Abstract
Two fundamental properties of stem cells are their ability to self-renew and to differentiate. Self-renewal is an integration of proliferation control with the maintenance of an undifferentiated state. Stem cell self-renewal is regulated by the dynamic interplay between transcription factors, epigenetic control, microRNA (miRNA) regulators, and cell-extrinsic signals from the microenvironment in which stem cells reside. Recent progress in defining specific roles for cell-intrinsic factors and extrinsic factors in regulating stem cell self-renewal starts to unfold the multilayered regulatory networks. This review focuses on cell-intrinsic regulators, including orphan nuclear receptor TLX, polycomb transcriptional repressor Bmi1, high-mobility-group DNA binding protein Sox2, basic helix-loop-helix Hes genes, histone modifying enzymes and chromatin remodeling proteins, and small RNA modulators, as well as cell-extrinsic signaling molecules, such as Wnt, Notch, Sonic hedgehog (Shh), TGFα, EGF, and FGF. Unraveling the mechanisms by which neural stem cells renew themselves will provide insights into both basic neurosciences and clinical applications of stem cell-based cell replacement therapies for neurodegenerative diseases.
Keywords: neural stem cells, self-renewal, transcriptional regulators, epigenetic control, microRNA, nuclear receptors, histone deacetylases, Wnt
1. Introduction
One of the most important issues in stem cell biology is to uncover molecular mechanisms underlying stem cell self-renewal. Self-renewal is essential for stem cells because it is required for all stem cells to perpetuate themselves. Neural stem cells are a subset of undifferentiated precursors that retain the ability to proliferate and self-renew, and have the capacity to give rise to both neuronal and glial lineages [1-4]. Although the functional properties of neural stem cells have been studied extensively, we have just begun to understand how self-renewal of neural stem cells is regulated. A complete understanding of neural stem cells requires the identification of molecules that determine the self-renewal character of these cells. This review touches on some of the recently characterized pathways that are involved in regulating this process.
2. Transcription Regulators
Batteries of transcription factors have been proposed to control neural stem cell self-renewal (Fig. 1). Orphan nuclear receptor TLX is an essential transcriptional regulator of neural stem cell maintenance and self-renewal in the adult brain [5]. Although TLX knockout mice are viable and appear normal at birth, the TLX gene has been shown to be required for the formation of superficial cortical layers and the zinc-containing cortical circuits in embryonic brains [6, 7], to regulate the timing of neurogenesis in the cortex [8], and to control patterning of lateral telencephalic progenitor domains during development [9]. Mature TLX knockout mice have significantly reduced cerebral hemispheres [10, 11] and severe retinopathies [12-15]. Behaviorally, adult TLX mutants exhibit increased aggressiveness, decreased copulation, progressively violent behavior, late onset epilepsy and reduced learning abilities [10, 11, 16, 17].
Fig. 1.
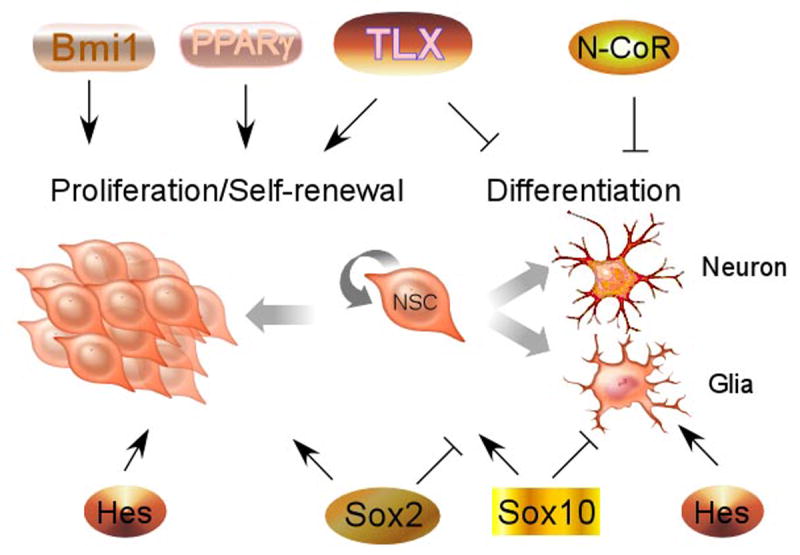
Transcriptional regulators of neural stem cell self-renewal. Transcription factors that are expressed in neural stem cells can potentiate cell proliferation and repress cell differentiation to maintain the undifferentiated and self-renewable state of neural stem cells. Due to space limitations, not all related transcription factors are listed here.
We have shown that TLX is an essential regulator of neural stem cell self-renewal [5]. TLX maintains adult neural stem cells in an undifferentiated and self-renewable state (Fig. 1). The TLX-expressing cells isolated from adult TLX-heterozygote brains can proliferate, self-renew and differentiate into all neural cell types in vitro. By contrast, TLX-null cells isolated from the brains of adult TLX-mutant mice fail to proliferate. Reintroducing TLX into TLX-null cells rescues their ability to proliferate and self-renew [5]. In vivo, TLX mutant mice show a loss of cell proliferation and reduced neural precursors in the neurogenic areas of adult brains. TLX represses the expression of astrocyte markers, such as GFAP, and tumor suppressor gene, pten, in neural stem cells, suggesting that transcriptional repression is crucial in maintaining the undifferentiated state of neural stem cells [5, 14]. Similarly, the Drosophila tailless acts as a dedicated repressor in the early Drosophila embryo to support normal embryonic development and establish accurate patterns of gene expression [18]. TLX could be a key regulator that acts by controlling the expression of a network of target genes to establish the undifferentiated and self-renewable state of neural stem cells. Elucidation of the network regulated by TLX in producing these outcomes would be a significant advance in understanding neural stem cell self-renewal and neurogenesis.
Recently, other nuclear receptors such as estrogen receptors (ER), thyroid hormone receptors (TR), and peroxisome proliferator-activated receptor γ (PPARγ), have also been shown to regulate neural stem cell proliferation and differentiation [19-24]. Significant neuronal loss has been detected in adult brains of ERβ knockout mice, suggesting an important role of ERβ in neuronal maintenance in the central nervous system [25]. Knockout of TRα inhibits progression of neural stem cells through cell cycles, suggesting a critical role of TRα in neurogenesis of mammalian adult brains [26]. Neural stem cells prepared from heterozygous PPARγ-deficient mouse brains have significantly reduced cell growth, which is also seen in PPARγ short hairpin RNA silenced or dominant negative PPARγ-treated neural stem cells, suggesting that PPARγ plays a role in neural stem cell proliferation control [27]. N-CoR, a nuclear receptor co-repressor, also plays a role in neural stem cell self-renewal. Deficiency in N-CoR leads to reduced neural stem cell self-renewal and premature differentiation into astrocytes [28].
Bmi1 is a polycomb family transcriptional repressor that has been shown to be required for post-natal maintenance of neural stem cells in the peripheral and central nervous system [29]. Deficiency in Bmi1 leads to progressive postnatal growth retardation and neurological defects [30]. Bmi1-null mice exhibit a post-natal self-renewal defect that leads to the depletion of stem cells by early adulthood [29]. One way in which Bmi1 promotes the maintenance of adult stem cells is by repressing the cyclin-dependent kinase inhibitors, p16Ink4a and p19Arf [31, 32].
The Sox family of high-mobility-group (HMG) DNA binding proteins plays a role in maintaining the undifferentiated state of neural stem cells in a context-dependent manner. In vertebrates, SoxB1 factors (Sox1, Sox2, and Sox3) are widely expressed in proliferating neural stem/progenitor cells, throughout development and adulthood [33-35]. SoxB1 factors have been shown to play a role in maintaining the undifferentiated state of embryonic neural progenitors [36]. Overexpression of Sox2 and/or Sox3 inhibits neuronal differentiation of neural progenitors and causes them to retain undifferentiated properties. In contrast, expression of a dominant negative form of Sox2 and/or Sox3 results in premature exit of neural progenitors from cell cycle and onset of neuronal differentiation [37, 38]. In addition to its function in early brain development [37-40], Sox2 is also necessary for the maintenance of neural stem cells in adult neurogenic areas [41, 42]. Regulatory mutations of Sox2 cause neurodegeneration and impaired adult neurogenesis [41]. In parallel, Sox10, a member of the Sox E subfamily, maintains the multipotency of neural crest stem cells in the peripheral nervous system [43].
Multiple basic helix-loop-helix (bHLH) genes also play a critical role in regulation of neural stem cell maintenance and differentiation [44-46]. Hes genes, homologs of Drosophila hairy and enhancer of split, are repressor-type bHLH genes. Among seven members of the Hes family, Hes1 and Hes5 are essential effectors of Notch signaling, the expression of which is upregulated by Notch activation [47, 48]. Hes1 and Hes5 are highly expressed by neural stem cells [49-51], mis-expression of which inhibits neuronal differentiation and maintains neural stem cells in the embryonic brain [52-54]. In contrast, neural progenitors underwent premature neuronal differentiation in Hes1 and Hes5 double knockout mice [48, 55-57], suggesting that Hes1 and Hes5 are essential for the maintenance and self-renewal of neural stem cells. Hes genes regulate neural stem cell self-renewal by repressing premature onset of the activator type bHLH genes such as Mash1, Math, and Neurogenin [46]. Hes-related bHLH genes, Hesr1 and Hesr2 are also expressed by neural stem cells in the embryonic brains and act as Notch signaling effectors. Hesrs regulate neural stem cell maintenance and self-renewal too, possibly through cooperative action with Hes [46]. Together, these data suggest that intrinsic transcription factors can work together to coordinate neural stem cell maintenance and self-renewal.
3. Epigenetic Control
Stem cell self-renewal and differentiation are the result of transcription control in concert with chromatin remodeling and epigenetic modifications. During central nervous system development in vertebrates, neural stem cell fate are strictly controlled under regional and temporal manners [58], accompanied by precise epigenetic control, including covalent histone modification and DNA methylation of CpG dinucleotides.
Histone modification includes histone acetylation, methylation, phosphorylation ubiquitylation, sumolyation, and ADP-ribosylation [59]. Histone acetylation, which is regulated by histone acetylases (HATs) and histone deacetylases (HDACs) (Fig. 2), has been best studied. HDACs can deacetylate the conserved acetylated lysine residues in histone tail, resulting in local condensation in chromatin and block access of transcriptional factors to their target genes. Mammalian HDACs can be classified based on their structure and homology to yeast HDACs. Class I HDACs (HDAC1, 2, 3, 8, and 11) are ubiquitously expressed, whereas Class II HDACs (HDAC4, 5, 7, and 9) display tissue-specific expression. Class II HDACS contain an amino-terminal extension that interacts with other transcriptional cofactors and confers responsiveness to extracellular signals. We have shown that HDAC-mediated transcriptional repression is essential for the maintenance and self-renewal of neural stem cells (Sun et al. unpublished results). In addition to self-renewal, HDACs also regulate neural cell differentiation. Treatment of adult neural stem cells with HDAC inhibitors induced neuronal differentiation [60], due in part to upregulating REST (RE1 silencing transcription factor, or NRSF)-regulated neuronal-specific genes. REST is a key transcriptional regulator of many neuronal genes through binding to a conserved 21 bp RE1 binding site [61, 62]. In non-neuronal cells, REST interacts with its co-factors, including Co-REST, N-CoR, and mSin3A, which then recruit HDAC complexes to repress neuronal gene expression through epigenetic regulation [61, 63, 64].
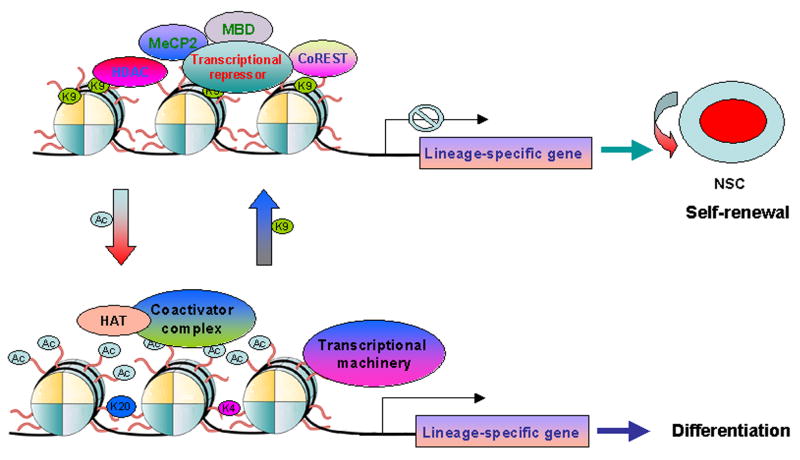
Fig. 2.
Epigenetic control of neural stem cell self-renewal. Maintenance of neural stem cells is normally associated with the repressive status of chromatin, which is represented by deacetylation of histones and methylation of histone H3 lysien 9 (K9). The repression is presumably resulted from recruitment of transcriptional corepressor complexes, including HDACs, MecP2, MBD, CoREST, to transcription factors at the promoter of target genes. Histone acetylation and methylation of histone H3 lysine 4 (K4) and lysine 20 (K20) are, on the other hand, involved in relaxation of chromatin structures and activation of lineage-specific gene expression, which in turn leads to differentiation of neural stem cells into mature neuron, astrocyte or oligodendrocyte. NSC stands for neural stem cells; Ac stands for histone acetylation.
More recently, histone methylation has gained attention as an epigenetic marker [65, 66]. Unlike histone acetylation, which only occurs on lysine (K) residues and is generally related with active transcription, methylation was detected on both lysine and arginine (R) residues and is linked to both transcriptional activation and repression [66]. For instance, histone H3 K9 methylation is associated with transcriptional silencing (Fig. 2). In contrast, methylation of histone H3 K4 and arginine residues of H3 and H4 leads to transcriptional activation [67]. Recent identification of LSD1 as a histone demethylase [68] indicates that similar to histone acetylation, histone methylation is also a dynamic process subject to regulation by both methyl transferases and demethylases. The degree of lysine methylation (mono-, di-, or trimethyl histones), as well as the residues modified, is tightly associated with neural cell differentiation [69]. For example, histones H3 trimethyl K9 and H4 monomethyl K20 were detected in proliferating neural stem cells, whereas histone H4 trimethyl K20 was enriched in differentiating neurons (Fig. 2).
The epigenetic state can also be regulated at the DNA level by DNA methylation. The most prominent form of DNA methylation in mammals is the symmetric methylation of cytosine in the 5’ position in CpG dinucleotides. DNA methylation and its related chromatin remodeling play critical roles in regulating gene transcription in response to neuronal activity [70]. For example, DNA methylation can repress astrocytic GFAP expression by preventing the binding of a transcription factor, signal transducer and activator of transcription 3 (STAT3), to the GFAP gene promoter at early stages of brain development [71]. The gene silencing effect of DNA methylation is mediated by a family of methyl-cytosine-binding proteins, including MeCP2, which is abundantly expressed in the central nervous system [72, 73]. MeCP2 is a member of a group of methylated-CpG binding proteins and is expressed at high levels in the postnatal brain. Mutations in the Mecp2 gene have been linked to a neurodevelopmental disorder, Rett syndrome [74], suggesting that MeCP2 may play a role in regulating neuronal function. MECP2 has been shown to be critical for embryonic neurogenesis for Xenopus [75]. Recent studies demonstrated that Mecp2 is involved in the maturation and maintenance of neurons at late stages of neuronal differentiation but not critical for embryonic neurogenesis in mammalian brains [76], consistent with the postnatal onset of symptoms in Rett Syndrome patients. Methyl-CpG binding protein MBD1 is also crucial for normal neural stem cell and brain functions. MBD1-/- neural stem cells exhibited reduced neuronal differentiation and increased genomic instability [77]. DNA methyltransferases are also expressed in the central nervous system and have a role in neurogenesis [78, 79]. DNA methyltransferase 1 (Dnmt1) deficiency mice displayed decreased neurogenesis. Conditional knockout of Dnmt1 in neural progenitor cells results in DNA hypomethylation and precocious astroglial differentiation [80]. Multiple layers of epigenetic modifications, therefore, regulate key transitions in the regulation of neural stem cell self-renewal and their differentiation.
4. miRNA Regulators
Many different classes of small non-coding RNAs are present in the brain, with diverse roles including RNA modification and chromatin remodeling [81]. Small double-stranded modulatory RNAs have been proposed to regulate the generation of neurons from adult neural stem cells by binding to REST [82]. miRNA is another recently identified large family of small non-coding RNAs, which are likely key post-transcriptional players in stem cell self-renewal and differentiation (Fig. 3). miRNAs are short 20-22 nucleotide RNA molecules that are expressed in a tissue-specific and developmentally-regulated manner and function as negative regulators of gene expression in a variety of eukaryotes. miRNAs are involved in numerous cellular processes including development, proliferation, and differentiation [83, 84].
Fig. 3.
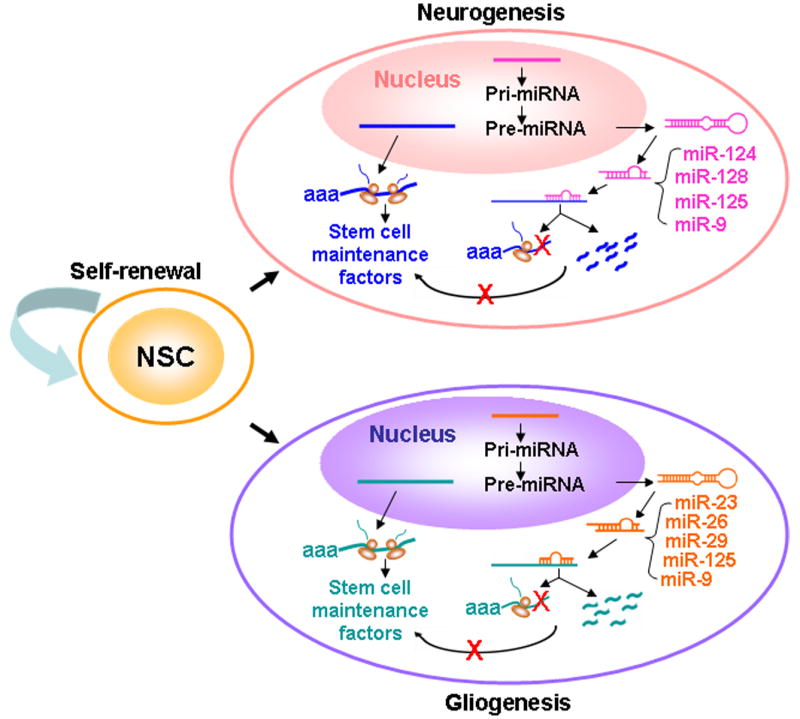
A model for miRNA action in neural stem cells. Molecules that are expressed in neural stem cells to maintain their self-renewable state are collectively referred to as stem cell maintenance factors. miRNAs are initially transcripted in the nucleus as pri-miRNAs. The pri-miRNAs are processed into pre-miRNAs (hairpin) and transferred to the cytosol, where it is processed into 22-24 nucleotide mature miRNAs. Upon differentiation, miRNAs, such as miR-124 and miR-128, are highly expressed in the neuronal lineage, whereas miR-23, miR-26, and miR-29 are upregulated in glia. MiR-9 and miR-125 are expressed in both neurons and glia. These miRNAs form incomplete base paring with their target mRNAs and direct the cleavage of these mRNAs or inhibit their translation, thus preventing the expression of the stem cell maintenance factors and enabling rapid neural differentiation.
miRNA genes belong to class II genes, which are transcribed by RNA polymerase II. A majority of miRNA loci are found in intronic regions of protein-coding or non-coding transcription units; others are found in exonic regions of non-coding transcription units [85]. The primary transcripts of miRNAs (normally > 1kb), called pri-miRNAs, are processed in the nucleus by RNase III endonuclease, Drosha, into 60 to 75 neocleotide hairpin-like precursors (pre-miRNAs). Pre-miRNAs are subsequently exported to the cytoplasm by exportin-5, a member of the Ran-dependent nuclear transport receptor family. In the cytoplasm, the hairpin precursors are cleaved into mature miRNAs by Dicer, a cytoplasmic RNase III-type protein. Mature miRNAs are then transferred to Argonaute proteins in the RNA-induced silencing complex (RISC), which direct the miRNAs to their target transcripts [86]. By base pairing with the target mRNA, miRNA functions as a guide molecule in post-transcriptional gene silencing, leading to mRNA cleavage or translational repression (Fig. 3).
miRNAs are especially attractive candidates for regulating stem cell self-renewal and cell fate decisions because of their ability to simultaneously regulate many target genes. Studies based on expression patterns, predicted targets, and overexpression analyses suggest that miRNAs are key regulators in stem cell biology. Distinct sets of miRNAs have been shown to be specifically expressed in embryonic stem cells [87, 88]. Loss of Dicer1 causes embryonic lethality and loss of stem cell populations [89, 90]. Argonaute family members, key components of RISC complexes, are required for maintaining germline stem cells in various species [91]. These observations together support a role for miRNAs in stem cell self-renewal.
Among miRNAs identified, around 70% of them are found to be expressed in mammalian brains [92, 93], suggesting possible roles of these miRNAs in neural function [94-97]. Indeed, miRNAs have been shown to regulate the development of chemosensory system of C. elegans [98] and brain morphogenesis of zebrafish [99]. Study of a set of highly expressed neural miRNA during mammalian brain development revealed significant differences in the onset and magnitude of induction for individual miRNAs and marked lineage specificity of the miRNAs [100]. During differentiation, progenitor cells express families of miRNAs sequentially, resulting in expression of lineage-specific genes. The most highly expressed miRNAs in adult brain, miR-124 and miR-128, were preferentially expressed in neurons; while miR-23 was restricted to astrocytes; miR-26 and miR-29 have stronger expression in astrocytes than neurons; whereas miR-9 and miR-125 were fairly evenly distributed [100]. Overexpression of miR-124, miR-128, and miR-9 in neural precursors decreased astrocyte differentiation. In contrast, inhibition of miR-9 expression alone or in combination with miR-124 led to reduced neurogenesis [101]. The regulation mediated by miR-9 and miR-124 is at least partly through modulating the STAT3 signaling pathway [101]. Let-7 family members are also highly represented in libraries of brain miRNAs, although not restricted to the nervous system [102]. Let-7 family members are highly expressed in nervous tissues of zebrafish [92] and in the developing mouse brains [103]. Both transcriptional activation and increased precursor processing activity led to significant induction of mature forms of let-7 family members during neural differentiation, suggesting a role of let-7 in neural cell specification [103].
Target prediction of miRNA indicates that more than one-third of animal genes may be regulated by miRNAs [104]. Each miRNA may suppress multiple targets, and one mRNA can be targeted by many miRNAs [84]. So far, some of mRNA targets have been validated and shown to control over a broad spectrum of cellular process [83]. MiR-124, the most abundant miRNA in adult mammalian brains [102], has its binding motif in more than 1100 genes as predicted computationally [104]. Introduction of miR-124 into HeLa cells down-regulated more than 100 genes [105] and promoted a neuronal-like mRNA profile [106]. Recently laminin gamma 1 and integrin beta1 were identified as targets of miR-124 in chicken neural tube, both of which are highly expressed in neural precursor cells and repressed upon neuronal differentiation [107]. In addition, miR-124 binds to 3’UTR of small C-terminal domain phosphatase 1 (SCP1), a phosphatase that plays a role in neural developement, to suppress its expression, further supporting a role of miR-124 in neurogenesis [108]. Senseless, a binary switch during sensory organ precursor selection in flies [109], has been identified as a target gene of miR-9 [110]. Loss of miR-9 in flies induced extra sense organs, whereas overexpression of miR-9 led to massive loss of sensory organ precursors [110]. Hunchback, a gene regulates the temporal identity of neuroblasts [111], is regulated by let-7 miRNA as a target gene [112, 113]. Identification of comprehensive miRNA targets in the neural system remains an important and non-trivial task that will help us to better understand the regulation of neural stem cell self-renewal and differentiation.
5. Cell-extrinsic signaling
Stem cell self-renewal and differentiation is also regulated by the specialized microenvironment, or niche, in which these cells reside [114-117]. Direct physical interactions between stem cells and their niches are critical for maintaining stem cell characters. Signaling molecules in the niche are composed of soluble factors, membrane bound molecules, and extracellular matrix, including Wnt, Notch, and Sonic hedgehog (Shh) [114]. Receptor tyrosine kinase (RTK) signaling has also been implicated in regulation of neural stem cell proliferation and self-renewal.
Genetic studies have implicated the Wnt/β-catenin pathway in neural stem cell self-renewal. In mice that express a stabilized β-catenin, the central nervous system is greatly enlarged. Progenitors exit the cell cycle less frequently and continue to proliferate in the brain [118]. In contrast, ablation of β-catenin results in a marked decrease of the overall size of the nervous system [119]. Mice with null alleles of LRP6, a required co-receptor for Wnt signaling, also showed reduced dentate granule cell production and failed expansion of a defined dentate granule precursor cell pool [120]. These studies indicate that the Wnt/β-catenin signaling plays an important role in the proliferation and self-renewal of neural precursors, presumably through its downstream target genes, such as cyclin D1 [121]. On the other hand, Wnt proteins have been shown to promote neuronal differentiation in neural stem cell culture and in adult hippocampus [122-124], suggesting that signaling by the canonical Wnt pathway has multiple functions in stem cells. Indeed, recently it has been shown that Wnt3a and Wnt5a could both increase proliferation and stimulate neuronal differentiation of neural progenitor cells isolated from postnatal mouse brains [125]. Thus, Wnt pathways are critical regulators of both neural stem cell self-renewal and neurogenesis.
Notch signaling is activated when a Notch receptor on one cell interacts with Notch ligands, such as Delta, on an adjacent cell. This interaction triggers proteolytic release of the Notch intracellular domain and its translocation to the nucleus, where it binds to transcriptional regulator CSL and induces downstream effector expression [126]. The best known Notch effectors are members of the Hes families, including Hes1 and Hes5 [47, 48, 127]. Studies using mouse models of the Notch pathway led to a prevailing view that Notch maintains the self-renewable state of neural stem cells [128]. One of the first Notch pathway genes to be knocked-out was Notch1 [129, 130]. Consistent with the view that Notch activity is needed for stem cell maintenance, increased neuronal differentiation was detected in Notch1 mutant brains. Conditional deletion of Notch1 in the neural progenitor pool was also found to result in precocious neuronal differentiation and earlier neural progenitor pool depletion [131, 132]. In addition to the receptor mutations, many Notch ligand mutations have been examined. One such study found that deleting Delta-like 1 (Dll1) led to a decrease in the radial glia progenitor marker RC2 and an increase in neuronal marker expression in embryonic brains, supporting the traditional view that Notch signaling inhibits neuronal differentiation in the developing central nervous system [128]. Notch pathway components are also expressed in postnatal and adult mouse brains, both in germinal zones and in neurons [133]. Notch signaling can be context-dependent. In addition to maintaining stem cell state in germinal regions, Notch can also promote terminal glial differentiation [134].
Shh is an important morphogen in development. In Shh null mice, the telencephalon is greatly dysmorphic and much reduced in size. Both dorsoventral patterning and general brain proliferation are significantly affected [135, 136]. Shh is also a potent mitogen for neural progenitor cells of adult brains [137]. Over-expression of Shh near the dentate gyrus increases proliferation and neurogenesis of hippocampal subgranular zone cells [137]. In vitro, Shh maintains proliferation of adult hippocampal neuronal progenitors [137] and increases subventricular zone cell proliferation [138]. The hedgehog signaling is mediated by the zinc finger-containing transcription factor Gli1, a direct transcriptional target of Shh signaling [139, 140], acting to both promote proliferation and maintain populations of neural progenitors in postnatal brains [141].
Epidermal growth factor (EGF), transforming growth factor α (TGFα), and fibroblast growth factor (FGF) are all extracellular ligands of RTKs and play critical roles in the proliferation of neural stem cells [142-145]. EGF and TGFα bind preferentially to the EGFR [146, 147], a member of the RTK family. EGFR is expressed in neurogenic regions such as the adult subventricular zone (SVZ) [148-150]. Targeted disruption of EGFR causes forebrain cortical dysgenesis at late embryonic and postnatal ages [151-153]. TGFα is considered the predominant endogenous EGFR ligand [150], the expression of which is temporally and spatially coordinated with EGFR expression. TGFα null mice exhibit decreased proliferation within the SVZ [154]. In vivo administration of EGF and TGFα via intraventricular infusion increased neural stem cell proliferation in the adult brain. In vitro experiments have also shown that SVZ-derived progenitor cells can be expanded by EGF and FGF administration [143]. FGF acts primarily through FGF receptors (FGFR), members of the RTK family also (Johnson and Williams 1993). Targeted deletion of FGFR1 causes defects in cell proliferation and embryonic lethality [155-157]. FGF2, the first member of the FGF family, is expressed in the dorsolateral cortical neuroepithelium of the forebrain and acts primarily through FGFR1. FGF2 null mice show significant reduction in cortical progenitor cell proliferation before neurogenesis begins [158]. Intraventricular delivery of FGF2 increased cell proliferation within the adult SVZ [159, 160]. Cyclin D2 has been recently suggested to be an effector of the FGF signaling [161], which promotes early G1 cell cycle progression, a function relevant to neural stem cell proliferation.
Neural stem cells in the brain are likely to be influenced by a convergence of extracellular signals, such as Wnt, Notch, and Shh, from many neighboring cell types, including astrocytes, neuroblasts, ependymal cells, and endothelial cells [117, 162, 163]. These extrinsic factors may interact with intrinsic regulators by various signaling cascades, including Wnt/β-catenin-cyclin D1 [121], Notch-Hes1/5 [47, 48, 127], and Shh-Gli1 [139, 140]. Understanding more about these signaling pathways will help reveal how the niches influence neural stem cell self-renewal and tissue development [164].
6. Conclusion
An emerging regulatory network controlling stem cell self-renewal and differentiation is defined by integration of cell-intrinsic regulators, including transcription factors, epigenetic controls, and small RNA regulators, with cell-extrinsic signals from stem cell niches. These mechanisms are coordinated to regulate the development, maintenance, self-renewal, and differentiation of stem cells. Unraveling how individual signaling cascades integrate into the global regulatory networks will be essential to better understand stem cell biology. It will also facilitate the development of new and targeted therapies using neural stem cells for a host of neurological disorders, including brain injuries, brain tumors, and neurodegenerative diseases such as Huntington’s, Alzheimer’s and Parkinson’s disease.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations and thank Rose Chavarin and Dr. Kamil Alzayady for proof-reading of the manuscript. GS is a Herbert Horvitz Fellow and RS is a NIH NIMH DPN Fellow. YS is a Kimmel Scholar. This work is supported by Whitehall Foundation, the Margaret E. Early Medical Research Trust, the Sidney Kimmel Foundation, and NIH NINDS.
Biography
Dr. Yanhong Shi is an assistant professor of Neurosciences in Beckman Research Institute of City of Hope, Duarte, CA. Dr. Shi’s research focuses on neural stem cells in the adult brain-specifically, she is interested in characterizing the molecular cascades that program these cells to be self-renewable. Dr. Shi received her PhD degree from Northwestern University, Evanston, IL, followed by a postdoctoral training in the Salk Institute, La Jolla, CA, where she gained valuable experiences in the gene expression laboratory and laboratory of Genetics under the guidance of Drs. Ronald M. Evans and Fred Gage. Dr. Shi is the recipient of a number of prestigious awards and has published her work extensively in respected scientific journals, including Nature, Genes and Development, and Proceedings of the National Academy of Sciences of the United States of America.
References
- 1.McKay R. Stem cells in the central nervous system. Science. 1997 Apr 4;276(5309):66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Buylla A, Temple S. Stem cells in the developing and adult nervous system. J Neurobiol. 1998 Aug;36(2):105–110. [PubMed] [Google Scholar]
- 3.Gage FH, Kempermann G, Palmer TD, Peterson DA, Ray J. Multipotent progenitor cells in the adult dentate gyrus. J Neurobiol. 1998 Aug;36(2):249–266. doi: 10.1002/(sici)1097-4695(199808)36:2<249::aid-neu11>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Weiss S, van der Kooy D. CNS stem cells: where’s the biology (a.k.a. beef)? J Neurobiol. 1998 Aug;36(2):307–314. doi: 10.1002/(sici)1097-4695(199808)36:2<307::aid-neu14>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Chichung Lie D, Taupin P, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004 Jan 1;427(6969):78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 6.Land PW, Monaghan AP. Expression of the transcription factor, tailless, is required for formation of superficial cortical layers. Cereb Cortex. 2003 Sep;13(9):921–931. doi: 10.1093/cercor/13.9.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Land PW, Monaghan AP. Abnormal development of zinc-containing cortical circuits in the absence of the transcription factor Tailless. Brain Res Dev Brain Res. 2005 Aug 8;158(12):97–101. doi: 10.1016/j.devbrainres.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy K, Kuznicki K, Wu Q, et al. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci. 2004 Sep 22;24(38):8333–8345. doi: 10.1523/JNEUROSCI.1148-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003 Nov 19;23(33):10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monaghan AP, Bock D, Gass P, et al. Defective limbic system in mice lacking the tailless gene. Nature. 1997 Dec 4;390(6659):515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 11.Chiang MY, Evans RM. Dissertation. Univ of California San Diego; La Jolla, CA: 1997. Reverse Genetic Analysis of Nuclear Receptors, RXRγ, RARβ, and TLX in Mice. [Google Scholar]
- 12.Yu RT, Chiang MY, Tanabe T, et al. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2621–2625. doi: 10.1073/pnas.050566897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyawaki T, Uemura A, Dezawa M, et al. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci. 2004 Sep 15;24(37):8124–8134. doi: 10.1523/JNEUROSCI.2235-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006 May 15;20(10):1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uemura A, Kusuhara S, Wiegand SJ, Yu RT, Nishikawa S. Tlx acts as a proangiogenic switch by regulating extracellular assembly of fibronectin matrices in retinal astrocytes. J Clin Invest. 2006 Feb;116(2):369–377. doi: 10.1172/JCI25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy K, Thiels E, Monaghan AP. Loss of the tailless gene affects forebrain development and emotional behavior. Physiol Behav. 2002 Dec;77(45):595–600. doi: 10.1016/s0031-9384(02)00902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young KA, Berry ML, Mahaffey CL, et al. Fierce: a new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav Brain Res. 2002 May 14;132(2):145–158. doi: 10.1016/s0166-4328(01)00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran E, Jimenez G. The tailless nuclear receptor acts as a dedicated repressor in the early Drosophila embryo. Mol Cell Biol. 2006 May;26(9):3446–3454. doi: 10.1128/MCB.26.9.3446-3454.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brannvall K, Korhonen L, Lindholm D. Estrogen-receptor-dependent regulation of neural stem cell proliferation and differentiation. Mol Cell Neurosci. 2002 Nov;21(3):512–520. doi: 10.1006/mcne.2002.1194. [DOI] [PubMed] [Google Scholar]
- 20.Ambrogini P, Cuppini R, Ferri P, et al. Thyroid hormones affect neurogenesis in the dentate gyrus of adult rat. Neuroendocrinology. 2005;81(4):244–253. doi: 10.1159/000087648. [DOI] [PubMed] [Google Scholar]
- 21.Fowler CD, Johnson F, Wang Z. Estrogen regulation of cell proliferation and distribution of estrogen receptor-alpha in the brains of adult female prairie and meadow voles. J Comp Neurol. 2005 Aug 22;489(2):166–179. doi: 10.1002/cne.20638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama K, Wada K, Nakajima A, Kamisaki Y, Mayumi T. Nuclear receptors as targets for drug development: the role of nuclear receptors during neural stem cell proliferation and differentiation. J Pharmacol Sci. 2005 Feb;97(2):171–176. doi: 10.1254/jphs.fmj04008x3. [DOI] [PubMed] [Google Scholar]
- 23.Kishi Y, Takahashi J, Koyanagi M, et al. Estrogen promotes differentiation and survival of dopaminergic neurons derived from human neural stem cells. J Neurosci Res. 2005 Feb 1;79(3):279–286. doi: 10.1002/jnr.20362. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Sun G, Stewart R. Nuclear receptors in stem cell biology. Critical Rev in Euk Gene Exp. 2006;16(2):1–11. doi: 10.1615/critreveukargeneexpr.v16.i2.50. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Andersson S, Warner M, Gustafsson JA. Morphological abnormalities in the brains of estrogen receptor beta knockout mice. Proc Natl Acad Sci U S A. 2001 Feb 27;98(5):2792–2796. doi: 10.1073/pnas.041617498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemkine GF, Raj A, Alfama G, et al. Adult neural stem cell cycling in vivo requires thyroid hormone and its alpha receptor. Faseb J. 2005 May;19(7):863–865. doi: 10.1096/fj.04-2916fje. [DOI] [PubMed] [Google Scholar]
- 27.Wada K, Nakajima A, Katayama K, et al. Peroxisome proliferator-activated receptor gamma-mediated regulation of neural stem cell proliferation and differentiation. J Biol Chem. 2006 May 5;281(18):12673–12681. doi: 10.1074/jbc.M513786200. [DOI] [PubMed] [Google Scholar]
- 28.Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002 Oct 31;419(6910):934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 29.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003 Oct 30;425(6961):962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Lugt NM, Domen J, Linders K, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994 Apr 1;8(7):757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 31.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005 Jun 15;19(12):1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005 Jun 15;19(12):1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003 Apr;4(4):509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 34.Zappone MV, Galli R, Catena R, et al. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000 Jun;127(11):2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- 35.Pevny L, Rao MS. The stem-cell menagerie. Trends Neurosci. 2003 Jul;26(7):351–359. doi: 10.1016/S0166-2236(03)00169-3. [DOI] [PubMed] [Google Scholar]
- 36.Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005 Feb;15(1):7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003 Aug 28;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 38.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003 Nov;6(11):1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 39.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003 Jan 1;17(1):126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci U S A. 2003 Sep 30;100(Suppl 1):11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferri AL, Cavallaro M, Braida D, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004 Aug;131(15):3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 42.Episkopou V. SOX2 functions in adult neural stem cells. Trends Neurosci. 2005 May;28(5):219–221. doi: 10.1016/j.tins.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003 Apr 10;38(1):17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 44.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002 Jul;3(7):517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 45.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003 Jul 3;39(1):13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 46.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005 Jun 10;306(2):343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995 Sep 28;377(6547):355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 48.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. Embo J. 1999 Apr 15;18(8):2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992 Dec;6(12B):2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 50.Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992 Oct 25;267(30):21879–21885. [PubMed] [Google Scholar]
- 51.Allen T, Lobe CG. A comparison of Notch, Hes and Grg expression during murine embryonic and post-natal development. Cell Mol Biol (Noisy-le-grand) 1999 Jul;45(5):687–708. [PubMed] [Google Scholar]
- 52.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. Embo J. 1994 Apr 15;13(8):1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirata H, Ohtsuka T, Bessho Y, Kageyama R. Generation of structurally and functionally distinct factors from the basic helix-loop-helix gene Hes3 by alternative first exons. J Biol Chem. 2000 Jun 23;275(25):19083–19089. doi: 10.1074/jbc.M001075200. [DOI] [PubMed] [Google Scholar]
- 54.Ohtsuka T, Sakamoto M, Guillemot F, Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 2001 Aug 10;276(32):30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- 55.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995 Dec 15;9(24):3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 56.Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000 Jun;127(11):2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- 57.Hatakeyama J, Bessho Y, Katoh K, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004 Nov;131(22):5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 58.Temple S. Stem cell plasticity--building the brain of our dreams. Nat Rev Neurosci. 2001 Jul;2(7):513–520. doi: 10.1038/35081577. [DOI] [PubMed] [Google Scholar]
- 59.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan 6;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 60.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004 Nov 23;101(47):16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005 May 20;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 62.Lunyak VV, Rosenfeld MG. No rest for REST: REST/NRSF regulation of neurogenesis. Cell. 2005 May 20;121(4):499–501. doi: 10.1016/j.cell.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Lunyak VV, Burgess R, Prefontaine GG, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002 Nov 29;298(5599):1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 64.Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005 Jan 28;307(5709):596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 65.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001 Jun;13(3):263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001 Sep 15;15(18):2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 67.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004 May 27;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 68.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004 Dec 29;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 69.Biron VL, McManus KJ, Hu N, Hendzel MJ, Underhill DA. Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev Biol. 2004 Dec 15;276(2):337–351. doi: 10.1016/j.ydbio.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 70.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003 Oct 31;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 71.Takizawa T, Nakashima K, Namihira M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001 Dec;1(6):749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 72.Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 73.Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999 Apr;9(2):158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 74.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999 Oct;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 75.Stancheva I, Collins AL, Van den Veyver IB, Zoghbi H, Meehan RR. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol Cell. 2003 Aug;12(2):425–435. doi: 10.1016/s1097-2765(03)00276-4. [DOI] [PubMed] [Google Scholar]
- 76.Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004 Nov;27(3):306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 77.Zhao X, Ueba T, Christie BR, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994 Apr;56(12):39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 79.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999 Oct 29;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 80.Fan Y, Nikitina T, Zhao J, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005 Dec 29;123(7):1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 81.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005 Apr 15;14(Spec No 1):R121–132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 82.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004 Mar 19;116(6):779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 83.Ambros V. The functions of animal microRNAs. Nature. 2004 Sep 16;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 84.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 85.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005 May;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 86.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005 Apr;15(2):200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 87.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003 Aug;5(2):351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 88.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004 Jun 15;270(2):488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 89.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003 Nov;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 90.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003 Nov;35(3):217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 91.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002 Nov 1;16(21):2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 92.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005 Jul 8;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 93.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005 Nov;132(21):4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 94.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003 Oct;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim J, Krichevsky A, Grad Y, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004 Jan 6;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miska EA, Alvarez-Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5(9):R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004 Aug 12;430(7001):785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 99.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005 May 6;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 100.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005 Mar;21(6):1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 101.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006 Apr;24(4):857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002 Apr 30;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 103.Wulczyn FG, Smirnova L, Rybak A, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. Faseb J. 2006 Dec 13; doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 104.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 105.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005 Feb 17;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 106.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007 Mar 1;21(5):531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007 Apr 1;21(7):744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jafar-Nejad H, Tien AC, Acar M, Bellen HJ. Senseless and Daughterless confer neuronal identity to epithelial cells in the Drosophila wing margin. Development. 2006 May;133(9):1683–1692. doi: 10.1242/dev.02338. [DOI] [PubMed] [Google Scholar]
- 110.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006 Oct 15;20(20):2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001 Aug 24;106(4):511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 112.Lin SY, Johnson SM, Abraham M, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003 May;4(5):639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 113.Abrahante JE, Daul AL, Li M, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003 May;4(5):625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 114.Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004 Mar 4;41(5):683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 115.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004 Mar 19;116(6):769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 116.Ohlstein B, Kai T, Decotto E, Spradling A. The stem cell niche: theme and variations. Curr Opin Cell Biol. 2004 Dec;16(6):693–699. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 117.Wurmser AE, Palmer TD, Gage FH. Neuroscience. Cellular interactions in the stem cell niche. Science. 2004 May 28;304(5675):1253–1255. doi: 10.1126/science.1099344. [DOI] [PubMed] [Google Scholar]
- 118.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002 Jul 19;297(5580):365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 119.Zechner D, Fujita Y, Hulsken J, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003 Jun 15;258(2):406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 120.Zhou CJ, Zhao C, Pleasure SJ. Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. J Neurosci. 2004 Jan 7;24(1):121–126. doi: 10.1523/JNEUROSCI.4071-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Willert K, Nusse R. Beta-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev. 1998 Feb;8(1):95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 122.Hirabayashi Y, Itoh Y, Tabata H, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004 Jun;131(12):2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 123.Muroyama Y, Kondoh H, Takada S. Wnt proteins promote neuronal differentiation in neural stem cell culture. Biochem Biophys Res Commun. 2004 Jan 23;313(4):915–921. doi: 10.1016/j.bbrc.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 124.Lie DC, Colamarino SA, Song HJ, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005 Oct 27;437(7063):1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 125.Yu JM, Kim JH, Song GS, Jung JS. Increase in proliferation and differentiation of neural progenitor cells isolated from postnatal and adult mice brain by Wnt-3a and Wnt-5a. Mol Cell Biochem. 2006 Aug;288(12):17–28. doi: 10.1007/s11010-005-9113-3. [DOI] [PubMed] [Google Scholar]
- 126.Lasky JL, Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005 May;57(5 Pt 2):104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- 127.Nyfeler Y, Kirch RD, Mantei N, et al. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. Embo J. 2005 Oct 5;24(19):3504–3515. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005 Jun;8(6):709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 129.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994 Mar 15;8(6):707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 130.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995 May;121(5):1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 131.Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004 Oct 27;24(43):9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004 May 1;269(1):81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 133.Stump G, Durrer A, Klein AL, Lutolf S, Suter U, Taylor V. Notch1 and its ligands Delta-like and Jagged are expressed and active in distinct cell populations in the postnatal mouse brain. Mech Dev. 2002 Jun;114(12):153–159. doi: 10.1016/s0925-4773(02)00043-6. [DOI] [PubMed] [Google Scholar]
- 134.Tanigaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001 Jan;29(1):45–55. doi: 10.1016/s0896-6273(01)00179-9. [DOI] [PubMed] [Google Scholar]
- 135.Chiang C, Litingtung Y, Lee E, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996 Oct 3;383(6599):407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 136.Rallu M, Machold R, Gaiano N. Corbin JG, McMahon AP, Fishell G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development. 2002 Nov;129(21):4963–4974. doi: 10.1242/dev.129.21.4963. [DOI] [PubMed] [Google Scholar]
- 137.Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003 Jan;6(1):21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 138.Palma V, Lim DA, Dahmane N, et al. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005 Jan;132(2):335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004 Aug 20;118(4):505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 140.Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005 Oct 6;437(7060):894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 141.Machold R, Hayashi S, Rutlin M, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003 Sep 11;39(6):937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- 142.Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992 Nov;12(11):4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992 Mar 27;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 144.Vescovi AL, Reynolds BA, Fraser DD, Weiss S. bFGF regulates the proliferative fate of unipotent (neuronal) and bipotent (neuronal/astroglial) EGF-generated CNS progenitor cells. Neuron. 1993 Nov;11(5):951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- 145.Gritti A, Parati EA, Cova L, et al. Multipotential stem cells from the adult mouse brain proliferate and self-renew in response to basic fibroblast growth factor. J Neurosci. 1996 Feb 1;16(3):1091–1100. doi: 10.1523/JNEUROSCI.16-03-01091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Massague J. Epidermal growth factor-like transforming growth factor. I. Isolation, chemical characterization, and potentiation by other transforming factors from feline sarcoma virus-transformed rat cells. J Biol Chem. 1983 Nov 25;258(22):13606–13613. [PubMed] [Google Scholar]
- 147.Marquardt H, Hunkapiller MW, Hood LE, Todaro GJ. Rat transforming growth factor type 1: structure and relation to epidermal growth factor. Science. 1984 Mar 9;223(4640):1079–1082. doi: 10.1126/science.6320373. [DOI] [PubMed] [Google Scholar]
- 148.Seroogy KB, Gall CM, Lee DC, Kornblum HI. Proliferative zones of postnatal rat brain express epidermal growth factor receptor mRNA. Brain Res. 1995 Jan 23;670(1):157–164. doi: 10.1016/0006-8993(94)01300-7. [DOI] [PubMed] [Google Scholar]
- 149.Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002 Dec 19;36(6):1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 150.Kornblum HI, Hussain RJ, Bronstein JM, Gall CM, Lee DC, Seroogy KB. Prenatal ontogeny of the epidermal growth factor receptor and its ligand, transforming growth factor alpha, in the rat brain. J Comp Neurol. 1997 Apr 7;380(2):243–261. doi: 10.1002/(sici)1096-9861(19970407)380:2<243::aid-cne7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 151.Threadgill DW, Dlugosz AA, Hansen LA, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995 Jul 14;269(5221):230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 152.Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995 Jul 14;269(5221):234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 153.Sibilia M, Steinbach JP, Stingl L, Aguzzi A, Wagner EF. A strain-independent postnatal neurodegeneration in mice lacking the EGF receptor. Embo J. 1998 Feb 2;17(3):719–731. doi: 10.1093/emboj/17.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tropepe V, Craig CG, Morshead CM, van der Kooy D. Transforming growth factor-alpha null and senescent mice show decreased neural progenitor cell proliferation in the forebrain subependyma. J Neurosci. 1997 Oct 15;17(20):7850–7859. doi: 10.1523/JNEUROSCI.17-20-07850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994 Dec 15;8(24):3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 156.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994 Dec 15;8(24):3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 157.Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997 Jul;124(14):2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- 158.Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000 Jul 1;20(13):5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Craig CG, Tropepe V, Morshead CM, Reynolds BA, Weiss S, van der Kooy D. In vivo growth factor expansion of endogenous subependymal neural precursor cell populations in the adult mouse brain. J Neurosci. 1996 Apr 15;16(8):2649–2658. doi: 10.1523/JNEUROSCI.16-08-02649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH. Epidermal growth factor and fibroblast growth factor-2 have different effects on neural progenitors in the adult rat brain. J Neurosci. 1997 Aug 1;17(15):5820–5829. doi: 10.1523/JNEUROSCI.17-15-05820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lobjois V, Benazeraf B, Bertrand N, Medevielle F, Pituello F. Specific regulation of cyclins D1 and D2 by FGF and Shh signaling coordinates cell cycle progression, patterning, and differentiation during early steps of spinal cord development. Dev Biol. 2004 Sep 15;273(2):195–209. doi: 10.1016/j.ydbio.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 162.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002 May 2;417(6884):39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- 163.Shen Q, Goderie SK, Jin L, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004 May 28;304(5675):1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 164.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001 Nov 1;414(6859):98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]