Summary of recent advances
Since their first description more than twenty years ago, Escherichia coli and Klebsiella pneumoniae possessing extended-spectrum class A beta-lactamases (ESBLs) continue to thwart our best clinical efforts. In the “early years” the most common beta-lactamases were of the TEM and SHV varieties. Now, CTX-M enzymes are being discovered though out the world and are becoming the most prevalent beta-lactamases found in clinical isolates. The Klebsiella pneumoniae carbapenemases (KPC) (ESBL type enzymes that confer resistance to extended spectrum cephalosporins and carbapenems) present the most significant challenge to date. Structural studies of ESBLs indicate that active site expansion and remodeling are responsible for this extended hydrolytic activity. Continuing questions still exist regarding the optimal detection method for ESBLs. Most relevant are the increasing concerns regarding the status of carbapenems as “best therapy” for ESBL producing bacteria in light of the emergence of carbapenemases.
Introduction
The development of extended-spectrum cephalosporins in the early 1980s was regarded as a major addition to our therapeutic armamentarium in the fight against beta-lactamase-mediated bacterial resistance [1–3]. Regrettably, the emergence of Escherichia coli and Klebsiella pneumoniae resistant to ceftazidime and other cephalosporins seriously compromised the efficacy of these life-saving antibiotics. The new bacterial beta-lactamases present in these common enteric bacilli (the parent TEM-1 and SHV-1 enzymes) demonstrated unique hydrolytic properties. Point mutations in the blaSHV and blaTEM genes that resulted in single amino acid changes (Gly238→Ser, Glu240→Lys, Arg164→Ser, Arg164→His, Asp179→Asn, and Glu(Asp)104→Lys) formed the basis of this remarkable resistance phenotype [4,5]. Currently, ESBLs are becoming a major threat for patients in the hospital, long-term care facilities, and community. It is our goal in this analysis to:
Review the classification and global epidemiology of ESBL-producing enteric bacteria; highlighting the emergence of community-acquired ESBLs and KPC enzymes as the next major public health dangers.
Summarize the atomic features of ESBLs so the reader may appreciate “structure-function relationships” in these enzymes;
Present the current “state of the art” regarding the laboratory detection of ESBLs.
Discuss the clinical impact and current therapy of infections caused by ESBL-producing E. coli, Proteus spp. and Klebsiella spp. Although Enterobacter spp. are not routinely tested for ESBL production, our discussion will include mention of this genus when appropriate.
What is an ESBL and why are they important?
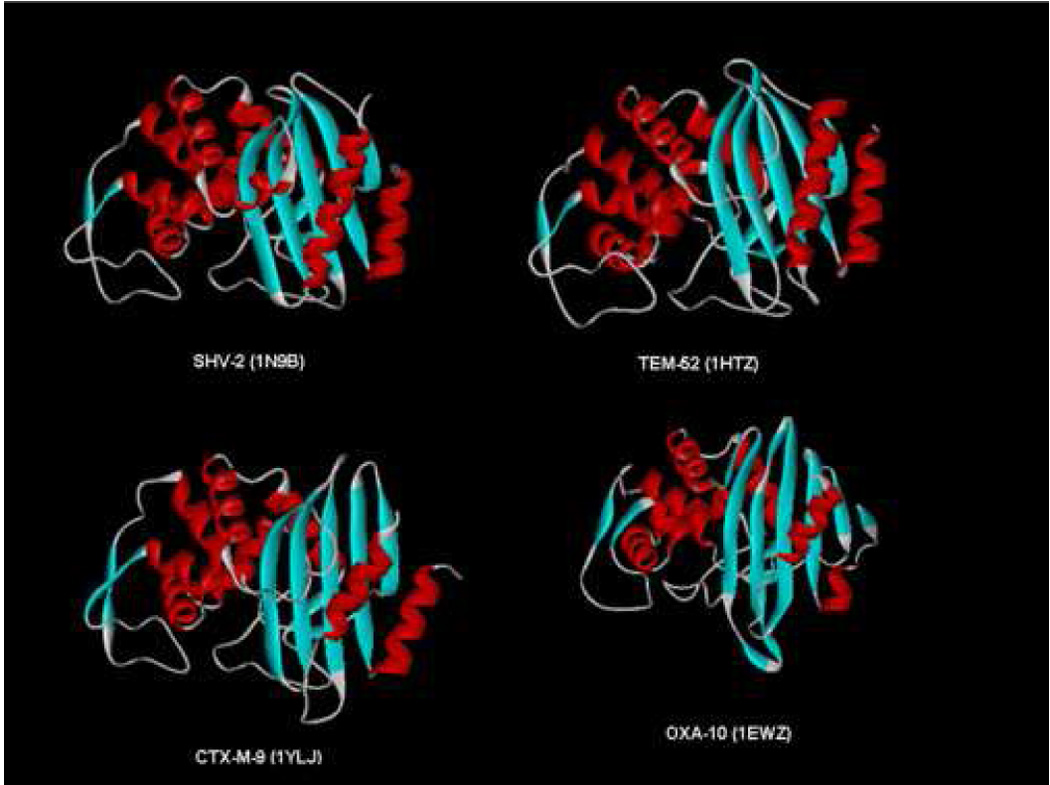
Beta-lactamases are among the most heterogeneous group of resistance enzymes. More than 700 distinct beta-lactamases are described (personal communication, Dr. Karen Bush). These globular proteins are composed of alpha–helices and beta–pleated sheets [6]. Despite a significant amount of amino acid sequence variability, beta-lactamases share a common overall topology (Figure 1).
Figure 1.
In general, ESBL are capable of hydrolyzing penicillins (e.g, ampicillin and piperacillin), cephalosporins of the first-, second-, third- and fourth-generations, and the monobactam aztreonam (but not the cephamycins or carbapenems) [7,8]. In contrast, ESBLs (particularly TEM and SHV family derivatives) are readily inhibited by the commercially available beta-lactamase inhibitors (i.e., clavulanic acid, tazobactam, or sulbactam). This unique property serves as an important phenotypic test that is conveniently exploited to identify ESBLs in bacteria [vide infra].
Two systems are commonly used to classify beta-lactamases: the Ambler scheme and the Bush-Medeiros-Jacoby system [7,9]. They are summarized in Table 1. Both systems are used interchangeably in the literature: ESBLs belong to group 2be in the Bush-Medeiros-Jacoby system and to class A in the Ambler system.
Table 1.
Classification of beta-lactamases.
| Bush-Jacoby-Medeiros system | Major subgroups | Ambler System | Main attributes |
|---|---|---|---|
| Group 1 cephalosporinases | – | C (cephalosporinases) | Usually chromosomal; Resistance to all β-lactams except carbapenems; Not inhibited by clavulanate |
| Group 2 penicillinases | 2a | A (serine β-lactamases) | Staphylococcal penicillinases |
| (clavulanic acid susceptible) | 2b | A | Broad-spectrum – TEM-1, TEM-2, SHV-1 |
| 2be | A | Extended-spectrum – TEM-3–160, SHV-2–101 | |
| 2br | A | Inhibitor resistant TEM (IRT) | |
| 2c | A | Carbenicillin-hydrolyzing | |
| 2e | A | Cephalosporinases inhibited by clavulanate | |
| 2f | A | Carbapenemases inhibited by clavulanate | |
| 2d | D(oxacillin-hydrolyzing) | Cloxacillin-hydrolyzing (OXA) | |
| Group 3 metallo-b- | 3a | B (metalloenzymes) | Zinc-dependent |
| lactamase | carbapenemases | ||
| 3b | B | ||
| 3c | B | ||
| Group 4 | Not classified | Miscellaneous enzymes, most not yet sequenced |
Rapid sequencing methods are enabling investigators the opportunity to identify many TEM and SHV family variants (see: www.lahey.org) as well as “non-TEM, non-SHV” type ESBLs that are relatively resistant to beta-lactamase inhibitors.
Global epidemiology of Enterobacteriaceae-producing ESBLs
First described in Germany (1983) and France (1985) among Klebsiella spp, ESBLs exist in every region of the world and in most genera of enterobacteria [8,10]. A contemporary perspective on the epidemiology of ESBLs reveals the widespread emergence of the CTX-M-type. These are now considered the most prevalent ESBLs worldwide [11–13]. PER- and OXA-type enzymes are more common in P. aeruginosa and Acinetobacter spp., but there have been sporadic reports of PER type ESBLs in Enterobacteriacea originating from France, Argentina and Italy [14–16]. Of tremendous import because of their ability to inactivate carbapenems, plasmid-borne KPC enzymes are emerging among K. pneumoniae and other enterobacteria in the Eastern seabord and the heartland of the U.S. and internationally [17]. Table 2 summarizes current global trends in beta-lactam resistance among enterobacteria.
Table 2.
Resistance rates of selected enterobacteria to third generation cephalosporins and carbapenems. Data from MYSTIC (www.mystic-data.org).
| Country | Year | Resistance rate in percentage (Total number of isolates tested) | |||
|---|---|---|---|---|---|
| E. coli | Klebsiella spp. | ||||
| †3GC | ††Carbapenems | 3GC | Carbapenems | ||
| United States | 2000 | 2 | 0 | 6.6 | 2.1 |
| (312) | (312) | (233) | (233) | ||
| 2006 | 7.2 | 0 | 16.5 | 9 | |
| (640) | (312) | (619) | (619) | ||
| Canada | 2000 | 15 | 0 | 0 | 0 |
| (33) | (33) | (17) | (35) | ||
| 2006 | 31.5 | 0 | 20.8 | 1 | |
| (73) | (73) | (120) | (120) | ||
| Mexico | 2000 | 31 | 0 | 15.4 | 0 |
| (39) | (39) | (52) | (52) | ||
| 2006 | 72 | 1 | 42.1 | 0 | |
| (108) | (108) | (76) | (76) | ||
| Brazil | 2000 | 8 | 0 | 54 | 0 |
| (24) | (24) | (24) | (24) | ||
| 2006 | 16.5 | 0 | 54 | 0.6 | |
| (139) | (139) | (170) | (170) | ||
| Australia | 2000 | 8 | 0 | 0 | 0 |
| (13) | (13) | (19) | (19) | ||
| 2006 | 0 | 0 | 4 | 0 | |
| (45) | (45) | (55) | (55) | ||
| Israel | 2000 | 15.8 | 0 | 43.5 | 0 |
| (19) | (19) | (23) | (23) | ||
| 2006 | 21 | 5 | 56.5 | 13 | |
| (19) | (19) | (23) | (23) | ||
| Turkey | 2000 | 27.6 | 1.5 | 61 | 2 |
| (203) | (203) | (194) | (194) | ||
| 2006 | 45.2 | 0.4 | 54.7 | 4 | |
| (272) | (272) | (245) | (246) | ||
| Russia | 2000 | 43 | 0 | 84 | 0 |
| (16) | (16) | (19) | (19) | ||
| 2006 | 20 | 0 | 85 | 0 | |
| (20) | (20) | (20) | (20) | ||
| Poland | 2000 | 47.4 | 0 | 95 | 0 |
| (19) | (19) | (22) | (22) | ||
| 2006 | 30 | 0 | 35 | 0 | |
| (20) | (20) | (40) | (40) | ||
| Czech Republic | 2000 | 0 | 0 | 39 | 0 |
| (19) | (19) | (18) | (18) | ||
| 2006 | 5 | 0 | 40 | 0 | |
| (20) | (20) | (20) | (20) | ||
| Belgium | 2000 | 5 | 0 | 19.3 | 0 |
| (166) | (166) | (140) | (140) | ||
| 2006 | 10.4 | 0 | 21 | 0 | |
| (182) | (182) | (196) | (196) | ||
| Germany | 2000 | 3 | 0 | 8.2 | 0 |
| (134) | (134) | (102) | (102) | ||
| 2006 | 13.2 | 1 | 16.2 | 1.3 | |
| (91) | (91) | (74) | (74) | ||
| Sweden | 2000 | 0 | 0 | 0 | 0 |
| (20) | (20) | (10) | (10) | ||
| 2006 | 2.7 | 0 | 1.2 | 1.2 | |
| (148) | (148) | (82) | (82) | ||
| United Kingdom | 2000 | 6.5 | 0 | 23.6 | 0 |
| (92) | (92) | (72) | (72) | ||
| 2006 | 15 | 1.5 | 17 | 0 | |
| (67) | (67) | (47) | (47) | ||
| Spain | 2000 | 3.7 | 0 | 1.8 | 0 |
| (63) | (63) | (56) | (56) | ||
| 2006 | 16 | 0 | 12.4 | 0 | |
| (113) | (111) | (137) | (137) | ||
3rd Generation Cephalosporins (e.g., cefotaxime, ceftazidime, ceftriaxone and aztreonam). Resistant phenotype defined as MIC ≥2 µg/ml.
Carbapenems (i.e., ertapenem, meropenem and ertapenem). Resistant phenotype defined as MIC ≥8 µg/ml.
The emerging threat of CTX-M beta-lactamases in the community
A recent development after the year 2000 is the identification of infections caused by bacteria harboring ESBLs in community dwellers. These are typically urinary tract infections (UTIs) caused by E. coli expressing CTX-M. These E. coli are also resistant to quinolones, aminoglycosides, and sulfonamides. In the Calgary Health Region of Canada, Pitout et al described the clonal spread of two closely related strains harboring CTX-M-14, isolated most often from urine samples [18,19]. Nationwide surveillance carried out in the United Kingdom saw the rise of several types of ESBLs in the community, among which clonally-spread E. coli carrying CTX-M-15 predominated, although CTX-M 9 was also represented [20].
Similarly, CTX-M-15 producing isolates appear with increased in frequency among clonally related E. coli strains across Italy, Portugal and France [21–24]. In Spain, the clonal spread of E. coli harboring CTX-M-15 has occurred as well, but the situation appears more complex [25]. Rodriguez-Bano et al, [26], described 49 patients in the region of Sevilla with community-acquired ESBL producing E. coli, 64% of which carried CTX-M-9. All the strains were clonally unrelated. A larger national study confirmed the high prevalence of CTX-M-9 and CTX-M-14, without evidence of clonal dissemination [27]. Further investigation suggested the dissemination of closely related plasmids harboring CTX-M-14 among clonally unrelated E. coli isolates from the community [28].
Outside of Europe, the occurrence of CTX-M ESBLs in the community has been reported as well. In Hong-Kong, 42/600 (7%) of community isolates of urinary E. coli were ESBL producers [29]. A low prevalence (1.8%) and great diversity of enzymes and bacterial species were found among community isolates in Brazil, in contrast with Bolivian and Peruvian isolates among which E. coli harboring CTX-M-15 and CTX-M-9 are predominant [30,31].
The initial observation of infections caused by bacteria harboring ESBLs in hospitals would suggest that CTX-Ms arose in the nosocomial setting and spread to the community. In fact, hospitals are the arena where the selective pressure of broad-spectrum antimicrobials and suboptimal infection control practices best conspire to foster the emergence and transmission of multidrug-resistant organisms. Nursing homes, in turn, may serve as reservoirs from which colonized and infected patients transfer to the community or back to the hospitals [8,32]. Prior to the rise of CTX-M, such a model seemed to have corresponded with the available data [33]. Even after the noted spread of bacteria producing CTX-M into the community, recent hospitalization, along with age and exposure to cephalosporins and/or quinolones, have consistently been identified as risk factors for infection with these organisms [8,34,35]. Furthermore, the above mentioned reports from Spain, Portugal and the United Kingdom demonstrate the clonal dissemination of E. coli producing CTX-M-15 in the community, with the concurrent spread of these clones into hospitals and long term care facilities [23,25,36].
Is it possible that there may be dissemination of ESBL harboring organisms from the community into hospitals? A recent report from Israel described high rates of patients with bacteremia and colonization with ESBL-producing Enterobacteriaceae on admission to the hospital [37]. Clinicians are already facing the tremendous challenges posed by this situation, in terms of the detection and isolation of patients to prevent further nosocomial expansion, and of the choice of empiric antibiotic therapy [38–40]. An additional potential reservoir of resistant bacteria and genetic determinants of resistance which intersects with the community is the food supply, as illustrated by the finding of diverse ESBL-producing bacteria, including CTX-M-15, in poultry and other farm animals [41–43]. Finally, the origin of CTX-M enzymes probably lies in beta-lactamases found in environmental species, like Kluyvera spp. [12], further supporting the notion of a community reservoir for these enzymes. The existence of resistant microorganisms predating the clinical use of antibiotics has given rise to the concept of the “antibiotic resistome”[44,45].
Structural properties of ESBLs
Important insights have emerged from the study of the atomic structures of class A ESBL enzymes. Overall, one appreciates that the first common theme to emerge is that the active site is selectively “remodeled and expanded” to accommodate the bulky R1 side chain of extended-spectrum cephalosporins [6]. This remodeling is observed in the atomic structures of Toho-1, TEM-52, TEM-64, the Gly238Ala ESBL in TEM, SHV-2, PER-1 and OXA-10 beta-lactamases [46–52]. Recently, the crystal structure of KPC-2 was determined, demonstrating modifications in the active site that allow access to carbapenems [53]. Understanding the three dimensional structure of these enzymes is essential for the future development of beta-lactams as pharmacological agents [54]. Our attention here will focus on the analysis of CTX-M ESBLs.
CTX-M ESBLs
Among the non-TEM, non-SHV ESBLs, the CTX-M beta-lactamases are the most prevalent. They can be divided into distinct clusters (see www.lahey.org). Unlike most (but not all) TEM-and SHV-derived ESBLs, CTX-M betalactamases hydrolyze cefotaxime and ceftriaxone better than they do ceftazidime. It also appears that CTX-M enzymes are more readily inhibited by tazobactam than they are by clavulanic acid.
CTX-M beta-lactamases are commonly found in K. pneumoniae, E. coli, thyphoidal and nonthyphoidal Salmonella, Shigella, Citrobacter freundii, Enterobacter, spp., and Serratia marcescens [12]. Of note, different genetic elements may be involved in the mobilization of blaCTX-M genes. Plasmids and Insertion Sequences (e.g., ISEcp1 or ISEcp1-like insertion sequences) have repeatedly been observed upstream of ORFs encoding the CTX-M-1, CTX-M-2, CTX-M-3, CTX-M-9, CTX-M-13, CTX-M-14, CTX-M-15, CTX-M-17, CTX-M-19, CTX-M-20, and CTX-M-21 enzymes.
From comparative sequence analyses, modeling, and mutagenesis studies, key amino acids involved in the substrate specificity for hydrolysis in the CTX-M beta-lactamases are idenitifed: Asn104, Asn132, Ser237, and Asp240 [12]. Unlike what is seen with TEM and SHV (enlargement of the active site), the crystal structures of CTX-M-14, CTX-M-27, CTX-M-9 and CTX-M-16 indicate that point substitutions leading to specific interactions may be responsible for the improved activity against ceftazidime and cefotaxime, rather than active site expansion.
In general, the remodeling of a “broad spectrum” beta-lactamase to an extended spectrum enzyme comes at a “price”. Many of these TEM and SHV family ESBLs are not as catalytically efficient as wild-type progenitors against natural substrates (especially against penicillins). One uniformly observes a decrease in kcat/Km ratio and decrease in penicillin MICs. In addition, these enzymes are less stable [55–58]. It is sobering to ponder all the amino acid changes that are permitted in a beta-lactamase that permit the evolution of the ESBL phenotype [59–61]. The implications for the novel design of future generations of cephalosporins are far reaching.
Detection of ESBLs
As ESBLs are found though out the world, numerous detection strategies have been developed. According to the Clinical Laboratory Standard Institute (CLSI) criteria, enterobacterial resistance to ceftriaxone, cefotaxime, ceftazidime, cefepime, and aztreonam is defined by MICs ≥ 16 µg/ml [62]. However, since several ESBL producers have MIC values for extended-spectrum cephalosporins and aztreonam below the standard breakpoints for resistance (e.g., between 2 and 8 µg/ml), the real prevalence of these organisms may be unappreciated.
Highly revealing studies performed in the United States and Europe by Tenover et al and Livermore et al, respectively, reported that errors in the detection of ESBL mediated resistance are frequently encountered with both automated and disk diffusion methods [63,64]. A contemporary analysis from Italy involving 60 independent clinical microbiology laboratories showed that only 25/60 (41.7%) correctly recognized and reported all the 5 ESBL-producing enterobacteria presented for analysis. Nearly 56% of the isolates were incorrectly characterized when testing cephalosporins and aztreonam; this was most worrisome in the case of CTX-M-1-producing E. coli and TEM-52-producing P. mirabilis [65].
Since the inaccurate identification of ESBL producers bears important clinical implications for antibiotic therapy and infection control measures, specific reporting guidelines are issued [[62] Health Protection Agency 2005, SFM 2007]. For all confirmed ESBL producers, the general consensus states that ESBL producers should be reported as resistant to all penicillins, cephalosporins (except for the cephamycins cefoxitin and cefotetan), and aztreonam irrespective of routine antimicrobial susceptibility results [62]. Notably, beta-lactam/beta-lactamase inhibitor combinations (e.g., piperacillin-tazobactam, amoxicillin-clavulanate, and ampicillin-sulbactam) are not affected by this rule and should be reported as obtained during routine susceptibility tests.
Screening for ESBL production
Both broth dilution and disk diffusion methods for screening for ESBL producers are advised by CLSI. It is recommended that E. coli, K. pneumoniae and K. oxytoca strains with MIC ≥ 8 µg/ml for cefpodoxime or MICs ≥ 2 µg/ml against ceftazidime, cefotaxime, ceftriaxone, or aztreonam should be investigated using specific phenotypic confirmatory tests for ESBL production. For P. mirabilis isolates, confirmatory tests should be performed if strains demonstrate MICs ≥ 2 µg/ml for ceftazidime, cefotaxime or cefpodoxime. The use of more than one of the above agents for screening improves the sensitivity of ESBL detection.
We recommend that if laboratories are using the disk diffusion method, E. coli, K. pneumoniae, K. oxytoca and P. mirabilis with a zone inhibition diameter lower than the following values should be investigated with confirmatory tests: ceftazidime (≤ 22 mm), cefotaxime and aztreonam (≤ 27 mm), ceftriaxone (≤ 25 mm). In the case of cefpodoxime, the “cutoff” for P. mirabilis is ≤ 22 mm, whereas in the remaining three species cited above it is ≤ 17 mm [62].
Criteria for screening for ESBL production in other Enterobacteriaceae have not been established by the CLSI [62]. In contrast, the British Society for Antimicrobial Chemotherapy (BSAC) maintains that all Enterobacteriaceae resistant to ceftazidime (MIC ≥ 4 µg/ml or zone inhibition ≤ 21 mm for E. coli and Klebiella spp. and ≤ 27 mm for the remaining species), cefotaxime (MIC ≥ 2 µg/ml or zone inhibition ≤ 29 mm), or cefpodoxime (MIC ≥ 2 µg/ml or zone inhibition ≤ 19 mm) should be evaluated by the ESBL confirmatory tests (Health Protection Agency, 2005). Similarly, the Société Française de Microbiologie (SFM) suggests the evaluation of all enterobacteria with the confirmatory test. However, criteria used to detect an ESBL producer are not closely related with MICs and zone diameter inhibition of cephalosporins and aztreonam. In addition, the ESBL confirmatory test should be performed when an isolate shows resistance to aminoglycosides (Comite de L’Antibiogramme de la Société Française de Microbiologie, SFM, 2007), since bla genes encoding ESBLs are frequently found in the same plasmid that encode resistance determinants to other classes of antibiotics such as aminoglycosides, tetracycline and sulfonamides [10].
Phenotypic confirmatory tests for ESBL production
According to the CLSI, ESBL confirmatory testing based upon phenotype requires the use of both ceftazidime and cefotaxime alone and in combination with clavulanate [62]. Several commercial vendors offer disks for use in this application (Oxoid, Becton-Dickinson and Mast). Typically, these discs contain 30 µg /disk of ceftazidime, cefotaxime with or without clavulanate (10 µg/disk). These discs have been rigorously tested and developed and are reported to have sensitivity and specificity of greater than 95% [66,67].
As mentioned above, screening and confirmatory tests for other Enterobacteriaceae, including those producing inducible AmpC chromosomal enzymes (e.g., Enterobacter spp., M. morganii, Providencia spp., Citrobacter freundii, and Serratia marcescens), have not yet been established by the CLSI [62]. Not detecting ESBLs in these pathogens may have a significant adverse impact on patients, especially those treated with extended-spectrum cephalosporins [68,69]. ESBLs are more difficult to detect in these organisms because AmpC enzymes may be induced by clavulanate (which inhibits them poorly) and may increase resistance to cephalosporins, overcoming the synergy arising from inhibition of the ESBL [70]. Therefore, the application of CLSI guidelines for ESBL detection in this group of enterobacteria is discouraged [71].
Paradoxically, the use of cefepime and cefpirome with clavulanate might improve the ability to detect ESBLs [72,73]. For this reason, the BSAC advises the use of cefpirome/clavulanate combination disks (in addition to cefpodoxime/clavulanate) for Enterobacter spp. and C. freundii as a confirmatory test (Health Protection Agency, 2005), whereas the SFM recommends to use cefepime or cefpirome with the double-disk diffusion test for all enterobacteria producing AmpC enzymes (CASFM, 2007).
Commercial methods for ESBL detection
The Etest method produced by AB Biodisk (Solna, Sweden) and Bio-Stat (Stockport, UK) is a plastic drug-impregnated strip, one end of which generates a stable concentration gradient of cephalosporin (i.e., ceftazidime 0.5–32 µg/ml, cefotaxime and cefepime 0.25–16 µg/ml) and the remaining end of which generates a gradient of cephalosporin (i.e., ceftazidime and cefepime 0.064–4 µg/ml, cefotaxime 0.016–1 µg/ml) plus a constant concentration of clavulanate (4 µg/ml). ESBL production is inferred if the MIC ratio for cephalosporin alone/cephalosporin plus clavulanate MIC is ≥ 8 (Health Protection Agency, 2005). Accurate and precise (but more expensive than combination disks) these tests are suggested by the BSAC as a confirmatory test for ESBL production. Significantly, BASC recommends the cefepime/clavulanate Etest for Enterobacter spp. (Health Protection Agency, 2005). In fact, neither ceftazidime/clavulanate nor cefotaxime/clavulanate Etest strips are able to detect the ESBLs in Enterobacter spp. In contrast, the cefepime/clavulanate strip is the only commercially available and highly reliable test that permits accurate detection of ESBLs within this group of organisms [73].
Automated method for bacterial identification and susceptibility testing are used in the detection of ESBL producing organisms. The BD Phoenix System (Becton-Dickinson Biosciences, Sparks, MD) uses its "expert software" to interpret the growth response to ceftazidime, cefotaxime, ceftriaxone and cefpodoxime, with or without clavulanate. Similarly, the Vitek 2 System (bioMérieux, Marcy L’Etoile, France) uses a "card" containing ceftazidime and cefotaxime alone and in combination with clavulanate. Ceftazidime or cefotaxime plus betalactamase inhibitors are also used in the MicroScan Walkaway-96 System (Dade Behring, Inc., West Sacramento, CA). The above three semi-automated systems were recently compared to the conventional phenotypic confirmatory tests with regard to their ability to detect ESBL production in well characterized Enterobacteriaceae including Enterobacter spp., C. freundii and S. marcescens. The system with the highest sensitivity was Phoenix (99%), followed by Vitek 2 (86%) and MicroScan (84%); however, specificity was more variable, ranging from 52% (Phoenix) to 78% (Vitek 2). In addition, the performance differed widely with the species investigated. In contrast, the three available Etest strips and four disks combination (including ceftazidime, cefotaxime, cefpodoxime and cefpirome) showed sensitivity of 94% and 93%, and specificity of 85% and 81%, respectively. The double-disk test with ceftazidime, cefotaxime, cefpodoxime and cefpirome showed the highest specificity and positive predictive value among all test methods (i.e., 97% and 98%, respectively) [67].
Clinical impact and implications for therapy
Patients with infection due to ESBL-producing enterobacteria tended to have less satisfactory outcomes than those infected by pathogens that do not produce ESBLs [74–79]. In a prospective multinational study analyzing bloodstream infections (BSIs) due to ESBL-producing K. pneumoniae isolates, cephalosporin monotherapy was associated with a 40% 14-day mortality rate [80]. A comparable mortality rate among patients treated empirically with cephalosporin monotherapy was observed in another study concerning BSIs due to CTX-M-type-producing E.coli isolates [39]. These data force us to conclude that extended-spectrum cephalosporin treatment is associated with high rate of treatment failure (>80%) and mortality (>35%) when the susceptibility of infecting strains is close to the CLSI breakpoints (i.e., MIC ≥ 16 µg/ml). Data regarding cephalosporin use in infections due to ESBL-producing Enterobacteriaceae other than E. coli and K. pneumoniae are insufficient [75].
One of the determinant factors in the outcome of infected patients is choosing the appropriate empiric therapy within the first 24–48 hours of presentation. In our opinion optimal therapy of infections due to ESBL producers should be based on studies of in vitro activity of antimicrobial agents, carefully performed pharmacokinetic/pharmacodynamic (PK/PD) studies, and clinical investigations conducted in a prospective fashion. In reality, these studies are logistically challenging and rare. The clinician is faced with the fact that many ESBL producing bacteria also are resistant to quinolones (e.g., ciprofloxacin), aminoglycosides (e.g, gentamicin and tobramycin), and sulfamethoxazole [10]. A “magic bullet” is indeed hard to find.
Cefepime
Many in vitro studies demonstrate that ESBL-producing enterobacteria are frequently susceptible to cefepime (MIC ≤ 8 µg/L). However, it is important to note that these isolates should be reported as resistant to cefepime, according to CLSI and BSAC criteria. Several studies analyze the outcomes of patients treated with cefepime therapy for infection with ESBL-producing, cefepime-susceptible E. coli and K. pneumoniae [81–83]. From these retrospective analyses, cefepime was associated with a failure rate of 23–83% especially when MICs against ESBL producing Gram-negative bacteria were > 1 µg/ml. A randomized, evaluator-blind, multicenter trial found imipenem/cilistatin (0.5g q6h by i.v. route) to be substantially better than cefepime (2g q8h by i.v. route) for treatment of nosocomial pneumonia among ICU-patients[84]. These results suggest that cefepime is not the optimal therapy in the treatment of ESBL-producing enterobacteria, particularly during serious infections (e.g., bacteremia and pneumonia). Is a higher dose regimen of cefepime more effective? In combination with amikacin and ciprofloxacin, 2 g of cefepime q8h is comparable to carbapenems in patients infected by E. aerogenes containing the TEM-24 ESBLs [85]. The administration of higher doses (i.e., 4–6g administered as a continuous infusion or 2g q6–8h with prolonged infusion) results in adequate time above the MIC (%T > MIC) for the infecting organism achieving clinical success [86].
Beta-lactam/beta-lactamases inhibitor combinations
Since ESBL-producing enterobacteria are frequently susceptible in vitro to beta-lactam/beta-lactamases inhibitor combinations, it is logical to assume these combinations would also be clinically effective. Clinicians must keep in mind that presence of a chromosomal AmpC enzymes that is normally resistant to inactivation by a beta-lactamase inhibitor may be present [70]. Based on this and the data obtained from microbiological and clinical observations, we do not regard beta-lactam/beta-lactamases inhibitor combinations as a suitable option for serious infections due to ESBL-producing enterobacteria [87,88].
Carbapenems
Carbapenems (e.g., imipenem, meropenem, and ertapenem) have the most consistent activity against ESBL-producing Enterobacteriaceae. Large scale surveillance studies demonstrate that >98% against ESBL-producing E. coli, Klebsiella spp, and P. mirabilis isolates are susceptible to carbapenems [89–91]. Based on retrospective and prospective analyses, carbapenems should be considered as the preferred treatment for infections due to ESBL-producing enterobacteria [87,92]. In an international study conducted by Paterson et al. that includes ESBL-producing K. pneumoniae bloodstream isolates, patients who were treated with imipenem/cilistatin during the 5-day period after onset of infections had a 14-day mortality of 4.8%, compared with 27.6% when another in vitro active beta-lactam was used [80].
Quinolones
If the pathogen producing an ESBL is in vitro susceptible to ciprofloxacin, a satisfactory clinical response can be achieved using quinolones. Unfortunately, epidemiological studies reveal a strong link between fluoroquinolone resistance and ESBL production in Enterobacteriaceae [89]. For example, a multicenter prospective study of K. pneumoniae bacteremia conducted during 1996–1997, detected ESBL production in 60% of ciprofloxacin-resistant isolates, compared with 16% of ciprofloxacin-susceptible strains [93]. We discourage the use of quinolones as empiric therapy, since resistance to these agents is increasing in the United States and throughout the world.
Aminoglycosides
As in the case with quinolones, aminoglycosides are effective therapy against ESBL producing pathogens provided the organism has a MIC appreciably lower than susceptibility breakpoints. Susceptibility to amikacin seems to be preserved, in contrast to gentamicin and tobramycin, thus justifying its use as empiric therapy [89].
Tigecycline
CLSI criteria to interpret susceptibility testing of tigecycline are not yet established [62]. The Food and Drug Administration (FDA) and European Committee an Antimicrobial Susceptibility Testing (EUCAST) have created provisional resistance breakpoints (MIC ≥ 4 µg/ml and ≥ 2 µg/ml, respectively). In vitro data supports the notion that tigecycline can be considered an alternative to carbapenems for treatment of infections due to ESBL-producing Enterobacteriaceae [94,95]. However, clinical experience with tigecycline is still evolving.
Fosfomycin
The excellent in vitro activity of fosfomycin against ESBL-producing E. coli and K. pneumoniae strains has been recently reported [96]. Further studies are required to assess the efficacy of fosfomycin for the treatment of UTIs caused by ESBL-producing enterobacteria [97].
Colistin
Although once considered a toxic antibiotic, clinicians have now turned to colistin as a last resort agent for the treatment of infections caused by multidrug resistant gram-negative bacteria, against which this cationic detergent-like compound remains active [98]. As mentioned above, carbapenem resistance mediated by KPC is emerging among enterobacteria. Their implication in outbreaks, as seen in hospitals in New York City, has created a context in which the empiric use of colistin is necessary [99,100].
Conclusions
ESBLs are paradigmatic as a mechanism of resistance because of the impact they have had on the therapy of infections and the insight they have offered on the relationship between structure and function, in antibiotics and in their determinants of resistance. In TEM and SHV family betalactamases, single amino acid substitutions allow the efficient hydrolysis of extended-spectrum cephalosporins, while lowering the kcat/Km for penicillins. In the CTX-M family, point substitutions leading to specific interactions between enzyme and substrate are responsible for the improved activity against cefotaxime in preference to ceftazidime. Usually encoded on mobile genetic elements that accelerate their dissemination, the epidemiology of ESBLs has transformed recently with the surge of KPC and CTX-M ESBLs. This illustrates the complex interactions between antibiotic use, selection and transmission of resistance, colonization and infection in different populations. The future development of i) novel beta-lactams resistant to hydrolysis by these versatile enzymes and ii) the discovery of highly potent “second generation” beta-lactamase inhibitors are eagerly awaited.
Aknowledgements
The Veterans Affairs Merit Review Program and the National Institutes of Health support the work of Robert A. Bonomo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bush K. The impact of beta-lactamases on the development of novel antimicrobial agents. Curr Opin Investig Drugs. 2002;3:1284–1290. [PubMed] [Google Scholar]
- 2.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medeiros AA. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis. 1997;24 Suppl 1:S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 4.Jacoby GA, Medeiros AA. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philippon ALR, Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knox JR. Extended-spectrum and inhibitor-resistant TEM-type beta-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford PA. Extende-Spectrum beta-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin Microbiol Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••11.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483.Provides a detailed description of the shift in the epidemiology of ESBLs within Europe, from predominant TEM and SHV-type enzymes among nosocomial isolates to epidemics of E. coli harboring CTX-M ESBLs in community patients.
- 12.Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Luzzaro F, Mezzatesta M, Mugnaioli C, Perilli M, Stefani S, Amicosante G, Rossolini GM, Toniolo A. Trends in production of extended-spectrum beta-lactamases among enterobacteria of medical interest: report of the second Italian nationwide survey. J Clin Microbiol. 2006;44:1659–1664. doi: 10.1128/JCM.44.5.1659-1664.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinteros M, Radice M, Gardella N, Rodriguez MM, Costa N, Korbenfeld D, Couto E, Gutkind G. Extended-spectrum beta-lactamases in enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrob Agents Chemother. 2003;47:2864–2867. doi: 10.1128/AAC.47.9.2864-2867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lartigue M, Fortineau N, Nordmann P. Spread of novel expanded-spectrum beta-lactamases in Enterobacteriaceae in a university hospital in the Paris area, France. Clin Microbiol Infect. 2005;11:588–591. doi: 10.1111/j.1469-0691.2005.01172.x. [DOI] [PubMed] [Google Scholar]
- •17.Queenan A, Bush K. Carbapenemases: the Versatile Beta-Lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07.Comprehensively reviews this emerging threat. It summarizes the characteristics, epidemiology and detection of the carbapenemases, including KPC.
- ••18.Pitout JD, Church DL, Gregson DB, Chow BL, McCracken M, Mulvey MR, Laupland KB. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob Agents Chemother. 2007;51:1281–1286. doi: 10.1128/AAC.01377-06.Takes advantage of the wealth of information available to a central laboratory in Canada serving a large metropolitan area. Describes the dynamic nature of CTX-M expansion in the community.
- 19.Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005;56:52–59. doi: 10.1093/jac/dki166. [DOI] [PubMed] [Google Scholar]
- 20.Livermore DM, Hawkey PM. CTX-M: changing the face of ESBLs in the UK. J Antimicrob Chemother. 2005;56:451–454. doi: 10.1093/jac/dki239. [DOI] [PubMed] [Google Scholar]
- 21.Brigante G, Luzzaro F, Perilli M, Lombardi G, Coli A, Rossolini GM, Amicosante G, Toniolo A. Evolution of CTX-M-type beta-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int J Antimicrob Agents. 2005;25:157–162. doi: 10.1016/j.ijantimicag.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Mugnaioli C, Luzzaro F, De Luca F, Brigante G, Perilli M, Amicosante G, Stefani S, Toniolo A, Rossolini GM. CTX-M-type extended-spectrum beta-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob Agents Chemother. 2006;50:2700–2706. doi: 10.1128/AAC.00068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendonca N, Leitao J, Manageiro V, Ferreira E, Canica M. Spread of extended-spectrum beta-lactamase CTX-M-producing escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob Agents Chemother. 2007;51:1946–1955. doi: 10.1128/AAC.01412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavigne JP, Marchandin H, Delmas J, Moreau J, Bouziges N, Lecaillon E, Cavalie L, Jean-Pierre H, Bonnet R, Sotto A. CTX-M beta-lactamase-producing Escherichia coli in French hospitals: prevalence, molecular epidemiology, and risk factors. J Clin Microbiol. 2007;45:620–626. doi: 10.1128/JCM.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oteo J, Navarro C, Cercenado E, Delgado-Iribarren A, Wilhelmi I, Orden B, Garcia C, Miguelanez S, Perez-Vazquez M, Garcia-Cobos S, et al. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J Clin Microbiol. 2006;44:2359–2366. doi: 10.1128/JCM.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Bano J, Navarro MD, Romero L, Martinez-Martinez L, Muniain MA, Perea EJ, Perez-Cano R, Pascual A. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J Clin Microbiol. 2004;42:1089–1094. doi: 10.1128/JCM.42.3.1089-1094.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez JR, Martinez-Martinez L, Canton R, Coque TM, Pascual A. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases in Spain. Antimicrob Agents Chemother. 2005;49:2122–2125. doi: 10.1128/AAC.49.5.2122-2125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velasco C, Romero L, Martinez JM, Rodriguez-Bano J, Pascual A. Analysis of plasmids encoding extended-spectrum beta-lactamases (ESBLs) from Escherichia coli isolated from non-hospitalised patients in Seville. Int J Antimicrob Agents. 2007;29:89–92. doi: 10.1016/j.ijantimicag.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 29.Ho PL, Poon WW, Loke SL, Leung MS, Chow KH, Wong RC, Yip KS, Lai EL, Tsang KW. Community emergence of CTX-M type extended-spectrum {beta}-lactamases among urinary Escherichia coli from women. J Antimicrob Chemother. 2007;60:140–144. doi: 10.1093/jac/dkm144. [DOI] [PubMed] [Google Scholar]
- 30.Minarini LA, Gales AC, Palazzo IC, Darini AL. Prevalence of Community-Occurring Extended Spectrum beta-Lactamase-Producing Enterobacteriaceae in Brazil. Curr Microbiol. 2007;54:335–341. doi: 10.1007/s00284-006-0307-z. [DOI] [PubMed] [Google Scholar]
- ••31.Pallecchi L, Bartoloni A, Fiorelli C, Mantella A, Di Maggio T, Gamboa H, Gotuzzo E, Kronvall G, Paradisi F, Rossolini GM. Rapid Dissemination and Diversity of CTX-M Extended-Spectrum {beta}-Lactamase Genes in Commensal Escherichia coli from Healthy Children from Low-Resource Settings of Latin America. Antimicrob Agents Chemother. 2007 doi: 10.1128/AAC.00026-07.The investigators study the presence of CTX-M ESBLs among commensal E. coli strains found in Peruvian and Bolivian children, healthy and from remote areas presumably isolated from the use of broad-spectrum antibiotics. They demonstrate the presence of a new variant of CTX-M-2 and report the new fluoroquinolone resistance gene aac(6′)-Ib-cr for the first time in South America.
- 32.Wiener J, Quinn JP, Bradford PA, Goering RV, Nathan C, Bush K, Weinstein RA. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. Jama. 1999;281:517–523. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]
- 33.Arpin C, Dubois V, Maugein J, Jullin J, Dutilh B, Brochet JP, Larribet G, Fischer I, Quentin C. Clinical and molecular analysis of extended-spectrum {beta}-lactamase-producing enterobacteria in the community setting. J Clin Microbiol. 2005;43:5048–5054. doi: 10.1128/JCM.43.10.5048-5054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calbo E, Romani V, Xercavins M, Gomez L, Vidal CG, Quintana S, Vila J, Garau J. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum beta-lactamases. J Antimicrob Chemother. 2006;57:780–783. doi: 10.1093/jac/dkl035. [DOI] [PubMed] [Google Scholar]
- 35.Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, Raz R. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis. 2004;23:163–167. doi: 10.1007/s10096-003-1084-2. [DOI] [PubMed] [Google Scholar]
- 36.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother. 2004;54:735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Ami R, Schwaber MJ, Navon-Venezia S, Schwartz D, Giladi M, Chmelnitsky I, Leavitt A, Carmeli Y. Influx of extended-spectrum beta-lactamase-producing enterobacteriaceae into the hospital. Clin Infect Dis. 2006;42:925–934. doi: 10.1086/500936. [DOI] [PubMed] [Google Scholar]
- 38.Helfand MS, Bonomo RA. Extended-spectrum beta-lactamases in multidrug-resistant Escherichia coli: changing the therapy for hospital-acquired and community-acquired infections. Clin Infect Dis. 2006;43:1415–1416. doi: 10.1086/508891. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Bano J, Navarro MD, Romero L, Muniain MA, de Cueto M, Rios MJ, Hernandez JR, Pascual A. Bacteremia due to extended-spectrum beta -lactamase-producing Escherichia coli in the CTX-M era: a new clinical challenge. Clin Infect Dis. 2006;43:1407–1414. doi: 10.1086/508877. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Bano J, Navarro MD, Romero L, Muniain MA, Perea EJ, Perez-Cano R, Hernandez JR, Pascual A. Clinical and molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli as a cause of nosocomial infection or colonization: implications for control. Clin Infect Dis. 2006;42:37–45. doi: 10.1086/498519. [DOI] [PubMed] [Google Scholar]
- 41.Duan RS, Sit TH, Wong SS, Wong RC, Chow KH, Mak GC, Yam WC, Ng LT, Yuen KY, Ho PL. Escherichia coli producing CTX-M beta-lactamases in food animals in Hong Kong. Microb Drug Resist. 2006;12:145–148. doi: 10.1089/mdr.2006.12.145. [DOI] [PubMed] [Google Scholar]
- 42.Mesa RJ, Blanc V, Blanch AR, Cortes P, Gonzalez JJ, Lavilla S, Miro E, Muniesa M, Saco M, Tortola MT, et al. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage) J Antimicrob Chemother. 2006;58:211–215. doi: 10.1093/jac/dkl211. [DOI] [PubMed] [Google Scholar]
- 43.Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec JY. CTX-M-1- and CTX-M-15-type beta-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int J Antimicrob Agents. 2006;28:402–407. doi: 10.1016/j.ijantimicag.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 44.D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- •45.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614.Very compelling data revealing the evolution of resistant microorganisms in the environment.
- 46.Ibuka A, Taguchi A, Ishiguro M, Fushinobu S, Ishii Y, Kamitori S, Okuyama K, Yamaguchi K, Konno M, Matsuzawa H. Crystal structure of the E166A mutant of extended-spectrum beta-lactamase Toho-1 at 1.8 A resolution. J Mol Biol. 1999;285:2079–2087. doi: 10.1006/jmbi.1998.2432. [DOI] [PubMed] [Google Scholar]
- 47.Ibuka AS, Ishii Y, Galleni M, Ishiguro M, Yamaguchi K, Frere JM, Matsuzawa H, Sakai H. Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry. 2003;42:10634–10643. doi: 10.1021/bi0342822. [DOI] [PubMed] [Google Scholar]
- 48.Shimamura T, Ibuka A, Fushinobu S, Wakagi T, Ishiguro M, Ishii Y, Matsuzawa H. Acylintermediate structures of the extended-spectrum class A beta-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J Biol Chem. 2002;277:46601–46608. doi: 10.1074/jbc.M207884200. [DOI] [PubMed] [Google Scholar]
- 49.Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Minasov G, Blazquez J, Caselli E, Prati F, Shoichet BK. Recognition and resistance in TEM beta-lactamase. Biochemistry. 2003;42:8434–8444. doi: 10.1021/bi034242y. [DOI] [PubMed] [Google Scholar]
- 51.Nukaga M, Mayama K, Hujer AM, Bonomo RA, Knox JR. Ultrahigh resolution structure of a class A beta-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J Mol Biol. 2003;328:289–301. doi: 10.1016/s0022-2836(03)00210-9. [DOI] [PubMed] [Google Scholar]
- 52.Tranier S, B A, Maveyraud L, Guillet V, Sougakoff W, Samana JP. The high resolution crystal structure for class A beta-lactamase PER-1 reveals the bases for tis increase in breadth of activity. Journal of Biological Chemistry. 2000;275:28075–28082. doi: 10.1074/jbc.M003802200. [DOI] [PubMed] [Google Scholar]
- •53.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. Crystal structure of KPC-2: insights into carbapenemase activity in class A beta-lactamases. Biochemistry. 2007;46:5732–5740. doi: 10.1021/bi700300u.Describes the molecular structure of these emerging ESBL. Illustrates how changes in the active sites of the enzyme allow for the access of bulkier substrates such as carbapenems.
- 54.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 55.Hujer AM, Hujer KM, Bonomo RA. Mutagenesis of amino acid residues in the SHV-1 beta-lactamase: the premier role of Gly238Ser in penicillin and cephalosporin resistance. Biochim Biophys Acta. 2001;1547:37–50. doi: 10.1016/s0167-4838(01)00164-9. [DOI] [PubMed] [Google Scholar]
- 56.Hujer AM, Hujer KM, Helfand MS, Anderson VE, Bonomo RA. Amino acid substitutions at Ambler position Gly238 in the SHV-1 beta-lactamase: exploring sequence requirements for resistance to penicillins and cephalosporins. Antimicrob Agents Chemother. 2002;46:3971–3977. doi: 10.1128/AAC.46.12.3971-3977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raquet X, Lamotte-Brasseur J, Fonze E, Goussard S, Courvalin P, Frere JM. TEM beta-lactamase mutants hydrolysing third-generation cephalosporins. A kinetic and molecular modelling analysis. J Mol Biol. 1994;244:625–639. doi: 10.1006/jmbi.1994.1756. [DOI] [PubMed] [Google Scholar]
- 58.Raquet X, Vanhove M, Lamotte-Brasseur J, Goussard S, Courvalin P, Frere JM. Stability of TEM beta-lactamase mutants hydrolyzing third generation cephalosporins. Proteins. 1995;23:63–72. doi: 10.1002/prot.340230108. [DOI] [PubMed] [Google Scholar]
- 59.Cantu C, 3rd, Huang W, Palzkill T. Cephalosporin substrate specificity determinants of TEM-1 beta-lactamase. J Biol Chem. 1997;272:29144–29150. doi: 10.1074/jbc.272.46.29144. [DOI] [PubMed] [Google Scholar]
- 60.Huang W, Petrosino J, Hirsch M, Shenkin PS, Palzkill T. Amino acid sequence determinants of beta-lactamase structure and activity. J Mol Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 61.Venkatachalam KV, Huang W, LaRocco M, Palzkill T. Characterization of TEM-1 beta-lactamase mutants from positions 238 to 241 with increased catalytic efficiency for ceftazidime. J Biol Chem. 1994;269:23444–23450. [PubMed] [Google Scholar]
- 62.Clinical and Laboratory Standard Institute, Wayne, PA. CLSI document. 2007. Clinical and Laboratory Standard Methods. Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement; p. M100-S17. [Google Scholar]
- 63.Tenover FC, Mohammed MJ, Gorton TS, Dembek ZF. Detection and reporting of organisms producing extended-spectrum beta-lactamases: survey of laboratories in Connecticut. J Clin Microbiol. 1999;37:4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livermore DM, Yuan M. Antibiotic resistance and production of extended-spectrum beta-lactamases amongst Klebsiella spp. from intensive care units in Europe. J Antimicrob Chemother. 1996;38:409–424. doi: 10.1093/jac/38.3.409. [DOI] [PubMed] [Google Scholar]
- 65.Luzzaro F, Gesu G, Endimiani A, Ortisi G, Malandrin S, Pagani L, Rossolini GM. Performance in detection and reporting beta-lactam resistance phenotypes in Enterobacteriaceae: a nationwide proficiency study in Italian laboratories. Diagn Microbiol Infect Dis. 2006;55:311–318. doi: 10.1016/j.diagmicrobio.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 66.Carter MW, Oakton KJ, Warner M, Livermore DM. Detection of extended-spectrum beta-lactamases in klebsiellae with the Oxoid combination disk method. J Clin Microbiol. 2000;38:4228–4232. doi: 10.1128/jcm.38.11.4228-4232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •67.Wiegand I, Geiss HK, Mack D, Sturenburg E, Seifert H. Detection of extended-spectrum beta-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J Clin Microbiol. 2007;45:1167–1174. doi: 10.1128/JCM.01988-06.Investigates the performance characteristics of automated systems for the detection of ESBLs.
- 68.Liu CP, Wang NY, Lee CM, Weng LC, Tseng HK, Liu CW, Chiang CS, Huang FY. Nosocomial and community-acquired Enterobacter cloacae bloodstream infection: risk factors for and prevalence of SHV-12 in multiresistant isolates in a medical centre. J Hosp Infect. 2004;58:63–77. doi: 10.1016/j.jhin.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 69.Manzur A, Tubau F, Pujol M, Calatayud L, Dominguez MA, Pena C, Sora M, Gudiol F, Ariza J. Nosocomial Outbreak Due To Extended-Spectrum Betalactamase- Producing Enterobacter cloacae in a Cardio-Thoracic Intensive Care Unit. J Clin Microbiol. 2007 doi: 10.1128/JCM.02546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanson ND. AmpC beta-lactamases: what do we need to know for the future? J Antimicrob Chemother. 2003;52:2–4. doi: 10.1093/jac/dkg284. [DOI] [PubMed] [Google Scholar]
- 71.Schwaber MJ, Raney PM, Rasheed JK, Biddle JW, Williams P, McGowan JE, Jr, Tenover FC. Utility of NCCLS guidelines for identifying extended-spectrum beta-lactamases in non-Escherichia coli and Non-Klebsiella spp. of Enterobacteriaceae. J Clin Microbiol. 2004;42:294–298. doi: 10.1128/JCM.42.1.294-298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of extended-spectrum beta-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol. 2000;38:542–546. doi: 10.1128/jcm.38.2.542-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sturenburg E, Sobottka I, Noor D, Laufs R, Mack D. Evaluation of a new cefepimeclavulanate ESBL Etest to detect extended-spectrum beta-lactamases in an Enterobacteriaceae strain collection. J Antimicrob Chemother. 2004;54:134–138. doi: 10.1093/jac/dkh274. [DOI] [PubMed] [Google Scholar]
- 74.Wong-Beringer A, Hindler J, Loeloff M, Queenan AM, Lee N, Pegues DA, Quinn JP, Bush K. Molecular correlation for the treatment outcomes in bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae with reduced susceptibility to ceftazidime. Clin Infect Dis. 2002;34:135–146. doi: 10.1086/324742. [DOI] [PubMed] [Google Scholar]
- 75.Endimiani A, Luzzaro F, Brigante G, Perilli M, Lombardi G, Amicosante G, Rossolini GM, Toniolo A. Proteus mirabilis bloodstream infections: risk factors and treatment outcome related to the expression of extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2005;49:2598–2605. doi: 10.1128/AAC.49.7.2598-2605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •76.Tumbarello M, Spanu T, Sanguinetti M, Citton R, Montuori E, Leone F, Fadda G, Cauda R. Bloodstream infections caused by extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae: risk factors, molecular epidemiology, and clinical outcome. Antimicrob Agents Chemother. 2006;50:498–504. doi: 10.1128/AAC.50.2.498-504.2006.Recent study that identifies the characteristics of infections with ESBL producing organisms.
- 77.Ho PL, Chan WM, Tsang KW, Wong SS, Young K. Bacteremia caused by Escherichia coli producing extended-spectrum beta-lactamase: a case-control study of risk factors and outcomes. Scand J Infect Dis. 2002;34:567–573. doi: 10.1080/00365540210147516. [DOI] [PubMed] [Google Scholar]
- 78.Kim YK, Pai H, Lee HJ, Park SE, Choi EH, Kim J, Kim JH, Kim EC. Bloodstream infections by extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in children: epidemiology and clinical outcome. Antimicrob Agents Chemother. 2002;46:1481–1491. doi: 10.1128/AAC.46.5.1481-1491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marra R, W S, Castelo A, et al. Nosocomial bloodstream infections caused by Klebsiella pneumoniae: impact of extended-spectrum beta-lactamase (ESBL) production on clinical outcome in a hospital with hight ESBL prevalence. BMC Infect Dis. 2006;6:24. doi: 10.1186/1471-2334-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, Bonomo RA, et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum beta-lactamases. Clin Infect Dis. 2004;39:31–37. doi: 10.1086/420816. [DOI] [PubMed] [Google Scholar]
- 81.Paterson D, Ko WC, Von Gottbert A, et al. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum beta-lactamases: implications for the clinical microbiology laboratory. J Clin Microbiol. 2001;39:2206–2212. doi: 10.1128/JCM.39.6.2206-2212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kotapati S, K J, Nightingale CH, Nicolau DP. Clinical implications of extended-spectrum beta-lactamase (ESBL) producing Klebsiella species and Escherichia coli on cefepime effectiveness. J Infect. 2005;51:211–217. doi: 10.1016/j.jinf.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 83.LaBombardi VJ, R A, Tran K. Use of cefepime for the treatment of infections caused by extended-spectrum beta-lactamases-producing Klebsiella pneumoniae and Escherichia coli. Diagn Microbiol Infect Dis. 2006;56:313–315. doi: 10.1016/j.diagmicrobio.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 84.Zanetti G, Bally F, Greub G, Garbino J, Kinge T, Lew D, Romand JA, Bille J, Aymon D, Stratchounski L, et al. Cefepime versus imipenem-cilastatin for treatment of nosocomial pneumonia in intensive care unit patients: a multicenter, evaluatorblind, prospective, randomized study. Antimicrob Agents Chemother. 2003;47:3442–3447. doi: 10.1128/AAC.47.11.3442-3447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goethaert K, VL M, Lammens C, et al. High-dose cefepime as an alternative treatment for infections caused by TEM-24 ESBL-proucing Enterobacter aerogenes in serverely-ill patients. Clin Microbiol Infect. 2006;12:56–62. doi: 10.1111/j.1469-0691.2005.01290.x. [DOI] [PubMed] [Google Scholar]
- 86.Ambrose PG, Bhavnani SM, Jones RN. Pharmacokinetics-pharmacodynamics of cefepime and piperacillin-tazobactam against Escherichia coli and Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases: report from the ARREST program. Antimicrob Agents Chemother. 2003;47:1643–1646. doi: 10.1128/AAC.47.5.1643-1646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhavnani SM, Ambrose PG, Craig WA, Dudley MN, Jones RN. Outcomes evaluation of patients with ESBL- and non-ESBL-producing Escherichia coli and Klebsiella species as defined by CLSI reference methods: report from the SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis. 2006;54:231–236. doi: 10.1016/j.diagmicrobio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 88.Gavin PJ, Suseno MT, Thomson RB, Jr, Gaydos JM, Pierson CL, Halstead DC, Aslanzadeh J, Brecher S, Rotstein C, Brossette SE, et al. Clinical correlation of the CLSI susceptibility breakpoint for piperacillin- tazobactam against extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella species. Antimicrob Agents Chemother. 2006;50:2244–2247. doi: 10.1128/AAC.00381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirakata Y, Matsuda J, Miyazaki Y, Kamihira S, Kawakami S, Miyazawa Y, Ono Y, Nakazaki N, Hirata Y, Inoue M, et al. Regional variation in the prevalence of extended-spectrum beta-lactamase-producing clinical isolates in the Asia-Pacific region (SENTRY 1998–2002) Diagn Microbiol Infect Dis. 2005;52:323–329. doi: 10.1016/j.diagmicrobio.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 90.Sader HS, Hsiung A, Fritsche TR, Jones RN. Comparative activities of cefepime and piperacillin/tazobactam tested against a global collection of Escherichia coli and Klebsiella spp. with an ESBL phenotype. Diagn Microbiol Infect Dis. 2007;57:341–344. doi: 10.1016/j.diagmicrobio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 91.Goossens H, Grabein B. Prevalence and antimicrobial susceptibility data for extended-spectrum beta-lactamase- and AmpC-producing Enterobacteriaceae from the MYSTIC Program in Europe and the United States (1997–2004) Diagn Microbiol Infect Dis. 2005;53:257–264. doi: 10.1016/j.diagmicrobio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 92.Wong-Beringer A. Therapeutic challenges associated with extended-spectrum, beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Pharmacotherapy. 2001;21:583–592. doi: 10.1592/phco.21.6.583.34537. [DOI] [PubMed] [Google Scholar]
- 93.Paterson DL, Mulazimoglu L, Casellas JM, Ko WC, Goossens H, Von Gottberg A, Mohapatra S, Trenholme GM, Klugman KP, McCormack JG, et al. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum beta-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin Infect Dis. 2000;30:473–478. doi: 10.1086/313719. [DOI] [PubMed] [Google Scholar]
- 94.Morosini MI, Garcia-Castillo M, Coque TM, Valverde A, Novais A, Loza E, Baquero F, Canton R. Antibiotic coresistance in extended-spectrum-beta-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob Agents Chemother. 2006;50:2695–2699. doi: 10.1128/AAC.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hope R, Warner M, Potz NA, Fagan EJ, James D, Livermore DM. Activity of tigecycline against ESBL-producing and AmpC-hyperproducing Enterobacteriaceae from south-east England. J Antimicrob Chemother. 2006;58:1312–1314. doi: 10.1093/jac/dkl414. [DOI] [PubMed] [Google Scholar]
- 96.de Cueto M, Lopez L, Hernandez JR, Morillo C, Pascual A. In vitro activity of fosfomycin against extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: comparison of susceptibility testing procedures. Antimicrob Agents Chemother. 2006;50:368–370. doi: 10.1128/AAC.50.1.368-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••97.Pullukcu H, Tasbakan M, Sipahi OR, Yamazhan T, Aydemir S, Ulusoy S. Fosfomycin in the treatment of extended spectrum beta-lactamase-producing Escherichia coli-related lower urinary tract infections. Int J Antimicrob Agents. 2007;29:62–65. doi: 10.1016/j.ijantimicag.2006.08.039.Investigation of a very appealing therapeutic option for this difficult to treat infections that could result in less use of carbapenems.
- •98.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1.A comprehensive review that corrects many common misconceptions of the pharmacologic characteristics of colistin.
- 99.Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med. 2005;165:1430–1435. doi: 10.1001/archinte.165.12.1430. [DOI] [PubMed] [Google Scholar]
- 100.Falagas ME, Rafailidis PI. When to include polymyxins in the empirical antibiotic regimen in critically ill patients with fever? A decision analysis approach. Shock. 2007;27:605–609. doi: 10.1097/01.shk.0000246899.73315.cb. [DOI] [PubMed] [Google Scholar]


