Abstract
Checkpoints maintain the order and fidelity of the eukaryotic cell cycle, and defects in checkpoints contribute to genetic instability and cancer. Much of our current understanding of checkpoints comes from genetic studies conducted in yeast. In the fission yeast Schizosaccharomyces pombe (Sp), SpRad3 is an essential component of both the DNA damage and DNA replication checkpoints. The SpChk1 and SpCds1 protein kinases function downstream of SpRad3. SpChk1 is an effector of the DNA damage checkpoint and, in the absence of SpCds1, serves an essential function in the DNA replication checkpoint. SpCds1 functions in the DNA replication checkpoint and in the S phase DNA damage checkpoint. Human homologs of both SpRad3 and SpChk1 but not SpCds1 have been identified. Here we report the identification of a human cDNA encoding a protein (designated HuCds1) that shares sequence, structural, and functional similarity to SpCds1. HuCds1 was modified by phosphorylation and activated in response to ionizing radiation. It was also modified in response to hydroxyurea treatment. Functional ATM protein was required for HuCds1 modification after ionizing radiation but not after hydroxyurea treatment. Like its fission yeast counterpart, human Cds1 phosphorylated Cdc25C to promote the binding of 14-3-3 proteins. These findings suggest that the checkpoint function of HuCds1 is conserved in yeast and mammals.
In eukaryotic cells, checkpoints operating in the G2 phase of the cell cycle block entry into mitosis in the presence of unreplicated or damaged DNA (1). In addition, an S phase DNA damage checkpoint slows replication in response to damaged DNA. In yeast, these checkpoints require phosphoinositide (PI)-3-like kinases including ScMec1 (2, 3) and SpRad3 (4–7). In Saccharomyces cerevisiae, ScMec1 has partially overlapping functions with ScTel1, a PI-3-like kinase involved in telomere maintenance (8–11). In Schizosaccharomyces pombe, SpRad3 and SpTel1 function coordinately to maintain telomere length (12). PI-3-like kinases related to ScMec1, ScTel1, SpRad3, and SpTel1 also play a role in mammalian checkpoints. These include ATM (ataxia-telangiectasia-mutated) (13) and ATR (ataxia-telangiectasia- and rad3+-related) (7, 14). Cells derived from ataxia-telangiectasia (AT) patients lacking functional ATM protein, are unable to arrest at the G1–S boundary after irradiation and have a characteristic inability to arrest DNA synthesis after irradiation. ATM has been shown to directly phosphorylate p53, and this may explain why loss of ATM leads to loss of G1 arrest after DNA damage (15, 16). A-T cells that are in G2 at the time of irradiation also fail to arrest before entering mitosis, but the regulatory pathway disrupted in this case is unknown (for review, see ref. 17). ATR, a PI-3-like kinase similar to ATM, is able to partially suppress the UV sensitivity of a mec1 mutant in S. cerevisiae, suggesting conservation of function in yeast and humans (7). In addition, ATR may function to transmit signals from certain types of DNA damage and may be a component of the DNA replication checkpoint (18).
Important mediators of the Mec1/Rad3-dependent checkpoints are ScRad53 and its structural homolog in fission yeast, SpCds1 (11, 19–21). ScRad53 is an essential component of both the DNA damage and replication checkpoints. In response to DNA damage, ScRad53 inhibits the expression of G1 cyclins (22) and activates another serine–threonine protein kinase, ScDun1 (23), which controls the transcriptional response to DNA damage. In fission yeast, loss of cds1+ leads to hydroxyurea (HU) sensitivity, but loss of both cds1+ and chk1+ results in a complete abrogation of the DNA replication checkpoint (21, 24, 25). These results establish a role for SpCds1 in the DNA replication checkpoint. In addition, a role for SpCds1 in the S phase DNA damage checkpoint has been proposed (20).
Recent studies have shed light on how checkpoints interface with cell cycle regulators to prevent entry into mitosis in response to DNA damage and replication blocks. In several species, the DNA damage and replication checkpoints block entry into mitosis by inactivating Cdc2, the cyclin-dependent protein kinase that initiates mitosis (26–28). This is accomplished in part by maintaining the Cdc25 phosphatase in a phosphorylated form that binds 14-3-3 proteins (25, 29, 30). The 14-3-3-bound form of Cdc25 is prevented from activating Cdc2, and cells arrest in the G2 phase of the cell cycle. Checkpoint kinases that regulate the interactions between Cdc25 and 14-3-3 proteins include SpCds1 (25, 31) and Chk1 [from Schizosaccharomyces pombe, Homo sapiens (Hu), and Xenopus (29, 32–35)].
To date, vertebrate homologs of ScRad53 and SpCds1 have not been identified; therefore, it is not known whether an ATM/ATR-dependent pathway similar to the yeast ScMec1–ScRad53 or SpRad3–SpCds1 pathways exists in mammals. Here we report the identification of a human kinase (denoted HuCds1) that shows sequence similarity to SpCds1 and ScRad53. HuCds1 was modified by phosphorylation and activated in response to ionizing radiation. HuCds1 was also modified in response to HU treatment. Phosphorylation of HuCds1 in response to ionizing radiation but not HU treatment required a functional ATM protein. Finally, HuCds1 phosphorylated human Cdc25C on Ser-216, the 14-3-3 binding site (29). These findings indicate that the checkpoint function of the Cds1 protein kinase is conserved in yeast and mammals.
EXPERIMENTAL PROCEDURES
Isolation of HuCds1 cDNAs.
A CLONTECH HeLa cDNA library in the yeast expression vector pGADGH was screened for human proteins that derepress S. cerevisiae telomeres by using a strategy described previously (36).
Northern and Western Analyses.
A human multiple tissue Northern blot (CLONTECH) was hybridized at 68°C in QuickHyb (Stratagene) with a probe derived from nucleotide 1198–1680 of HuCds1 cDNA. The human multiple tissue Western blot (Geno Technology, St. Louis) was immunoblotted with affinity-purified antibody against HuCds1.
Expression of Strep-Tagged HuCds1 in Tissue Culture Cells.
The expression vector for Strep-epitope tagged HuCds1, which contains the amino acid sequence MGWSHPQFEKNSARAHAVV (Strep epitope in bold) before the first methionine of HuCds1, was constructed in plasmid pRetro-Off (CLONTECH) and transiently transfected into Phoenix-Ampho cells, a 293-based retrovirus packaging cell line (American Type Culture Collection, Inventory no. SD 3443), by using Lipofectamine (GIBCO/BRL). Although Phoenix-Ampho is a retroviral packaging cell line, it was used here strictly for transient expression and not for retroviral production. After 2 days in a medium containing 1 μg/ml doxycycline (Sigma), the cells were grown in a medium without doxycycline for 3 more days to induce Strep-HuCds1 protein. Strep-HuCds1 and the endogenous HuCds1 proteins were detected by immunoblotting with affinity-purified anti-HuCds1 antibody.
Quantitation of HuCds1 Kinase Activity After Irradiation.
One hour after irradiation (20 and 100 Gy), HeLa S3 cells were washed twice in ice-cold PBS (pH 7.4). Lysis was performed on ice for 30 minutes in lysis buffer (50 mM Tris⋅HCl, pH 7.4/150 mM NaCl/2 mM EGTA/1 mM EDTA/25 mM sodium fluoride/0.2% Triton X-100/0.3% Nonidet P-40/1 mM sodium orthovanadate and protease inhibitors). Lysates were clarified by centrifuging at 14,000 × g for 15 minutes. Lysates containing 400 μg of protein were precleared with 10 μl of immobilized Protein A (Pierce) for 10 minutes at 4°C and immunoprecipitated with 13 μg of antibody specific for BC3 peptide (see HuCds1 Antibody Production) and 10 μl of immobilized Protein A at 4°C for 1 hour with or without 2 μg of competing peptide BC3. Beads were washed three times with lysis buffer and twice with kinase buffer (10 mM Hepes, pH 7.5/75 mM KCl/5 mM MgCl2/0.5 mM EDTA). Kinase buffer supplemented with 2 mM DTT, 100 μM cold ATP, 15 μCi [γ-32P]ATP (>3,000 Ci/mmol; 1 Ci = 37 GBq) and 2 μg of glutathione S-transferase (GST)-Cdc25C(200–256) (37) was added to the washed beads and incubated at 30°C for 30 minutes. Proteins were separated by SDS/PAGE and visualized by Coomassie blue staining and autoradiography. Incorporation of 32P into GST-Cdc25C substrates was quantitated with a PhosphorImager (Molecular Dynamics). The activity of HuCds1 kinase was induced to the same extent after treatment of HeLa 53 cells with 20 or 100 Gy irradiation.
Construction of Catalytically Inactive HuCds1(K249R).
An EcoRI-XhoI HuCds1 fragment was subcloned into plasmid pUC19. A Morph mutagenesis kit (5 Prime → 3 Prime) was used to introduce two nucleotide changes in the kinase domain of HuCds1 such that Lys-249 was changed to Arg. The mutagenic oligonucleotide was: 5′-CATGTAAGAAAGTAGCCATAAGAATCATCAGCAAAAGGAAGTTTGC-3′. The mutation was verified by sequencing.
Production of GST-HuCds1 in Schizosaccharomyces pombe.
To express GST-HuCds1, an EcoRI-XhoI fragment containing the HuCds1 cDNA (WT or K249R) was blunted and ligated into the SmaI site located immediately 3′ of the GST domain in plasmid pESP-1 (Stratagene). The plasmid was transformed and expressed in Schizosaccharomyces pombe. Cells were disrupted at 4°C in sorbitol buffer containing 0.5% Triton X-100 and protease inhibitors (Boehringer Mannheim mini tablets) by using a French press, and the extract was purified over a glutathione-Sepharose column to obtain GST-HuCds1 (WT or K249R).
Production of Strep-HuCds1 in Insect Cells.
Recombinant baculovirus encoding HuCds1 was produced by using a MaxBac 2.0 transfection kit (Invitrogen). Recombinant baculovirus encoding GST-HuCds1 was generated by inserting an NdeI-XhoI fragment encoding GST-HuCds1 (see above) into the NheI-XhoI sites of pBlueBac 4.5 (Invitrogen) after the NdeI site was converted to an NheI site. To replace the GST domain with the Strep epitope tag, oligonucleotides CL55 (5′-CTAGCCACCATGGGCTGGTCCCACCC CCAGTTCGAAAAGG-3′) and CL56 (5′-GATCCCTTTTCGAACTGGGGGTGGGACC AGCCCATGGTGG-3′) were annealed and exchanged with the existing NheI-BamHI fragment encoding the GST domain to generate pBacStrepWT. Sf9 insect cells were cotransfected with pBacStrepWT and Bac-N-Blue DNA by using InsectinPlus insect cell-specific liposomes (Invitrogen). The recombinant virus was produced by the procedure accompanying the MaxBac 2.0 transfection kit (Invitrogen). Strep-HuCds1 protein from the virus-infected cells was then purified by using a StrepTactin Sepharose column (Genosys, The Woodlands, TX). Sf9 insect cells were maintained in Grace’s insect medium supplemented with 10% fetal bovine serum and 10 μg/ml gentamicin (TNM-FH medium) at 27°C.
HuCds1 Antibody Production.
Purified GST-HuCds1 (wild type) protein (100 μg) was injected into New Zealand white rabbits. Affinity-purified antibodies against HuCds1 were prepared by preclearing the sera with immobilized GST followed by binding to immobilized GST-HuCds1. After washing, the HuCds1-specific antibody was eluted with 0.1 M glycine (pH 3.0) and dialyzed against PBS. Peptide antibody against HuCds1 was raised in rabbits against peptide BC3 (AQPSTSRKRPREGE) which was derived from the carboxyl terminus of HuCds1. The antibody was then affinity purified.
HuCds1–Green Fluorescent Protein (GFP) Expression.
A 1.64-kb EcoRI-PvuII HuCds1 cDNA fragment which contains all of the translated region of HuCds1 except for 21 base pairs (encoding the last 7 amino acids) at the 3′ end was ligated into EcoRI-SmaI-digested pEGFP-NI (CLONTECH). The resulting construct joins the GFP-coding region to the 3′ end of the HuCds1 cDNA. HeLa cells were transfected with 10 μg of either the pHuCds1-GFP fusion construct or pEGFP-N1 by using the calcium phosphate transfection method. The GFP signal was visualized 48 hours after transfection with a fluorescent microscope and photographed (×40).
Phosphatase Treatment.
In a 50-μl reaction volume, 5 μl of λ-phosphatase buffer (500 mM Tris⋅HCl, pH 7.5/50 mM DTT), 5 μl of 20 mM MnCl2, and 1,200 units of λ-phosphatase (New England Biolabs) were incubated with 37 μl of cell lysate for 30–60 minutes at 30°C.
Mapping Studies.
GST-HuCds1 and GST-SpCds1 (25) were purified in soluble form from Schizosaccharomyces pombe and insect cells, respectively. GST-Cdc25C(200–256) (37) and GST-Cdc25C (38) were purified from bacteria. Kinase reactions were performed in 50-μl reaction volumes containing complete kinase buffer (50 mM Tris, pH 7.4/10 mM MgCl2/2 mM DTT/10 μM ATP) and 10 μCi [γ-32P]ATP (>3,000 Ci/mmol) and were incubated at 30°C for 25 minutes. Proteins were resolved on a 4–12% SDS gel and then transferred to nitrocellulose. The nitrocellulose containing radiolabeled protein was excised, blocked with 0.5% poly(vinylpyrrolidone) (PVP-40) in 100 mM acetic acid for 30 min at 37°C, washed, and digested with trypsin (Worthington) at a final concentration of 30 mg/ml in 0.1 M NH4HCO3 (pH 8.0). In some cases, further digestion was performed with 2 units of proline-specific endopeptidase (ICN) in 0.1 M sodium phosphate/5 mM EDTA (pH 7.4) at 37°C for 16 h. HPLC analysis and manual Edman degradation were performed as described (29).
RESULTS AND DISCUSSION
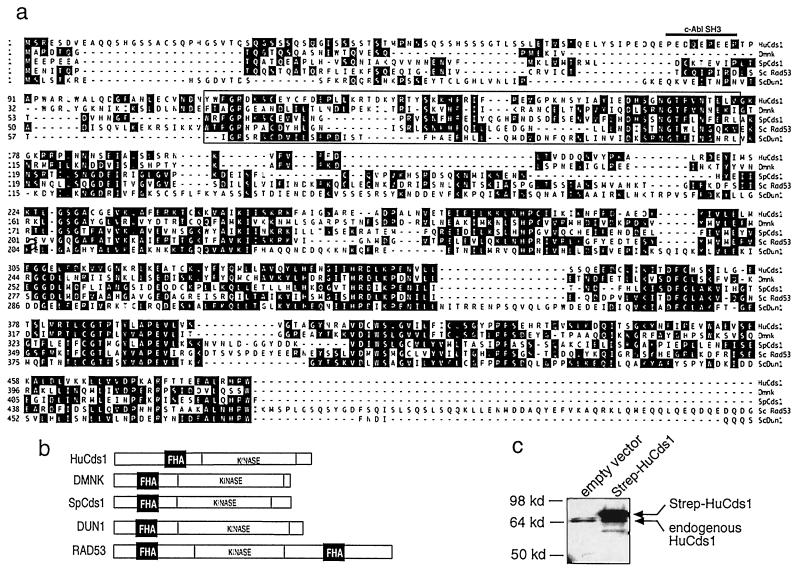
In Saccharomyces cerevisiae, the chromatin near the telomeres is in a condensed state like that of heterochromatic regions in higher eukaryotes and as a result, transcription of genes located near the telomeres is repressed (39). If the telomeric length or chromatin structure is perturbed, the genes located near the telomere are derepressed (36, 40–43). In a screen to identify human cDNAs that derepress the telomeric URA3 gene when overproduced in S. cerevisiae, we isolated a cDNA that possessed this property. Its sequence was closely related to SpCds1 (31), ScRad53 (44) and ScDun1 (23), a group of protein kinases that regulate the cellular response to DNA damage and replication blocks in yeast, as well as to the Drosophila melanogaster protein Dmnk (45) (Fig. 1a). The sequence of the longest cDNA (1,902 bp) predicted a translation product of 543 aa with a molecular mass of 61 kDa. The predicted protein is 34% identical to Dmnk, 28% identical to SpCds1, 23% identical to ScRad53, and 24% identical to ScDun1.
Figure 1.
Sequence of human Cds1. (a) Alignment of HuCds1 (up to amino acid 485) with Dmnk, SpCds1, ScRad53, and ScDun1. The FHA domain (boxed) and the c-Abl SH3 consensus site (thick line, for HuCds1 only) are indicated. The GenBank accession no. for HuCds1 is AF096279. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (b) Domain structures of HuCds1, Dmnk, SpCds1, ScRad53, and ScDun1. FHA domains (black box) and kinase domains are shown. (c) Cellular lysates prepared from 293 cells or 293 cells induced to express a Strep epitope-tagged form of HuCds1 (Strep-HuCds1, see Experimental Procedures) were resolved by using SDS/PAGE. HuCds1 protein was detected by immunoblotting with affinity-purified HuCds1 antibody.
The yeast kinases ScRad53, ScDun1 and SpCds1 contain a signature motif called the fork head-associated domain (FHA) that was first identified in several transcription factors with fork head DNA-binding domains (Fig. 1b)(46, 47). ScRad53 is the largest of the kinases and is unique in that it contains two FHA domains, one in the amino-terminal region (FHA1) and one in the carboxyl-terminal region (FHA2). ScRad53 integrates signals from some forms of DNA damage by binding to phosphorylated ScRad9 through its FHA2 domain (48). However, this FHA domain is not essential for the response of ScRad53 to replication blocks. Instead, deletion of the FHA1 domain of ScRad53p confers sensitivity both to HU and to UV light (49). The newly identified human kinase contains a single amino-terminal FHA domain that may function to integrate signals emanating from damaged and/or unreplicated DNA. Because of its size, sequence, and structural similarity to SpCds1, we have named this kinase HuCds1. A feature that is unique to HuCds1 is the presence of a c-Abl SH3-consensus binding sequence (PXXXXPXXP) upstream of the FHA domain (50). The c-Abl tyrosine kinase is a downstream target of phosphorylation and activation by ATM in the cellular response to ionizing radiation (51, 52). Because c-Abl is activated and HuCds1 is modified in response to ionizing radiation (see Fig. 3a), there might be a physical and functional link between HuCds1 and c-Abl in response to DNA damage.
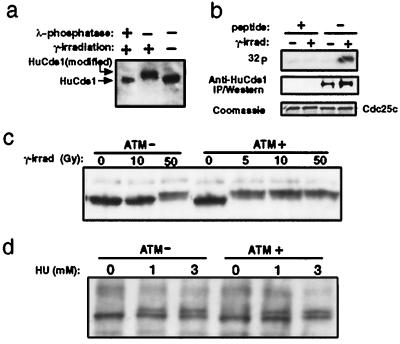
Figure 3.
HuCds1 is downstream of ATM in the cellular response to DNA damage but not replication blocks. (a) HeLa cells were mock-irradiated or exposed to 20 Gy or 100 Gy (shown) γ-irradiation. After 1 hour, cell extracts were prepared and treated with buffer containing λ-phosphatase or buffer alone. HuCds1 protein was detected by immunoblotting with affinity-purified HuCds1 antibody after SDS/PAGE. (b) HeLa cells were mock-irradiated or irradiated as described above. HuCds1 was immunoprecipitated with BC3 peptide-specific antibody in the presence (peptide: +) or absence (peptide: −) of competing BC3 peptide. Kinase assays were performed in vitro in the presence of GST-hCdc25C(200–256). 32P incorporation into GST-hCdc25C(200–256) was detected by using autoradiography. Levels of HuCds1 in the immunoprecipitates were detected by immunoblotting, and levels of GST-hCdc25C(200–256) were detected by Coomassie blue staining. (c) ATM (+) and ATM (−) cells were exposed to 5 to 50 Gy γ-irradiation. One hour after irradiation, cell extracts were prepared and HuCds1 was detected by immunoblotting with affinity-purified HuCds1 antibody. (d) ATM (+) and ATM (−) cells were exposed to HU (0, 1, and 3 mM) for 27 hours. HuCds1 protein was detected by immunoblotting as above.
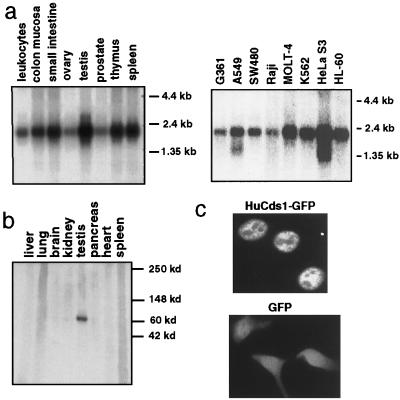
Affinity-purified antibodies to HuCds1 recognized an endogenous 65-kDa protein in 293 cells and a 65-kDa protein in cells transfected with a plasmid encoding untagged HuCds1 (Fig. 1c and data not shown), suggesting that the HuCds1 cDNA contains the full open reading frame. Cells induced to express a Strep-tagged (53) version of HuCds1, which has 19 additional amino acids, produced a protein of ≈68 kDa (Fig. 1c). Northern blot analyses demonstrated that HuCds1 mRNA is ubiquitously expressed in human tissues and in all cells lines tested (Fig. 2a). In contrast, HuCds1 protein was detected only in testis when several human tissues were analyzed by immunoblotting (Fig. 2b). This indicates that HuCds1 mRNA is subjected to tissue-specific translation or that the HuCds1 protein is unstable in other tissues. The presence of Cds1 in testis suggests a meiotic role for the human kinase. Of interest, many of the yeast checkpoint kinases have been shown to have a meiotic function (54–57). Furthermore, the Drosophila Dmnk protein kinase is expressed exclusively in germ cell nuclei (45), and the human ATM, ATR, and Chk1 kinases have been localized to meiotic chromosomes (58, 59). A fusion protein between HuCds1 and GFP localized exclusively to the nucleus of transfected HeLa cells, suggesting that HuCds1 is a nuclear kinase in human cells (Fig. 2c).
Figure 2.
Expression of HuCds1 mRNA and protein. (a) Northern blots containing polyadenylated RNA (2 μg) from the indicated tissues (Left) and cell lines (Right) were probed with HuCds1 cDNA. (b) Cell extracts from different human tissues (80 μg each lane) were electrophoresed on an SDS gel (4–20%), transferred to a nitrocellulose filter, and immunoblotted with affinity-purified rabbit polyclonal antibody raised against HuCds1. (c) Plasmids expressing GFP fused to the carboxyl terminus of HuCds1 (Upper) or GFP alone (Lower) were transiently transfected into HeLa cells by the calcium phosphate method and 48 hours later, the GFP signal was detected and photographed (×40).
ScRad53, ScDun1, and SpCds1 are phosphorylated in response to DNA damage and replication blocks in yeast (11, 19, 23, 24). To determine whether HuCds1 was similarly modified, we examined the electrophoretic mobility of HuCds1 in response to ionizing radiation (Fig. 3a). When HeLa (Fig. 3a) and MCF7 (data not shown) cells were γ-irradiated, the electrophoretic mobility of HuCds1 decreased. When extracts of irradiated cells were treated with phosphatase, HuCds1 mobility reverted to that seen in unirradiated cell extracts, demonstrating that the mobility shift was due to phosphorylation. The change in the electrophoretic mobility of HuCds1 after DNA damage occurred in all phases of the cell cycle (data not shown) unlike that of SpCds1, which is S phase-specific (24). Kinase assays were performed to determine the effects of ionizing radiation on the enzymatic activity of HuCds1 (Fig. 3b). A 56-aa region of human Cdc25C containing Ser-216 was used as a substrate for HuCds1 in vitro (see Fig. 4). The kinase activity of HuCds1 was found to increase by 5- to 6-fold after γ-irradiation (Fig. 3b), suggesting that phosphorylation stimulates the kinase activity of HuCds1. Phosphoamino acid analysis demonstrated that HuCds1 phosphorylated itself, HuCdc25C, and acid-denatured enolase on serine residues and to a lesser extent on threonine residues. Phosphotyrosine was not detected in these assays (data not shown).
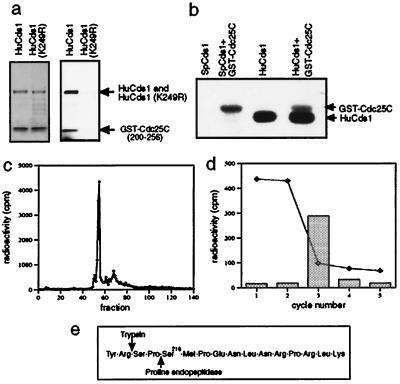
Figure 4.
HuCds1 phosphorylates Cdc25C on Ser-216, the 14-3-3 binding site. (a) Recombinant GST-HuCds1 or catalytically inactive GST-HuCds1 (K249R) were incubated with GST-hCdc25C (200–256), and kinase assays were performed in vitro. After electrophoresis, proteins were visualized by Coomassie blue staining (Left) followed by autoradiography (Right). (b) GST-SpCds1 and Strep-HuCds1 were tested for their ability to autophosphorylate or to phosphorylate full-length GST-Cdc25C by performing kinase assays in vitro. Radiolabeled proteins were resolved on a 7% gel and visualized by autoradiography. (c) Radiolabeled GST-Cdc25C(200–256) was digested with trypsin and the tryptic peptides were resolved by reverse-phase HPLC. Column fractions were collected and monitored for the presence of radioactivity. (d) Manual Edman degradation of tryptic phosphopeptide present in fraction 54. The dotted line indicates radioactivity remaining bound to the sequencing membrane at the end of each cycle, and bars represent radioactivity released from the membrane. (e) Amino acids inclusive of and surrounding Ser-216 showing amino-terminal trypsin and proline endopeptidase cleavage sites.
ScRad53, ScDun1, and SpCds1 function downstream of the PI-3-like kinases ScMec1, ScTel1, and SpRad3 in yeast (11, 19–21). The mammalian PI-3-like kinase ATM is an essential component of the DNA damage checkpoint (15–17). To determine whether the DNA damage-induced phosphorylation of HuCds1 required functional ATM, immortalized fibroblasts derived from an AT patient, stably transfected with either the ATM expression vector [ATM (+)] or an empty vector [ATM (−)], were γ-irradiated (60). The electrophoretic mobility of HuCds1 was then monitored by immunoblotting (Fig. 3c). In ATM (+) cells, the mobility of HuCds1 was retarded in response to 5 Gy irradiation, whereas in ATM (−) cells, a similar change in mobility was not seen until 50 Gy irradiation. The observation that HuCds1 is modified with 50 Gy irradiation in ATM− cells suggests that in the presence of extensive DNA damage, other pathways, perhaps involving ATR or DNA-dependent protein kinase, may activate HuCds1 (61, 62).
We next examined whether HuCds1 was also modified on activation of the DNA replication checkpoint (Fig. 3d). ATM (+) and ATM (−) cells were incubated with HU, and the electrophoretic mobility of HuCds1 was monitored by immunoblotting. In contrast to the ATM dependence observed for γ-irradiation, HuCds1 was modified by HU treatment in an ATM-independent manner. Thus, HuCds1 lies downstream of ATM in the checkpoint response to DNA damage but not to replication blocks. ATR, rather than ATM, may function upstream of HuCds1 during a replication block. It is also possible that ATM and ATR have overlapping functions in the replication checkpoint.
In fission yeast, SpCds1 and SpChk1 phosphorylate Cdc25 within 14-3-3 binding sites. This is required for a wild-type response to unreplicated DNA (25). In addition, both the human and fission yeast Chk1 kinases phosphorylate human Cdc25C on Ser-216, the 14-3-3 binding site (29, 34). The Cdc25C–14-3-3 interaction is required for a normal checkpoint response in both human and fission yeast cells (25, 29). We therefore tested whether the fission yeast and human Cds1 kinases would phosphorylate human Cdc25C in vitro. GST-HuCds1, but not a catalytically inactive mutant of GST-HuCds1 (K249R), phosphorylated a 56-aa region of human Cdc25C, containing Ser-216 (Fig. 4a). Both SpCds1 and HuCds1 kinases also phosphorylated full-length Cdc25C fused to GST (Fig. 4b). Trypsin digestion followed by reverse-phase HPLC analysis gave rise to a predominant phosphopeptide that eluted in fraction 54 for both GST-Cdc25(200–256) (Fig. 4c) and full-length Cdc25C (data not shown). Sequencing of the tryptic fragment gave rise to a cycle 3 release (Fig. 4d). This analysis unequivocally identified Ser-216 as the site of Cdc25C phosphorylation in vitro and suggests that one function of HuCds1 may be to regulate the Cdc25C–14-3-3 interaction in human cells as does SpCds1 in fission yeast (25). Ser-216 phosphorylation and 14-3-3 binding do not detectably alter the activity of Cdc25C but rather are proposed to functionally sequester Cdc25C and thereby prevent activation of Cdc2 (29).
The identification and characterization of HuCds1 suggest that cell cycle checkpoints and DNA damage and replication block responses are conserved from yeast to man (Fig. 5). Our studies place HuCds1 downstream of ATM in the cellular response to DNA damage, but of interest, we isolated HuCds1 based on its ability to derepress telomeric transcription, suggesting a telomeric function for HuCds1 as well. The fact that HuCds1 is also modified in response to DNA replication blocks suggests a possible role in the DNA replication checkpoint. In addition, like its fission yeast counterpart, one function of HuCds1 may be to regulate the interactions between Cdc25 and 14-3-3 proteins to induce cell cycle arrest (25, 29). Finally, many of the conserved checkpoint kinases have either been demonstrated or proposed to have roles in meiotic recombination based on genetic analysis or localization studies (54–59). Our finding that HuCds1 is primarily localized in adult testis suggests a role for HuCds1 in regulating meiosis in humans. Taken together, these findings underscore the conservation of structure and function of components of the signaling cascades that regulate cellular responses to DNA damage and replication blocks in eukaryotic organisms. The finding that loss of ATM leads to tumorigenesis emphasizes the importance of checkpoints in maintaining organismal homeostasis. We expect future studies of HuCds1 to further our understanding of human checkpoints.
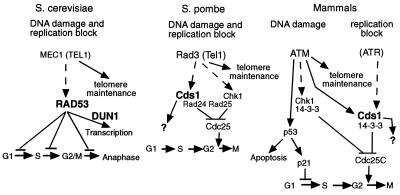
Figure 5.
Evolutionary conservation of the DNA damage and the replication checkpoints. In mammalian systems, the DNA damage pathway reaches HuCds1 through ATM, but the DNA replication block pathway reaches HuCds1 through another kinase, possibly ATR (see text for details). Because the proposed role of ATR in the replication block pathway has not been demonstrated, it is tentatively placed in parentheses. With severe DNA damage, PI-3 kinases such as DNA-PK or ATR may activate HuCds1 as well (see text for details). Kinases containing FHA domains are indicated with large bold characters.
Acknowledgments
We thank Yosef Shiloh for the ATM (− and +) cells, Dan Gottschling for S. cerevisiae strain UCC3505, and Dominique Griffith for assistance in construction of plasmids. Janis Gales and Mary Stephenson are thanked for excellent technical assistance and Yan Zeng is thanked for assistance with the yeast studies. This work was supported in part by the National Institutes of Health. H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HU
hydroxyurea
- AT
ataxia telangiectasia
- ATM
AT mutated
- ATR
AT- and rad3+-related
- FHA
fork head associated
- PI
phosphoinositide
- GFP
green fluorescent protein
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF096279).
References
- 1.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 3.Kato R, Ogawa H. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez G, Yucel J, Rowley R, Subramani S. Proc Natl Acad Sci USA. 1992;89:4952–4956. doi: 10.1073/pnas.89.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seaton B L, Yucel J, Sunnerhagen P, Subramani S. Gene. 1992;119:83–89. doi: 10.1016/0378-1119(92)90069-2. [DOI] [PubMed] [Google Scholar]
- 6.Enoch T, Carr A M, Nurse P. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 7.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 8.Lustig A J, Petes T D. Proc Natl Acad Sci USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 10.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 12.Naito T, Matsuura A, Ishikawa F. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 13.Savitsky K, Sfez S, Tagle D A, Ziv Y, Sartiel A, Collins F S, Shiloh Y, Rotman G. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 14.Cimprich K A, Shin T B, Keith C T, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 16.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 17.Jeggo P A, Carr A M, Lehmann A R. Trends Genet. 1998;14:312–316. doi: 10.1016/s0168-9525(98)01511-x. [DOI] [PubMed] [Google Scholar]
- 18.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 20.Rhind N, Russell P. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boddy M N, Furnari B, Mondesert O, Russell P. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 22.Sidorova J M, Breeden L L. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, Elledge S J. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay H D, Griffiths D J, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Nature (London) 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 26.Enoch T, Nurse P. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- 27.Jin P, Gu Y, Morgan D O. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye X S, Fincher R R, Tang A, O’Donnell K, Osmani S A. EMBO J. 1996;15:3599–3610. [PMC free article] [PubMed] [Google Scholar]
- 29.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai A, Yakowec P S, Dunphy W G. Mol Biol Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami H, Okayama H. Nature (London) 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 32.Walworth N, Davey S, Beach D. Nature (London) 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 33.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Lehmann A R, Carr A M. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 35.Kumagai A, Guo Z, Emami K H, Wang S X, Dunphy W G. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer M S, Gottschling D E. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 37.Ogg S, Gabrielli B, Piwnica-Worms H. J Biol Chem. 1994;269:30461–30469. [PubMed] [Google Scholar]
- 38.Peng C-Y, Graves P R, Ogg S, Thoma R S, Byrnes M J, Wu Z, Stephenson M, Piwnica-Worms H. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 39.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 40.Runge K W, Zakian V A. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aparicio O M, Billington B L, Gottschling D E. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 42.Kyrion G, Liu K, Liu C, Lustig A J. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J S, Ling X, Grunstein M. Nature (London) 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 44.Stern D F, Zheng P, Beidler D R, Zerillo C. Mol Cell Biol. 1991;11:987–1001. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oishi I, Sugiyama S, Otani H, Yamamura H, Nishida Y, Minami Y. Mech Dev. 1998;71:49–63. doi: 10.1016/s0925-4773(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 46.Weigel D, Jackle H. Cell. 1990;63:455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann K, Bucher P. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 48.Sun Z, Hsiao J, Fay D S, Stern D F. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 49.Fay D S, Sun Z, Stern D F. Curr Genet. 1997;31:97–105. doi: 10.1007/s002940050181. [DOI] [PubMed] [Google Scholar]
- 50.Feller S M, Ren R, Hanafusa H, Baltimore D. Trends Biochem Sci. 1994;19:453–458. doi: 10.1016/0968-0004(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 51.Baskaran R, Wood L D, Whitaker L L, Canman C E, Morgan S E, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M B, Wang J Y. Nature (London) 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 52.Shafman T, Khanna K K, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Nature (London) 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt T G, Koepke J, Frank R, Skerra A. J Mol Biol. 1996;255:753–766. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- 54.Weinert T. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 55.Lydall D, Nikolsky Y, Bishop D K, Weinert T. Nature (London) 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 56.McKim K S, Jang J K, Theurkauf W E, Hawley R S. Nature (London) 1993;362:364–366. doi: 10.1038/362364a0. [DOI] [PubMed] [Google Scholar]
- 57.Weber L, Byers B. Genetics. 1992;131:55–63. doi: 10.1093/genetics/131.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keegan K S, Holtzman D A, Plug A W, Christenson E R, Brainerd E E, Flaggs G, Bentley N J, Taylor E M, Meyn M S, Moss S B, et al. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 59.Flaggs G, Plug A W, Dunks K M, Mundt K E, Ford J C, Quiggle M R, Taylor E M, Westphal C H, Ashley T, Hoekstra M F, Carr A M. Curr Biol. 1997;7:977–986. doi: 10.1016/s0960-9822(06)00417-9. [DOI] [PubMed] [Google Scholar]
- 60.Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen T J, Tsarfati I, Shiloh Y. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
- 61.Hartley K O, Gell D, Smith G C, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 62.Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]