Abstract
Import of DNA into mammalian nuclei is generally inefficient. Therefore, one of the current challenges in human gene therapy is the development of efficient DNA delivery systems. Here we tested whether bacterial proteins could be used to target DNA to mammalian cells. Agrobacterium tumefaciens, a plant pathogen, efficiently transfers DNA as a nucleoprotein complex to plant cells. Agrobacterium-mediated T-DNA transfer to plant cells is the only known example for interkingdom DNA transfer and is widely used for plant transformation. Agrobacterium virulence proteins VirD2 and VirE2 perform important functions in this process. We reconstituted complexes consisting of the bacterial virulence proteins VirD2, VirE2, and single-stranded DNA (ssDNA) in vitro. These complexes were tested for import into HeLa cell nuclei. Import of ssDNA required both VirD2 and VirE2 proteins. A VirD2 mutant lacking its C-terminal nuclear localization signal was deficient in import of the ssDNA–protein complexes into nuclei. Import of VirD2–ssDNA–VirE2 complexes was fast and efficient, and was shown to depended on importin α, Ran, and an energy source. We report here that the bacterium-derived and plant-adapted protein–DNA complex, made in vitro, can be efficiently imported into mammalian nuclei following the classical importin-dependent nuclear import pathway. This demonstrates the potential of our approach to enhance gene transfer to animal cells.
Agrobacterium tumefaciens delivers bacterial DNA, complexed with bacterial proteins, into plant cells where it is (i) protected from degradation, (ii) directed to the nucleus, (iii) transiently expressed at high level, and (iv) integrated, frequently as a single copy. To exploit these properties for DNA transfer to human cells, we tested whether the proteins responsible for entry of the bacterial DNA into plant nuclei could be used for import of DNA into mammalian nuclei.
The plant pathogen A. tumefaciens transfers T-DNA (transferred DNA; 10–20 kb in size), derived from the bacterial plasmid, pTi (tumor-inducing), into plant cells. The T-DNA is exported to the plant cell as a nucleoprotein complex composed of single-stranded DNA (ssDNA) and bacterial proteins VirD2 and VirE2 (for reviews, see refs. 1–3). VirD2 is covalently attached to the 5′ terminus of the DNA, and VirE2 is bound along the DNA, because of its ssDNA binding activity. Once in the plant cell, the T-DNA–protein complex enters the nucleus, and the T-DNA can be transiently expressed. Finally, T-DNA is integrated into the plant cell genome (for reviews, see refs. 1–6). Agrobacterium-mediated T-DNA transfer to plant cells is widely used for plant transformation.
With the exception of the border sequences flanking the T-DNA, no T-DNA sequence requirements have been observed for Agrobacterium-mediated plant transformation. Thus, nuclear import of T-DNA likely depends on proteins accompanying it into the plant. The bacterial proteins VirD2 and VirE2 both contain bipartite nuclear localization signals (NLSs) that target them into plant nuclei (7–9). Furthermore, the C-terminal NLS of the VirD2 protein is required for efficient transfer of the bacterial T-DNA to the plant nucleus (10–12). Because mammalian NLSs have been shown to be recognized in plant systems (13–16), these target sequences may be universal.
We reconstituted in vitro the T-DNA complexes composed of ssDNA and virulence proteins VirD2 and VirE2 and tested them for import into the nuclei of permeabilized HeLa cells. We found that both virulence proteins, VirD2 and VirE2, are required for efficient import of the DNA to animal nuclei. A VirD2 mutant lacking its C-terminal NLS was deficient in import of the ssDNA–protein complexes into nuclei. Furthermore, this transfer was absolutely dependent on mammalian import factors, indicating that import of the T-DNA complex is mediated by the classical importin-dependent nuclear import pathway.
MATERIALS AND METHODS
Preparation of Fluorescently Labeled Proteins.
Proteins (VirD2, VirE2, BSA, and BSA-NLS) were labeled directly with Cy3.5 reagent (from Cy3.5 mAb labeling kit; Amersham Pharmacia) by using the protocol recommended by the supplier for labeling of mAb.
Site-Specific ssDNA Cleavage Activity of VirD2.
For overexpression of the VirD2 wild-type protein (VirD2wt), the previously described plasmid pFSVirD2 was used (17). The plasmid pFSVirD2NruI (for overexpression of the VirD2ΔNLS mutant protein) was obtained by exchanging a BamHI-EcoRI fragment from the plasmid pFSVirD2 with a BamHI-EcoRI fragment of pVD44 (11). VirD2wt and VirD2ΔNLS mutant proteins were purified as described (18). The efficiency of the cutting reaction by VirD2 proteins was tested on rhodamine-labeled 1-kb ssDNA. DNA was first dephosphorylated with shrimp alkaline phosphatase (Amersham Pharmacia) and then labeled with [γ-32P]ATP at the 5′ end by using polynucleotide kinase (Boehringer Mannheim). Radiolabeled DNA (0.15 μg) was reacted with 7.2 μg of VirD2wt or VirD2ΔNLS mutant protein for 1 h at 37°C in 15 μl of TNM buffer (20 mM Tris⋅HCl, pH 8.8/50 mM NaCl/5 mM MgCl2). The reaction was terminated by the addition of formamide to a final concentration of 30%, the mixture was loaded on a 20% polyacrylamide/8 M urea denaturing gel, and electrophoresis was performed for 2 h at 40 V/cm. Autoradiography was performed by using a PhosphorImager system (Molecular Dynamics), and the efficiency of the cleavage reaction was calculated as the ratio of the radioactivity of the cleavage product (17-mer) to the total amount of the radioactivity in the reaction (1-kb DNA + 17-mer).
ssDNA-Binding Activity of VirE2.
VirE2 protein was purified as described by Ziemienowicz et al. (unpublished data). The purified protein was tested for its ssDNA-binding properties as described (19–21). A 250-bp double-stranded DNA fragment was amplified by PCR using 10 μCi (1 Ci = 37 GBq) of 32P-labeled dATP and 0.25 mM of dNTP. The incorporation rate was measured, and the quantity of labeled dsDNA was determined. The binding assay was performed in buffer E1 (50 mM Tris⋅HCl, pH 8.5/100 mM NaCl). To 1 ng of heat-denatured labeled DNA, 1 mg of BSA and increasing amounts of purified VirE2 protein were added (final volume: 10 μl). Incubation was performed at 4°C for 30 min. The complex was then loaded on a 4% polyacrylamide gel, and electrophoresis was performed at 5 V/cm at 4°C. VirE2 protein (60 ng) was able to coat 1 ng of 250-base long ssDNA completely, amounting to about one molecule of VirE2 per 5 nucleotides of ssDNA. According to recently reported calculations (22), a ≈3.6-fold excess of VirE2 was needed to fully coat the DNA in our experimental system.
Production of Fluorescently Labeled ssDNA.
Rhodamine-labeled 1-kb DNA was produced by using PCR amplification with pUC18 plasmid DNA as a template and primers containing the border sequence of pTiA6 (underlined) with VirD2 cleavage site (∧) and template sequence: primer p3 (5′-CCACGGTATATATCCTG∧CCAGGGTATTTCACACCGCATATGG-3′) and primer p4 (5′-CAACGGTATATATCCTG∧CCAGGCTAGAGTAAGTAGTCGCC-3′). The reaction mixture of 50 μl final volume contained 0.1 mM each dATP, dCTP, dGTP, 0.09 mM dTTP, 0.01 mM tetramethylrhodamine-6-dUTP, 0.2 μM each primer, 0.04 μg of pUC18 DNA, and 5 units of Taq DNA polymerase (Boehringer Mannheim) in standard PCR buffer. The amplification procedure was denaturation for 5 min at 94°C and 30 cycles of 1 min at 94°C, 1 min at 60°C, and 45 sec at 72°C, followed by a final elongation step for 10 min at 72°C. The PCR product was purified and concentrated by using the Wizard DNA Clean-Up System (Promega). ssDNA was obtained by heat denaturation for 10 min at 94°C and fast cooling on ice.
Formation of the VirD2–ssDNA–VirE2 Complex.
For the formation of VirD2–ssDNA–VirE2 complex 0.15 μg of rhodamine-labeled 1-kb ssDNA was reacted with 7.2 μg of VirD2wt or VirD2ΔNLS protein in TNM buffer (20 mM Tris⋅HCl, pH 8.8/50 mM NaCl/5 mM MgCl2) for 1 h at 37°C (15 μl final volume). The full complex was made by addition of 0.5 μg of VirE2 protein and incubation of the mixture (20 μl final volume) for 30 min on ice.
Assay of in Vitro Nuclear Import in Permeabilized HeLa Cells.
Nuclear import assay using permeabilized HeLa cells and HeLa-derived import factors was performed as described (23). First, the fluorescently labeled import substrate (proteins, DNA, or protein–DNA complexes) was mixed with 25.5 pmol of Rch1 (importin α) and 7 pmol of importin β. Then Ran/TC4, an energy regeneration system, permeabilized cells, and buffer were added to a final volume of 10 μl in the amounts indicated in ref. 23. Import reaction mixes were incubated for 20 min at room temperature in the dark. Nuclei were then stained with fluorescent dye SYTO 13 (Molecular Probes) and analyzed by confocal microscopy using FITC filter for the dye and tetramethylrhodamine B isothiocyanate filter for rhodamine- or Cy3.5-labeled samples. No transmission between these two channels was observed. Signal detection (sensitivity) and processing of the scanned images were identical within each experimental series.
RESULTS
VirD2 and VirE2 Are Imported into Nuclei of Permeabilized HeLa Cells.
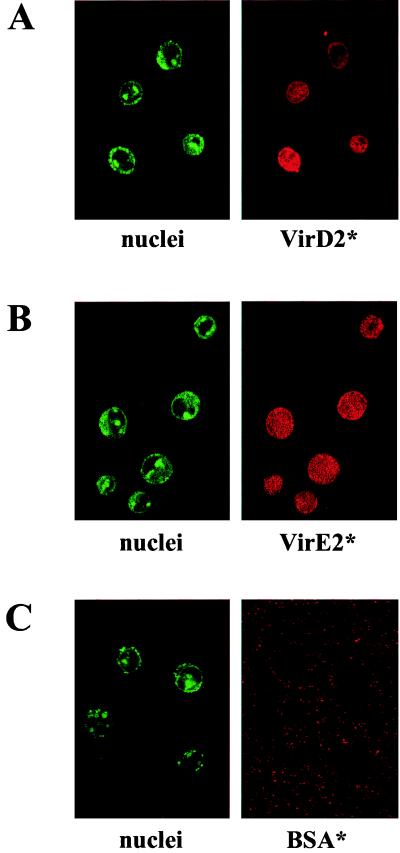
It has been shown recently that the VirD2 protein expressed in human cells localizes in the nucleus (24). To test whether the NLSs of plant-adapted proteins are recognized in vitro as well, fluorescently labeled VirD2 and VirE2 proteins were assayed for import into nuclei of permeabilized HeLa cells. In our system, BSA was not found in the nuclei, whereas BSA-NLS, VirD2, and VirE2 were found to localize to the mammalian nuclei (Fig. 1, Table 1). The kinetics of import of both VirD2 and VirE2 proteins was comparable to the import of BSA-NLS (data not shown). These results show that the NLSs of VirD2 and VirE2 are functional in permeabilized HeLa cells, consistent with previously shown nuclear localization of VirD2 in transfected human cells (24). Intracellular localization of VirE2 in human cells has not been analyzed thus far. VirE2 was shown to localize to the nuclei of embryonic Drosophila cells and Xenopus oocytes, but the particular protein used in these experiments had to be slightly modified in its NLS to resemble the NLS of nucleoplasmin (25).
Figure 1.
Localization of VirD2 and VirE2 proteins in HeLa nuclei. Cy3.5-labeled proteins (red fluorescence) were incubated with digitonin-permeabilized HeLa cells for 20 min at room temperature in the dark. The nuclei were stained with SYTO 13 dye (green fluorescence). (A) Localization of VirD2 protein. (B) Localization of VirE2 protein. (C) Localization of BSA.
Table 1.
Nuclear import of ssDNA depends on VirD2 and VirE2
| Import substrate | Import |
|---|---|
| BSA | − |
| BSA-NLS | + |
| VirE2 | + |
| VirD2 | + |
| ssDNA | − |
| ssDNA–VirD2 | − |
| ssDNA–VirE2 | − |
| ssDNA–VirD2–VirE2 | + |
| ssDNA–VirD2ΔNLS–VirE2 | − |
Import of substrates into mammalian nuclei was tested in vitro using permeabilized HeLa cells and HeLa-derived import factors. Each experiment was repeated at least three times, and for each sample at least 100 nuclei were scored. +, 99–100% of nuclei showed red fluorescence; −, 0–1% of nuclei showed red fluorescence.
Reconstruction of the T-DNA Complex in Vitro.
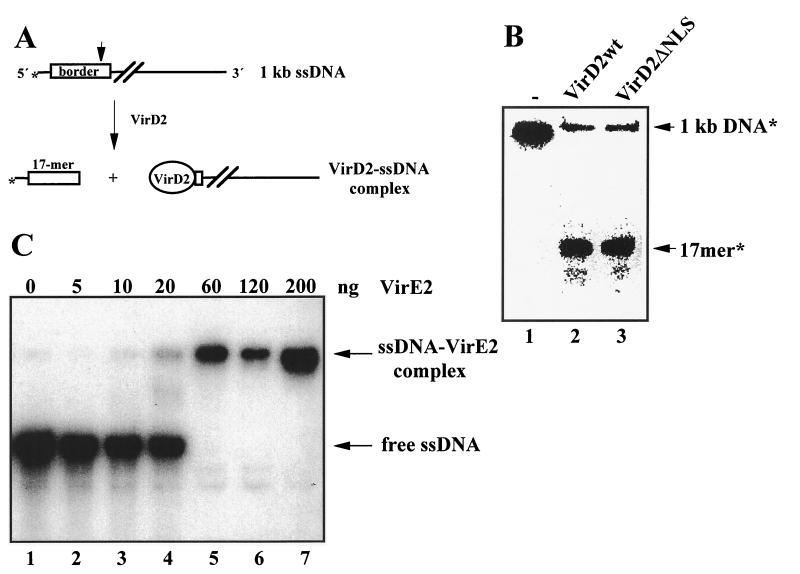
The reconstitution of the T-DNA complex was monitored by activity assays: site-specific cleavage for VirD2 and ssDNA binding for VirE2. Cleavage of border sequence containing single-stranded oligonucleotides or denatured double-stranded DNA by purified VirD2 protein has been reproduced in vitro (18, 26). Cleavage occurred at the same position as identified in Agrobacterium cells, resulting in a DNA-VirD2 complex in which VirD2 is bound via Tyr-29 to the 5′ end of the DNA (18, 26). We tested the efficiency of the cutting reaction on 1-kb rhodamine-labeled ssDNA (Fig. 2 A and B). The reaction was monitored by the VirD2-cleavage-dependent release of a 17-base labeled oligonucleotide. The cleavage was precise and efficient (73% of DNA was cleaved), and VirD2 remained covalently attached to the 5′ terminus of the cleaved border sequence (data not shown; see refs. 18 and 26). Cleavage by VirD2 lacking the C-terminal NLS was comparable in efficiency and precision to cleavage by the wild-type protein (Fig. 2B, lanes 2 and 3).
Figure 2.
Formation of T-DNA complex in vitro. (A) Scheme of the cleavage assay. (B) Cleavage of 1-kb ssDNA by VirD2wt (lane 2) or mutant VirD2ΔNLS protein (lane 3), control without protein (lane 1). The efficiency of the cutting reaction by VirD2 proteins was tested on rhodamine-labeled 1-kb ssDNA. (C) ssDNA binding activity of VirE2. Increasing amounts of VirE2 protein were added to ssDNA in amounts indicated on the picture.
The cooperativity of the binding of the purified VirE2 protein to ssDNA was tested in a gel-shift experiment (19–21). A change in the migration of the labeled ssDNA, indicating formation of the fully coated DNA (Fig. 2C, lanes 5–7 vs. lanes 1–4), could be observed in response to a small increase in the concentration of VirE2 protein. To prepare an artificial T-DNA complex, rhodamine-labeled DNA was first cleaved by VirD2, and the resulting ssDNA–VirD2 complex was then incubated with an excess of VirE2 to form the complete ssDNA–VirD2–VirE2 complex.
Nuclear Import of the Protein–DNA Complex in Permeabilized HeLa Cells.
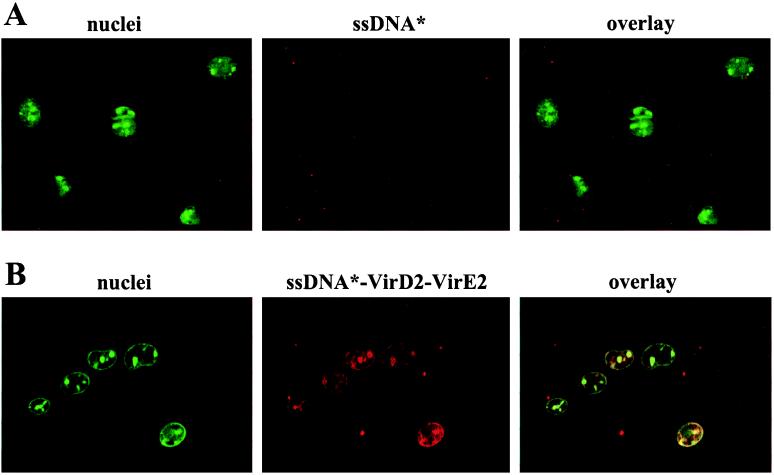
In vitro-reconstituted T-DNA complex, along with partial complexes, was tested for nuclear import in permeabilized HeLa cells complemented with HeLa-derived cytosolic import factors. We found that 1-kb fluorescently labeled ssDNA was neither imported into HeLa nuclei nor passively accumulated in them when used alone (Fig. 3A, Table 1). Surprisingly, VirD2 protein alone covalently attached to the ssDNA was not able to render the DNA nuclear (Table 1). Moreover, we did not observe efficient import of the 1-kb ssDNA as a complex with only the VirE2 protein (Table 1). It has been shown previously that modified VirE2 protein (see above) was able to target ssDNA into the nuclei of microinjected Drosophila embryos and Xenopus oocytes (25). Cytosol from embryonic Drosophila or Xenopus cells could contain endogenous factors that can cooperate with VirE2 and bypass a requirement for VirD2. In addition, T-DNA transfer to yeast nuclei in vivo was reduced only to 10% in the absence of VirE2 (27), also suggesting different requirements for nuclear import of T-DNA in different systems. However, when we used the complete ssDNA–VirD2–VirE2 complex, import of the protein–DNA complex into the HeLa nuclei could be observed (Fig. 3B). The import of VirD2–ssDNA–VirE2 complexes was very efficient and fast (occurring with a kinetics similar to that of the proteins tested alone: 90% of import in 10 min, 99–100% in 20 min; data not shown). The nuclear import of the T-DNA complex depended on the C-terminal NLS sequence of VirD2, because we could not detect any import of the VirD2ΔNLS–ssDNA–VirE2 complex (Table 1). Thus, the import of the T-DNA to the nuclei of permeabilized HeLa cells requires both the VirE2 protein and the NLS-carrying VirD2 protein.
Figure 3.
Import of 1-kb ssDNA into HeLa nuclei requires both VirD2 and VirE2 proteins. Different ssDNA–protein complexes containing rhodamine-labeled ssDNA (red fluorescence) were incubated with permeabilized HeLa cells for 20 min at room temperature in the dark. (A) ssDNA alone. (B) VirD2–ssDNA–VirE2 complex. The position of the nuclei is indicated by staining with SYTO 13 dye (green fluorescence). The fluorescently labeled component is indicated by ∗.
Requirements for Nuclear Import of VirD2–ssDNA–VirE2 Complexes.
In the classical, importin-dependent pathway, import of proteins is carried out by cytosolic factors such as importins α and β (which form a trimeric complex with an NLS-containing protein) and by Ran and pp15, which promote the translocation of the cargo–importins complex through the channel of the nuclear pore complex (for reviews, see refs. 28–31). Nuclear import of proteins in the mammalian in vitro system has been shown to be inhibited by wheat germ agglutinin (ref. 32) and by the nonhydrolyzable analog guanosine 5′-[γ-thio]triphosphate (33). To test whether import of T-DNA complex and import of proteins resembles each other mechanistically, requirements for cytosolic factors and effects of known inhibitors for protein import were tested. Import of ssDNA–VirD2–VirE2 complexes required importin α (Rch1), Ran, and (as an energy source) GTP/ATP (Table 2). Furthermore, import of the ssDNA–VirD2–VirE2 complex into the mammalian nuclei was inhibited in the presence of guanosine 5′-[γ-thio]triphosphate or wheat germ agglutinin (Table 2). Nuclear import of the T-DNA complex in permeabilized HeLa cells is thus mediated by the classical importin-dependent pathway for proteins.
Table 2.
Import of T-DNA complex into mammalian nuclei occurs via the classical NLS-dependent import pathway
| Import substrate | Inhibitor | Lacking factor | Import |
|---|---|---|---|
| ssDNA–VirD2–VirE2 | — | — | + |
| ssDNA–VirD2–VirE2 | — | Rch1 | − |
| ssDNA–VirD2–VirE2 | — | Ran | − |
| ssDNA–VirD2–VirE2 | — | GTP/ATP | − |
| ssDNA–VirD2–VirE2 | WGA | — | − |
| ssDNA–VirD2–VirE2 | GTPγS | — | − |
The import assay of fluorescently labeled T-DNA complex (ssDNA–VirD2–VirE2) into mammalian nuclei was done in vitro using permeabilized HeLa cells. Each experiment was repeated at least three times, and for each sample at least 100 nuclei were scored. +, 99–100% of nuclei showed red fluorescence signal; −, 0–1% of nuclei showed red fluorescence signal. GTPγS, guanosine 5′−[γ−thio]triphosphate. WGA, wheat germ agglutinin.
DISCUSSION
One of the rate-limiting steps in gene therapy is the low efficiency of gene transfer. We report here efficient transfer of a characterized protein–nucleic acid complex into HeLa nuclei in vitro. This finding may have important implications for gene transfer to animals and gene therapy. Currently, two types of vector systems are used for gene transfer—viral and nonviral. Although problems such as immunogenicity, toxic responses, and limitations in insert size are associated with viral vectors (34), they are more effective than nonviral systems in achieving high-efficiency gene transfer. Only those vectors that are able to overcome the intracellular barrier of the nuclear membrane can be used for transformation of nondividing cells such as brain, muscle, liver, and lung cells. The best examples are lentiviruses including HIV. HIV-1 DNA is presumably transported to the nucleus via NLSs of viral proteins, in the form of the preintegration complex (35–37). Nonviral vectors such as liposome–DNA conjugates are nonpathogenic but, unfortunately, less effective in gene transfer than viral vectors. Approaches to improve the efficiency of nonviral vectors led to the design of peptide–DNA complexes in which DNA was covalently linked to a peptide containing an intracellular localization signal. For example, covalent linkage of an SV40 large tumor antigen NLS-containing peptide to DNA led to nuclear accumulation of the conjugated DNA in digitonin-permeabilized HeLa cells (38). However, the covalent attachment of the NLS peptide completely abolished expression of the marker gene.
We report here efficient import of the DNA–protein complex into the nuclei of permeabilized HeLa cells. Considering the behavior of the T-DNA complex in plants, we foresee several advantages of our system for gene therapy. In the method reported here, the DNA in the complex is protected from degradation by nucleases because of the presence of bacterial proteins, VirD2 and VirE2, as has been shown both in vitro (23, 39) and in vivo (40). In addition, the use of the “agrobacterial” protein–DNA complex as vector should not be limited by the size of the transfecting DNA, because Agrobacterium-mediated transfer of a very long DNA (up to 170 kb) was observed (41, 42). Furthermore, the VirD2 and VirE2 proteins insure that the DNA is efficiently transported into mammalian nuclei. This implies the possibility of transformation of not only dividing but also nondividing cells, as has been achieved with lentiviral vectors. Because of their protective and targeting roles, the agrobacterial virulence proteins may also improve the efficiency of transient and/or stable transformation of mammalian cells; plants transformed by Agrobacterium show a high level of transient expression of genes present on the transferred DNA (43). Increase in efficiency of transient transformation may improve the chance for successful gene therapy. In addition to efficient nuclear targeting of DNA, our method may provide the advantage of VirD2- and VirE2-dependent precise DNA integration (17, 40).
Acknowledgments
We thank Christopher Marshallsay for kind donation of fluorescently labeled BSA-NLS, Serge Kocher for his help with the confocal microscope, and Véronique Gloeckler for excellent technical assistance. We also thank Drs. Philippe Crouzet, Witold Filipowicz, and Nancy Hynes for critical reading of the manuscript.
ABBREVIATIONS
- ssDNA
single-stranded DNA
- NLS
nuclear localization signal
References
- 1.Lartey R, Citovsky V. Genet Eng (NY) 1997;19:201–214. doi: 10.1007/978-1-4615-5925-2_11. [DOI] [PubMed] [Google Scholar]
- 2.Zupan J, Zambryski P. Crit Rev Plant Sci. 1997;16:279–295. [Google Scholar]
- 3.Rossi L, Tinland B, Hohn B. In: The Rhiziobiaceae. Spaink H, Hooykaas P, Kondorosi A, editors. Boston: Kluwer; 1998. pp. 302–330. [Google Scholar]
- 4.Tinland B, Hohn B. Genet Eng (NY) 1995;17:209–229. [PubMed] [Google Scholar]
- 5.Tinland B. Trends Plant Sci. 1996;1:178–184. [Google Scholar]
- 6.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard E A, Zupan J R, Citovsky V, Zambryski P C. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 8.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 9.Tinland B, Koukolikova-Nicola Z, Hall M N, Hohn B. Proc Natl Acad Sci USA. 1992;89:7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shurvinton C E, Hodges L, Ream W. Proc Natl Acad Sci USA. 1992;89:11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi L, Hohn B, Tinland B. Mol Gen Genet. 1993;239:345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 12.Narasimhulu S B, Deng X B, Sarria R, Gelvin S B. Plant Cell. 1996;8:873–886. doi: 10.1105/tpc.8.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Krol A R, Chua N H. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lassner M W, Jones A, Daubert S, Comai L. Plant Mol Biol. 1991;17:229–234. doi: 10.1007/BF00039497. [DOI] [PubMed] [Google Scholar]
- 15.Varagona M J, Raikhel N V. Plant J. 1994;5:207–214. doi: 10.1046/j.1365-313x.1994.05020207.x. [DOI] [PubMed] [Google Scholar]
- 16.Hicks G R, Raikhel N V. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 17.Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel A M, Hohn B. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Citovsky V, Wong M L, Zambryski P. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zupan J R, Citovsky V, Zambryski P. Proc Natl Acad Sci USA. 1996;93:2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen P, Pazour G J, Anderson D, Das A. J Bacteriol. 1989;171:2573–2580. doi: 10.1128/jb.171.5.2573-2580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citovsky V, Guralnick B, Simon M N, Wall J S. J Mol Biol. 1997;271:718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- 23.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 24.Relic B, Andjelkovic M, Rossi L, Nagamine Y, Hohn B. Proc Natl Acad Sci USA. 1998;95:9105–9110. doi: 10.1073/pnas.95.16.9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guralnick B, Thomsen G, Citovsky V. Plant Cell. 1996;8:363–373. doi: 10.1105/tpc.8.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jasper F, Koncz C, Schell J, Steinbiss H H. Proc Natl Acad Sci USA. 1994;91:694–698. doi: 10.1073/pnas.91.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P J. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Görlich D. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 29.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 30.Moroianu J. Crit Rev Eukaryotic Gene Expression. 1997;7:61–72. doi: 10.1615/critreveukargeneexpr.v7.i1-2.40. [DOI] [PubMed] [Google Scholar]
- 31.Pemberton L F, Blobel G, Rosenblum J S. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 32.Finlay D R, Newmeyer D D, Price T M, Forbes D J. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melchior F, Paschal B, Evans J, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma I M, Somia N. Nature (London) 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 35.Goldfarb D S. Curr Biol. 1995;5:570–573. doi: 10.1016/s0960-9822(95)00111-4. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson M. Trends Cell Biol. 1996;6:9–14. doi: 10.1016/0962-8924(96)81032-4. [DOI] [PubMed] [Google Scholar]
- 37.Whittaker G R, Helenius A. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 38.Sebestyén M G, Ludtke J J, Bassik M C, Zhang G, Budker V, Lukhtanov E A, Hagstrom J E, Wolff J A. Nat Biotechnol. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 39.Dürrenberger F, Crameri A, Hohn B, Koukolikova-Nicola Z. Proc Natl Acad Sci USA. 1989;86:9154–9158. doi: 10.1073/pnas.86.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi L, Hohn B, Tinland B. Proc Natl Acad Sci USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miranda A, Janssen G, Hodges L, Peralta E G, Ream W. J Bacteriol. 1992;174:2288–2297. doi: 10.1128/jb.174.7.2288-2297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton C M, Frary A, Lewis C, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janssen B J, Gardner R C. Plant Mol Biol. 1990;14:61–72. doi: 10.1007/BF00015655. [DOI] [PubMed] [Google Scholar]