together, we conclude that Gadd45 is required in human cells for a normal G2/M checkpoint in response to certain kinds of DNA damage.
The transition from G2 to M is regulated in part by the mitosis-promoting factor, consisting of cyclin B1 and a G2-specific cyclin-dependent kinase Cdc2 (for review, see ref. 57). The level of Cdc2 is relatively high and remains constant through the cell cycle, whereas the level of cyclin B1 is strictly dependent on the phase of the cell cycle and largely increases during the G2/M transition. In mammalian cells, Cdc2 can be phosphorylated by the Cdc2-activating kinase, a complex containing cyclin H and CDK7/MO15. This in turn activates the mitosis-promoting factor and promotes mitosis. Cdc2 also can be negatively regulated by phosphorylation at Tyr-15 by the WEE1 kinase. Cdc25C, one of the three cell cycle-specific protein phosphatases in mammalian cells, may activate the mitosis-promoting factor by dephosphorylating this site. Recent studies provide evidence indicating the involvement of Cdc25C in the IR-induced G2/M checkpoint through activation of a protein kinase Chk1 (27–29). This pathway, conserved from yeast to human, requires the activation of Chk1 followed by inactivation of Cdc25C through binding to 14-3-3 to initiate a G2/M checkpoint (27–29). However, multiple G2/M checkpoints may exist in mammalian cells. Analysis of the Gadd45-deficient cells in response to various DNA damaging agents reveal that these cells still retain a functional G2/M checkpoint after IR treatment, yet they are deficient in either UV- or MMS-induced G2/M arrest. Thus, the IR-mediated pathway differs from the UV- and the MMS-mediated pathways in initiating the G2/M checkpoint. UV radiation and IR induce different types of DNA damage, and the cellular responses also are probably different. For example, the p53 response to IR is rapid but very transitory compared with UV (58), and UV-induced p53 accumulation is partially dependent on UV-induced oxyradicals (59). One difference between IR and UV radiation is that UV causes less DNA strand breaks than does IR. It is known that cells from AT patients are defective in their response to IR, but show a normal response to UV radiation, indicating that the AT-related cell-cycle checkpoint deficiency is limited to IR.
p53 has been implicated in controlling a G2/M cell-cycle checkpoint (30–34). However, an IR-induced G2/M checkpoint is generally p53-independent (30, 48, 60). By using the oncogenic HPV-16 E6 gene to inactivate p53, we show here that p53 may be necessary for either the UV- or MMS-induced G2/M checkpoint but is not required for an IR-induced G2/M checkpoint. In contrast to previous work suggesting that Gadd45 may be a component of the p53 signaling pathway, our data indicate that p53 may be necessary for Gadd45 to execute the G2/M checkpoint because no G2/M arrest by Gadd45 expression or by UV radiation was observed in p53-null cells. It is possible that Gadd45 may cooperate with p53 and activate p53 to promote G2/M arrest, similar to the action of p33ING1 (61). However, no evidence so far indicates that Gadd45 directly binds to p53 (data not shown). Alternatively, Gadd45 and p53 may activate separate downstream targets that cooperate to execute the G2/M block. We favor this model because Gadd45 physically interacts with Cdc2 but not cyclin B1 in vivo and in vitro (62). In addition, a purified recombinant Gadd45 protein inhibits the H1-kinase activity associated with Cdc2/cyclin B1 in vitro through dissociation of Cdc2/cyclin B1 complex (62). Thus, Gadd45 may be involved in a G2/M checkpoint at least in part through direct inactivation of Cdc2/cyclin B1 kinase activity. The Gadd45-mediated G2/M checkpoint depends on p53 function. We hypothesize that p53 participates in this pathway by direct binding to Cdc2 (63–65). Because the level of cyclin B1 is critical during the G2-to-M transition, we also hypothesize that p53 also inhibits Cdc2/cyclin B1 kinase activity by either transrepressing cyclin B1 expression or enhancing its degradation. Another candidate to inhibit G2/M progression is 14-3-3σ (35). p53 transcriptionally up-regulates 14-3-3σ, which in turn may inhibit nuclear transport of Cdc25C by sequestering it in the cytoplasm (35). Consistent with this model are our findings that the expression of either cyclin B1 or Cdc25C only partially attenuates Gadd45-mediated G2/M arrest and that the coexpression of both genes results in a complete abrogation. Thus, the Gadd45 induction of a G2/M checkpoint is independent from the IR-induced pathway that requires ATM and Chk1. Our results indicate a multiplicity of G2/M checkpoint pathways linked to different types of DNA damage in mammalian cells.
Cell-cycle checkpoint genes are generally conserved from human to yeast, including the human ATM and Chk1 genes. Because a yeast gene homologous to either human Gadd45 or p53 has not yet been identified, Gadd45 or p53 may share a unique function in multicellular organisms to control the cell-cycle checkpoints and to maintain genomic stability.
Acknowledgments
We thank Ron Balczon, David Beach, Paul Nurse, Hiroto Okayama, James Roberts, Paul Russell, Michael Tainsky, Bert Vogelstein, Todd Waldman, and Juyin Yuan for their generosities in providing plasmids, antibodies, and cell lines. We thank Michael Mancini and James Resau for their expertise in Confocal imaging analysis, John Chen for his assistance with flow cytometry; Seung Chang, Pat Phelps, and Ben Trump for their excellent assistance with time-lapse video analysis; Michael Kasten, Steve Linke, and Geoffery Wahl for invaluable comments and critical reading of the manuscript; and Amy Hancock, Marshonna Forgues, Elisa Spillare, and Lynne Elmore for their advice and assistance. Portions of this work were completed by J.D.C. in partial fulfillment of requirement for a Master of Science at Hood College, Frederick, MD.
ABBREVIATIONS
- DAPI
4′,6-diamidino-2-phenylindole
- IR
ionizing radiation
- MMS
methyl methanesulfonate
- BrdUrd
bromodexoyuridine
- β-gal
β-galactosidase
- AT
ataxia telangiectasia
References
- 1.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Cox L S, Lane D P. BioEssays. 1995;17:501–508. doi: 10.1002/bies.950170606. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb T M, Oren M. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 4.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 5.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 6.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 8.Noda A, Ning Y, Venable S F, Pereira-Smith O M, Smith J R. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 9.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 11.Deng C X, Zhang P M, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 12.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 13.Levine A J, Momand J, Finlay C A. Nature (London) 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 14.Shiohara M, El-Deiry W S, Wada M, Nakamaki T, Takeuchi S, Yang R, Chen D L, Vogelstein B, Koeffler H P. Blood. 1994;84:3781–3784. [PubMed] [Google Scholar]
- 15.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 16.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell L H, Kastan M B. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 18.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 19.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 20.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 21.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 22.al-Khodairy F, Carr A M. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 24.Beamish H, Lavin M F. Int J Radiat Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- 25.Papathanasiou M A, Kerr N C, Robbins J H, McBride O W, Alamo I, Jr, Barrett S F, Hickson I D, Fornace A J., Jr Mol Cell Biol. 1991;11:1009–1016. doi: 10.1128/mcb.11.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artuso M, Esteve A, Bresil H, Vuillaume M, Hall J. Oncogene. 1995;11:1427–1435. [PubMed] [Google Scholar]
- 27.Furnari B, Rhind N, Russell P. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 29.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal M L, Agarwal A, Taylor W R, Stark G R. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 32.Guillouf C, Rosselli F, Krishnaraju K, Moustacchi E, Hoffman B, Liebermann D A. Oncogene. 1995;10:2263–2270. [PubMed] [Google Scholar]
- 33.Powell S N, DeFrank J S, Connell P, Eogan M, Preffer F, Dombkowski D, Tang W, Friend S. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]
- 34.Stewart N, Hicks G G, Paraskevas F, Mowat M. Oncogene. 1995;10:109–115. [PubMed] [Google Scholar]
- 35.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 36.Bunz F, Dutriaux C, Lengauer T, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 37.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 38.Fornace A J, Jr, Nebert D W, Hollander M C, Luethy J D, Papathanasiou M, Fargnoli J, Holbrook N J. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith M L, Chen I T, Zhan Q, Bae I, Chen C Y, Gilmer T M, Kastan M B, O’Connor P M, Fornace A J., Jr Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 40.Chen I T, Smith M L, O’Connor P M, Fornace A J., Jr Oncogene. 1995;11:1931–1937. [PubMed] [Google Scholar]
- 41.Hall P A, Kearsey J M, Coates P J, Norman D G, Warbrick E, Cox L S. Oncogene. 1995;10:2427–2433. [PubMed] [Google Scholar]
- 42.Kearsey J M, Coates P J, Prescott A R, Warbrick E, Hall P A. Oncogene. 1995;11:1675–1683. [PubMed] [Google Scholar]
- 43.Shivji M K, Kenny M K. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 44.Zhan Q, Lord K A, Alamo I, Jr, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Fornace A J., Jr Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X W, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers J H J, Harris C C. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 46.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 47.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 48.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 49.Huang L C, Clarkin K C, Wahl G M. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 51.Shaw P, Bovey R, Tardy S, Sahli R, Sordat B, Costa J. Proc Natl Acad Sci USA. 1992;89:4495–4499. doi: 10.1073/pnas.89.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Ko L J, Jayaraman L, Prives C. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 53.Balczon R, West K. Cell Motil Cytoskeleton. 1991;20:121–135. doi: 10.1002/cm.970200205. [DOI] [PubMed] [Google Scholar]
- 54.Gavrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith M L, Chen I T, Zhan Q, O’Connor P M, Fornace A J., Jr Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 56.Smith M L, Kontny H U, Zhan Q, Sreenath A, O’Connor P M, Fornace A J., Jr Oncogene. 1996;13:2255–2263. [PubMed] [Google Scholar]
- 57.King R W, Jackson P K, Kirschner M W. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 58.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 59.Renzing J, Hansen S, Lane D P. J Cell Sci. 1996;109:1105–1112. doi: 10.1242/jcs.109.5.1105. [DOI] [PubMed] [Google Scholar]
- 60.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garkavtsev I, Grigorian I A, Ossovskaya V S, Chernov M V, Chumakov P M, Gudkov A V. Nature (London) 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 62.Zhan, Q., Antinore, M. J., Wang, X. W., Carrier, F., Smith, M. L., Harris, C. C. & Fornace, A. J., Jr. (1998) Oncogene, in press. [DOI] [PubMed]
- 63.Milner J, Cook A, Mason J. EMBO J. 1990;9:2885–2889. doi: 10.1002/j.1460-2075.1990.tb07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bischoff J R, Friedman P N, Marshak D R, Prives C, Beach D. Proc Natl Acad Sci USA. 1990;87:4766–4770. doi: 10.1073/pnas.87.12.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sturzbecher H W, Maimets T, Chumakov P, Brain R, Addison C, Simanis V, Rudge K, Philp R, Grimaldi M, Court W, Jenkins J R. Oncogene. 1990;5:795–801. [PubMed] [Google Scholar]