Abstract
The generation of transport vesicles at the endoplasmic reticulum (ER) depends on cytosolic proteins, which, in the form of subcomplexes (Sec23p/Sec24p; Sec13p/Sec31p) are recruited to the ER membrane by GTP-bound Sar1p and form the coat protein complex II (COPII). Using affinity chromatography and two-hybrid analyses, we found that the essential COPII component Sec24p, but not Sec23p, binds to the cis-Golgi syntaxin Sed5p. Sec24p/Sed5p interaction in vitro was not dependent on the presence of [Sar1p⋅GTP]. The binding of Sec24p to Sed5p is specific; none of the other seven yeast syntaxins bound to this COPII component. Whereas the interaction site of Sec23p is within the N-terminal half of the 926-aa-long Sec24p (amino acid residues 56–549), Sed5p binds to the N- and C-terminal halves of the protein. Destruction by mutagenesis of a potential zinc finger within the N-terminal half of Sec24p led to a nonfunctional protein that was still able to bind Sec23p and Sed5p. Sec24p/Sed5p binding might be relevant for cargo selection during transport-vesicle formation and/or for vesicle targeting to the cis-Golgi.
Proteins passing the membrane-enclosed compartments of the biosynthetic and the endocytic pathway are transported in vesicular intermediates. Budding, targeting, and fusion of the vesicular carriers require complex machines (1) made up of numerous proteins, many of which are highly conserved from yeast to man (for reviews, see refs. 2–5).
In yeast, budding of endoplasmic reticulum (ER)-derived transport vesicles and the selection of soluble and membrane-bound cargo molecules destined to reach the Golgi complex require the coat protein complex II (COPII), which is assembled from the subcomplexes Sec23p/Sec24p and Sec13p/Sec31p (6, 7). Studies in vitro have shown that the small GTPase Sar1p in its GTP-bound conformation first recruits the Sec23p/Sec24p subcomplex to the ER membrane, and this is followed by the association of the Sec13p/Sec31p subcomplex and vesicle fission (7, 8). The vesicular type II integral membrane SNAP receptors (v-SNAREs Bet1p, Bos1p, and Sec22p) involved in anterograde protein traffic are specifically concentrated early during vesicle formation through their interaction with [Sar1p⋅GTP] and the Sec23p/Sec24p complex (7, 9). With purified proteins it was shown that the cytoplasmic domains of Bet1p and Bos1p bind directly and in a [Sar1p⋅GTP]-dependent fashion to the Sec23p/Sec24p complex, but not to either Sec23p or Sec24p alone (10).
It is thought (4, 6) that the vesicular coat is shed soon after budding of vesicles from the ER to expose the v-SNAREs for the interaction with their Golgi target membrane receptor (t-SNARE), the syntaxin-related Sed5 protein (11). Although at steady state, Sed5p appears to be localized primarily to an early Golgi compartment, recent studies show that this t-SNARE cycles between the ER and the Golgi (12). The same holds true for the mammalian Sed5p homologue syntaxin-5 (13). Therefore, as with the v-SNAREs, Sed5p is expected to directly or indirectly interact with COPII to enter vesicles at the ER membrane for anterograde traffic to the Golgi.
Here we report that in contrast to what has been described for the v-SNAREs Bet1p and Bos1p, the t-SNARE Sed5p binds to Sec24p alone and, apparently, in a Sar1p-independent fashion.
METHODS
Yeast Strains and Growth Conditions.
Yeast strains used in this study are listed in Table 1. They were constructed by standard methods and grown in standard yeast extract/peptone/dextrose or minimal medium as described (14). Yeast transformations were carried out by using a modified lithium acetate method (15).
Table 1.
Yeast strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| MSUC1A3D | MATa/α ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 LYS2/lys2 ade8/ADE8 | This laboratory |
| MSUCIA | MATa ura3 leu2 his3 trp1 ade8 | This laboratory |
| MSUC3D | MATα ura3 leu2 his3 trp1 lys2 | This laboratory |
| RPY5 | MATa/α ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 LYS2/lys2 ade8/ADE8 SEC24/sec24∷KanMX4 | This study |
| RPY18 | MATa sec24-11 ura3 leu2 his3 trpI ade8 | This study |
| RH227-3A | MATa sec23-1 ura3 his4 leu2 | H. Riezman (Biocenter, Basel) |
| CI3-ABYS-86 | MATα ura3-Δ5 leu2-3,112 his3 pra1-1 prb-1 prc1-1 cps1-3 canR | D. H. Wolf (University of Stuttgart) |
| Y190 | MATa gal4 gal80 his3 trpI ura3 leu2 ade2 URA3∷GAL →lacZ LYS2∷GAL → HIS cyhr | S. J. Elledge (Baylor College of Medicine, Houston) |
Plasmids.
Plasmids used in this study were constructed as follows. pUC19-SEC24 contains a 4.5-kb XbaI/HindIII SEC24 fragment isolated from a subgenomic library. pRS316-SEC24, pRS315-SEC24, and pRS13-SEC24 carry the entire SEC24 gene as a 3.6-kb of Bsu36I/HindIII fragment. To produce glutathione S-transferase (GST)-Sec24p, the BamHI/HindIII fragment of SEC24 first was inserted into pEG-KT (16). The missing N-terminal 32 aa were compensated for by inserting two synthetic oligonucleotides, thus producing pEG-KT-SEC24 with the entire SEC24 coding region fused to GST. pEG-KT-SEC23 contained an NcoI/HindIII SEC23 fragment from pTYY121 (6) on pEG-KT. The coding regions of Sed5p (amino acids 1–319), Ufe1p (1–326), Vam3p (1–261), Sso1p (1–264), Sso2p (1–267), Tlg1p (1–204), and Tlg2p (1–317) lacking the C-terminal transmembrane domains were amplified by PCR as NcoI-XhoI fragments (except for Sso1, where an XbaI-XhoI fragment was made) and expressed as GST fusion proteins from pGEX-TT (Amersham Pharmacia). The N- and C-terminal Sec24p fragments were expressed as GST fusion proteins from pEG-KT, a vector allowing GST-fusion protein expression in yeast. GST-Pep12 and GST-Sly1 fusions were constructed as described previously (16, 17). His6-Sec23p was expressed from pQE-30 (Qiagen, Chatsworth, CA) containing the whole coding region of SEC23. GST-Syntaxin-5 was expressed in Escherichia coli from pGEX-KG-Syntaxin-5, kindly provided by R. Jahn (Max Planck Institute for Biophysical Chemistry). For high expression in yeast, the genes encoding Sec24 wild-type and mutant proteins and the Sec23p were cloned into the 2 μ-based multicopy vector pRS326 (18).
The plasmids used for yeast two-hybrid assays were the following. pAS2-SEC24 contained the coding region of the GAL4 DNA-binding domain fused in-frame with the entire SEC24-coding region and 326 bp of the 3′ flanking region. SED5 (codons 1–319) and UFE1 (codons 1–326) were cloned into two-hybrid vectors pAS2 and pACTII as NcoI-XhoI fragments. pACTII-SLY1 contained the entire SLY1 gene as an NcoI-XhoI fragment amplified by PCR. pACTII-SEC24N was created by cloning a 1.47-kb NcoI fragment of SEC24 (codons 56–549) into pACTII. pACTII-SEC24C was created by cloning into pACTII a 1.46-kb NcoI/XhoI fragment containing SEC24 (codons 550–926) and 326 bp of the 3′ flanking region.
Antibodies.
A fragment of Sec24p (amino acids 720–926) was expressed as a His6-tagged protein from pET19b (Novagen) and used for the generation of polyclonal antibodies in rabbits. Antibodies against Sed5p were produced and affinity-purified as described previously (16). Anti-Sec23p antibodies were a kind gift from R. Schekman (University of California, Berkeley).
Purification of Recombinant Proteins.
Protease-deficient yeast strain cl3-ABYS-86 (19) was transformed with pEG-KT-derived plasmids carrying different gene fusions. Transformants first were grown in PM-glucose medium (0.67% yeast nitrogen base with ammonium sulfate/0.5% peptone 140/2% glucose) (16) to midlog phase, and then shifted to PM-galactose medium and grown for an additional 8–10 h. Cells were broken with glass beads in lysis buffer (50 mM Tris⋅HCl, pH 7.5/150 mM KCl/2 mM EDTA/protease inhibitors cocktail). GST fusion proteins were purified from the 100,000 × g supernatant with glutathione-Sepharose 4B (Amersham Pharmacia), and beads were washed four times with lysis buffer containing 1 M KCl. Purified GST fusion proteins immobilized to glutathione-Sepharose were eluted with lysis buffer containing 20 mM reduced glutathione or used directly for protein-binding assays. GST fusion or His6-tagged yeast proteins were expressed in E. coli as described previously (16, 17).
In Vitro Mutagenesis.
Substitutions to serine of cysteine-231 or cysteine-231/-234 were generated by site-directed mutagenesis using the QuikChange kit (Stratagene). Two sets of oligonucleotides were used: C231S, 5′-GAAGATGGACTTATTGTCCGTTCTCGTCGTTGTCGT-3′ and 5′-ACGACAACGACGAGAACGGACAATAAGTCCATCTTC-3′, and C231S/C234S, 5′-GGACTTATTGTCCGTTCTCGTCGTTCTCGTTCTTACATGAAC-3′ and 5′-GTTCATGTAAGAACGAGAACG-ACGAGAACGGACAATAAGTCC-3′. The mutations were verified by DNA sequencing.
Affinity-Binding Assay.
E. coli or Saccharomyces cerevisiae strains expressing the desired recombinant proteins were lysed and proteins were solubilized in lysis buffer (25 mM Hepes⋅KOH, pH 7.0/150 mM KOAc/5 mM MgCl2/1 mM DDT/1 mM EDTA/1% Triton X-100/protease inhibitor mix). Proteins (250–500 μg) of the 100,000 × g supernatant were incubated at 4°C for 1 h in lysis buffer with GST or GST fusion proteins immobilized on glutathione-Sepharose 4B beads (30 μl). The beads were washed four times (1 ml each) using the same buffer. Proteins on beads were denatured by boiling in SDS buffer, separated by SDS/PAGE, and visualized by Coomassie blue staining or immunoblotting using the enhanced chemiluminescence system (Amersham Pharmacia).
Yeast Two-Hybrid Assays.
S. cerevisiae strain Y190 was transformed simultaneously with pAS2- and pACTII-derived constructs harboring the different gene fusions. Transformants were selected on synthetic complete (SC) medium lacking tryptophan and leucine. 5-Bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) filter lift assay for β-galactosidase activity was carried out by growing transformants at 30°C overnight on nylon filter discs (Schleicher & Schuell) on SC-selective medium, followed by cell lysis in liquid nitrogen and incubation at 30°C with Z buffer containing 0.30 mg/ml X-Gal. Quantitative β-galactosidase activity assays were performed by selecting three different colonies from each transformation, assaying in duplicate each colony essentially as described (20), except that the cells were permeabilized by freezing in liquid nitrogen, followed by quick thawing. Units of β-galactosidase are expressed as 1,000 × OD420/OD600 of the culture × culture volume (ml) × reaction time (min).
RESULTS
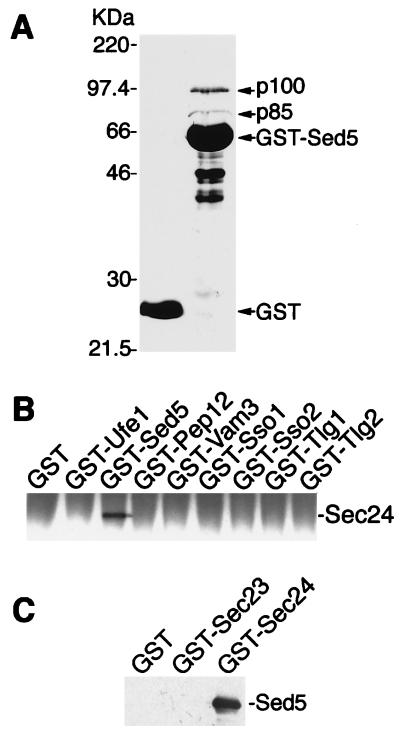
In studying the biochemical parameters of Sed5/Sly1 protein interaction (16), a GST-Sed5 fusion protein lacking the C-terminal membrane-spanning domain was immobilized on glutathione-Sepharose 4B and used as an affinity matrix with S. cerevisiae proteins solubilized in the presence of 2% Triton X-100. Two proteins larger than the 63-kDa fusion protein and exhibiting a molecular mass of about 85 and 100 kDa, respectively, were observed repeatedly to bind to GST-Sed5p but not to GST alone (Fig. 1A). After excision from polyacrylamide gels, both proteins were subjected to proteolytic digestion and peptide sequencing. From p85, six peptide sequences 6–16 aa in length were obtained that identified the protein as the COPII component Sec23p (21). Two peptide sequences were obtained from p100 (DHRASALNNL and DGEATGTIVLPQPINATSSL), identifying this protein as Sec24p (ORF YIL109C on chromosome 9). The data obtained suggested that Sec24p and/or Sec23p, the COPII proteins that are complexed largely in vivo (22), can bind to the cis-Golgi t-SNARE Sed5p.
Figure 1.
Specific binding of Sec24p to the syntaxin-5 homologue Sed5p. (A) Proteins from detergent-lysed yeast cells were incubated at 4°C for 2 h with GST alone (lane 1) or GST-Sed5 (lane 2) immobilized on glutathione-Sepharose 4B. Beads were washed three times, and the bound proteins were analyzed by SDS/PAGE and Coomassie blue staining. p85 and p100 were identified as Sec23p and Sec24p, respectively, after elution from a preparative gel, endoproteinase Asp-N digestion, and peptide sequencing. The positions of molecular mass markers are indicated to the left. (B) Triton X-100-solubilized proteins (250 μg) from protease-deficient yeast cells were incubated in 200 μl lysis buffer with 1 μM GST or GST fusion proteins previously immobilized on glutathione-Sepharose beads. The bound proteins were fractionated by SDS/PAGE and detected by immunoblotting with a polyclonal anti-Sec24 antibody. (C) GST-Sec24, GST-Sec23, or GST (0.5 μM) was immobilized on glutathione-Sepharose and, after washing with 1 M KCl, was incubated with 60 μg of E. coli-soluble proteins containing His6-Sed5 lacking its membrane anchor. Proteins bound to beads were fractionated by SDS/PAGE, followed by immunoblotting with anti-Sed5 antibodies.
To see whether the observed interaction of Sec24p and Sed5p was t-SNARE-specific, we constructed GST fusions with all known yeast syntaxins, the ER-localized Ufe1p (23), the plasma membrane t-SNAREs Sso1p and Sso2p (24), the prevacuolar Pep12p (25), the vacuolar Vam3p (26), and the syntaxins Tlg1p and Tlg2p, which appear to be localized to endosomal and/or the most distal Golgi compartments (27–29). Detergent-solubilized proteins from wild-type cells were incubated with Sepharose beads coupled with either GST or its fusions with the different t-SNAREs. As shown by immunoblot analysis using an anti-Sec24p antibody, Sec24p was retained by the GST-Sed5p matrix only (Fig. 1B). Under identical experimental conditions we did not observe binding of Sec24p to either the mammalian Sed5p-related syntaxin-5 (30) or the Sed5p-bound Sly1 protein (16, 31, 32).
To exclude the possibility that the specific binding of Sec24p to the t-SNARE Sed5p was mediated by Sec23p, GST and its fusions with either Sec24p or Sec23p were used in an affinity approach to bind Sed5p. For this experiment, GST and the fusion proteins were produced in yeast, extracted without the use of detergent, purified on glutathione-Sepharose 4B beads (Fig. 1C), and, after a wash with 1M KCl, incubated with bacterially produced, His6-tagged Sed5p lacking its C-terminal membrane-spanning domain. As can be seen in Fig. 1C, Sed5p was bound to Sec24p but not to Sec23p. Notably, Sed5p binding to Sec24p could be readily observed without the addition of Sar1p and/or GTP.
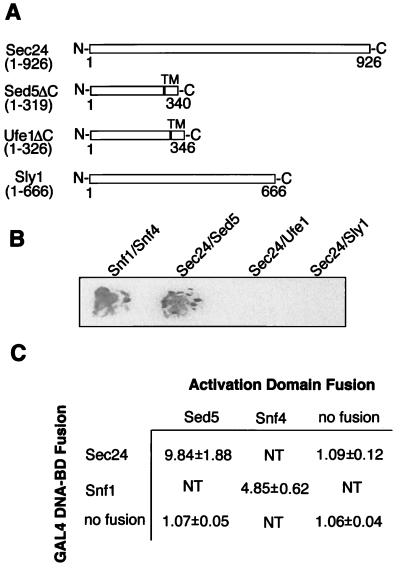
In another approach to show the direct and specific interaction of Sec24p and Sed5p, the yeast two-hybrid system (33) was used. The full-length Sec24 protein was fused to the Gal4 DNA-binding domain and probed for its interaction with either Sed5p, the ER-localized t-SNARE Ufe1p, or the Sed5p-bound Sly1p. The t-SNAREs lacked the C-terminal membrane anchors (Fig. 2A). As shown in Fig. 2 B and C, interaction of Sec24p in this in vivo system was observed only with Sed5p. The strength of the interaction resembled that of the control between the protein kinase Snf1p and its activating subunit, Snf4p. In line with the biochemical data described above, a two-hybrid interaction of Sed5p and Sec23p could not be observed (Fig. 3C).
Figure 2.
Sec24p and Sed5p interact in the two-hybrid system. (A) Schematic representation of the proteins fused to either the Gal4 DNA-binding domain (Sec24p) or the Gal4 transcription-activation domain (Sly1p or the t-SNAREs Sed5p and Ufe1p lacking the transmembrane domain, TM). (B) Colonies of yeast cells expressing the indicated Gal4-domain fusions were grown on nylon filters and subjected to the β-galactosidase color assay by using 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) as substrate. The interaction of the protein kinase Snf1 and its subunit Snf4 served as positive control. (C) Yeast cells expressing the indicated Gal4 fusion proteins were subjected to β-galactosidase activity measurement using O-nitrophenyl-β-d-galactoside as substrate. Numbers represent the mean values obtained with three individual transformants. NT, not tested.
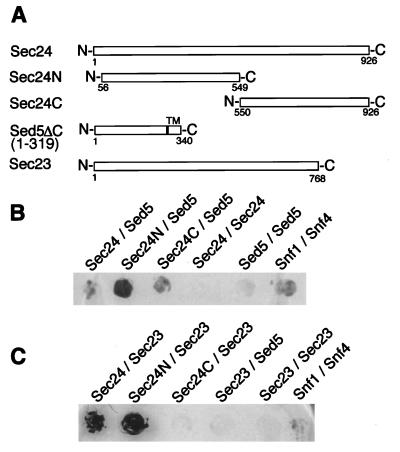
Figure 3.
The N-terminal and C-terminal regions of Sec24p bind Sed5p. The two-hybrid assays were performed as described in the legend to Fig. 2. (A) Schematic representation of the proteins used. (B) Interactions between Sed5p and the N- or C-terminal half of Sec24p. (C) Interactions between Sec23p and the N- or C-terminal half of Sec24p.
Previous two-hybrid analyses had shown that Sec24p exhibits a complex interaction pattern with the COPII components Sec23p and Sec31p and the ER-associated Sec16 protein (34, 35). In these studies it was shown that the binding site(s) for Sec23p lie within the N-terminal 666 aa of Sec24p. It was therefore of interest to determine the regions of Sec24p required for its binding to the t-SNARE Sed5p. Two Sec24p half-molecules comprising amino acid residues 56–549 (Sec24N) and 550–926 (Sec24C) were prepared for a two-hybrid study (Gal4 fusions) and for an affinity-binding analysis (GST fusions) to investigate their interaction with the Sed5 syntaxin. As can be seen in Fig. 3B, both the N-terminal and the C-terminal half of Sec24p interacted with Sed5p in a two-hybrid analysis. In contrast, only the N-terminal half of Sec24p interacted with Sec23p (Fig. 3C). The two-hybrid data were corroborated by affinity-binding experiments performed with matrix-bound GST-Sec24 fusion proteins. As can be seen in Fig. 4B, the full-length Sec24p as well as its N- and C-terminal halves bound His6-tagged Sed5p produced in E. coli, suggesting that the COPII component has at least two Sed5p-interaction sites. In contrast, bacterially produced Sec23p was retained by Sec24p and its N-terminal half but not by the C-terminal Sec24p fragment (Fig. 4C).
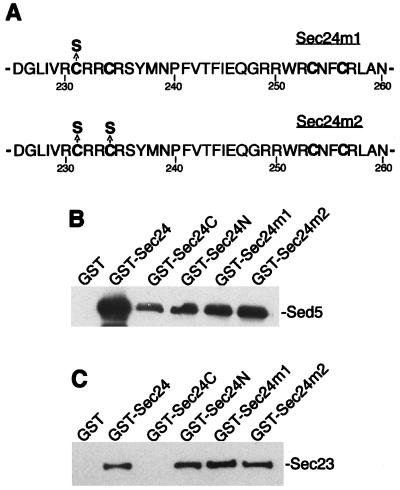
Figure 4.
Mutations in the putative zinc finger of Sec24p do not affect in vitro binding to Sed5p and Sec23p. The binding in vitro of Sed5p and Sec23p to GST and GST fusion proteins was performed as described in the legend to Fig. 1. (A) Amino acid substitutions in the putative zinc-finger of Sec24p. (B) Purified GST, GST-Sec24 (amino acids 1–926), GST-Sec24C (amino acids 550–926), GST-Sec24N (amino acids 56–549), GST-Sec24 m1 (amino acids 56–549), and GST-Sec24 m2 (56–549) (0.5 μM of each) were immobilized on glutathione-Sepharose beads and incubated with protein extract from E. coli expressing either His6-Sed5 (B) or His6-Sec23 (C). The bound proteins were separated by SDS/PAGE, followed by immunoblotting with antibody against Sed5p (B) or Sec23p (C).
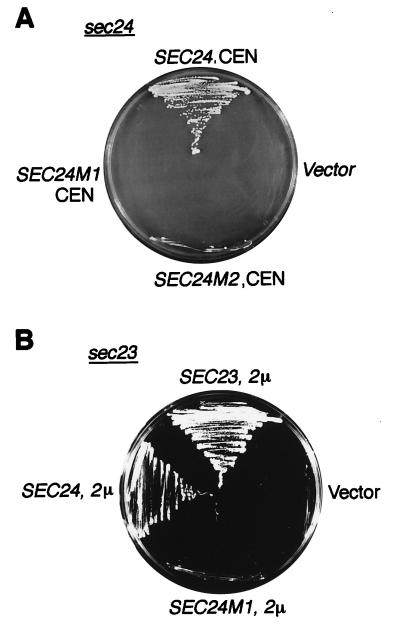
Sec24p is an essential protein, and it harbors within its N-terminal region a domain with a zinc finger-like motif (CysX2CysX18CysX2Cys) (Fig. 4A). This potential Zn2+-binding region could be of importance for stabilizing the structure of Sec24p or for serving as an interaction site with other proteins. To inquire into the possibility that the presumptive Zn2+-binding segment of Sec24p might be required for Sed5p or Sec23p interaction, two mutant proteins were created by substituting with serine either one (C231S; Sec24 m1) or two cysteines (C231S/C234S; Sec24 m2). The mutant genes were used first to replace one SEC24 wild-type allele in diploid strains (RPY5). After sporulation and tetrad dissection of the respective transformants, it was found that haploids carrying either of the two mutant sec24 alleles were inviable. The loss of function of the two mutant Sec24 proteins also was demonstrated by their inability to rescue a sec24 mutant from its growth defect at high temperature (35°C) (Fig. 5A). The mutant Sec24 proteins also lost the ability of wild-type Sec24p to suppress, at high intracellular concentration, the growth defect of a temperature-sensitive sec23 mutant. This clearly showed that the substitution of only one of the potential Zn2+-coordinating cysteine residues results in a complete loss of Sec24p function.
Figure 5.
Loss of function of Sec24p mutated in the putative zinc finger. The temperature-sensitive mutants sec24–11 (A) and sec23–1 (B) were transformed with the multicopy 2 μ-based vector pRS326 (2 μ) or the single copy, centromere-containing vector pRS316 (CEN) carrying the genes indicated. Transformants were grown on agar plates in selective media for 3 days at 35°C (nonpermissive temperature for both mutants). Only wild-type genes could rescue the mutants.
The two Sec24 mutant proteins then were tested for their interaction in vitro (affinity chromatography) with the syntaxin Sed5p and with the COPII subunit companion Sec23p. Surprisingly, the destruction of the presumptive zinc finger did not interfere with Sed5p or Sec23p binding of the otherwise nonfunctional mutant Sec24 COPII proteins (Fig. 4).
DISCUSSION
By means of affinity chromatography and two-hybrid analyses we have shown here that the Golgi t-SNARE Sed5p physically interacts with the vesicle coat protein Sec24p in vitro and in vivo. This interaction is specific considering none of the other known yeast syntaxins could be observed to bind to Sec24p under identical experimental conditions. We also excluded the possibility that Sec24p binding to Sed5p might be mediated by Sec23p, the COPII component bound to Sec24p in vivo (22), or by the Sec1p-related Sly1 protein that is tightly bound to Sed5p on Golgi membranes (16, 32). Binding to these proteins could not be observed either by affinity chromatography or by two-hybrid assays.
The Golgi syntaxin Sed5p recently has been shown to cycle through the ER as the v-SNAREs with which it associates in the preparation for vesicle fusion (12). Because the Sec23p/Sec24p COPII subcomplex participates in the recruitment of the v-SNAREs to the site of vesicle formation at the ER membrane (7), it appears likely that the binding of Sec24p to Sed5p serves a function in the process of cargo selection. Notably, in affinity-binding experiments similar to ours, COPII binding to v-SNAREs was not detected with either Sec23p or Sec24p alone but only with the complex of these two proteins and the additional presence of the GTP-bound Sar1 GTPase (10). Although in the study mentioned it could not be decided to which COPII component the v-SNAREs Bet1p or Bos1p bind, it now appears most likely that the v-SNAREs, as the t-SNARE Sed5p, directly interact with Sec24p. The failure to detect binding of the ER → Golgi v-SNAREs Sec22p and Ykt6p to the Sec23p/Sec24p COPII subcomplexes in vitro has been attributed to either the weakness of these interactions or their binding to other cargo molecules (7, 10). Given the apparent strength of the Sec24p/Sed5p binding that we have observed and the previous findings that Sec22p (36) and Ykt6p (37) bind to the cis-Golgi t-SNARE Sed5p, the Golgi syntaxin could well serve a function to recruit these v-SNAREs, in addition to Sec24p, to the site of vesicle generation at the ER membrane. This possible scenario, for instance, could be addressed experimentally by using the in vitro vesicle budding system with purified COPII and SNARE components (7, 10).
In contrast to similar studies aimed at analyzing COPII/v-SNARE interactions in vitro (10), the binding of Sec24p to Sed5p in vitro was not dependent on [Sar1p⋅GTP]. Binding of the two proteins was observed without the addition of the GTPase or GTP. Because the affinity matrix with the bound GST-Sec24p fusion proteins produced in yeast was washed with 1 M salt before adding E. coli-produced Sed5p, it is most unlikely that Sar1p was present in appreciable amounts during the binding reactions. Furthermore, Sar1p is not a nuclear protein and is unlikely to mediate the observed Sec24p/Sed5p two-hybrid interactions that take place in the nucleus. Binding of Sec24p to Sed5p also was independent of Sec23p.
In previous studies it was shown that the 103.5-kDa Sec24 protein has binding sites for the COPII components Sec23p and Sec31p and for the ER-associated Sec16 protein that appears to have a function in organizing the vesicle-budding site (35). The binding region for Sec23p was found to reside within the N-terminal 666 aa of Sec24p. In our study, we could delineate further the Sec23p-binding site to amino acid residues 56–549 of Sec24p. Importantly, we observed binding of Sed5p to the same N-terminal region and, in addition, to the C-terminal half of Sec24p. This indicates that Sec24p furnishes two distinct binding sites for the cytoplasmic domain of the cis-Golgi syntaxin Sed5p. They might be recognized by distinct sequence regions of the t-SNARE. So far, we have not attempted to delineate further the binding sites on either Sec24p or Sed5p. We have, however, excluded the possibility that a potential zinc finger within the N-terminal half of Sec24p is required for Sed5p (or Sec23p) binding. Although the exchange in this putative zinc finger of a single cysteine rendered Sec24p nonfunctional, Sed5p or Sec23p binding in vitro was undisturbed.
Although the binding of Sed5p to the Sec24 vesicle coat protein is likely to be of importance for cargo selection during vesicle formation at the ER membrane, ongoing investigations from this laboratory indicate that Sec24p might have an additional function at a later step in ER-to-Golgi trafficking. In this context, one might also envisage the possibility that an interaction of Golgi membrane-bound Sed5p with the transport vesicle-associated Sec24p initiates COPII removal from the vesicular membrane. A dual role in protein trafficking of a yeast COPII component would not be without precedent because Sec13p, in addition to acting in vesicle budding at the ER as part of the second COPII subcomplex (Sec13p/Sec31p), appears also to have a function in the sorting of newly synthesized plasma membrane proteins at a late Golgi compartment (38).
Acknowledgments
We thank K. Weber and U. Plessmann for help with the protein sequence analysis, R. Schekman for providing anti-Sec23p antibodies, C. Barlowe for plasmid pTYY121, S. J. Elledge for two-hybrid plasmids, and R. Jahn for a plasmid harboring the GST-Syntaxin-5 fusion. We thank P. Mienkus, U. Welscher-Altschäffel, and H. Behr for technical assistance and I. Balshüsemann for secretarial help. This work was supported by the Max Planck Society and by grants to D.G. from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
ABBREVIATIONS
- ER
endoplasmic reticulum
- COPII
coat protein complex II
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Bennett M K, Scheller R H. Proc Natl Acad Sci USA. 1993;90:2559–2563. doi: 10.1073/pnas.90.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 4.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 5.Pfeffer S R. Annu Rev Cell Dev Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- 6.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 7.Kuehn M J, Herrmann J M, Schekman R. Nature (London) 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 8.Matsuoka K, Orci L, Amherdt M, Bednarek S Y, Hamamoto S, Schekman R, Yeung T. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka K, Morimitsu Y, Uchida K, Schekman R. Mol Cell. 1998;2:703–708. doi: 10.1016/s1097-2765(00)80168-9. [DOI] [PubMed] [Google Scholar]
- 10.Springer S, Schekman R. Science. 1998;281:698–700. doi: 10.1126/science.281.5377.698. [DOI] [PubMed] [Google Scholar]
- 11.Hardwick K G, Pelham H R B. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wooding S, Pelham H R B. Mol Biol Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowe T, Dascher C, Bannykh S, Plutner H, Balch W E. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- 14.Guthrie, C. & Fink, G. R. (1991) Methods Enzymol.194. [DOI] [PubMed]
- 15.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabowski R, Gallwitz D. FEBS Lett. 1997;411:169–172. doi: 10.1016/s0014-5793(97)00720-5. [DOI] [PubMed] [Google Scholar]
- 17.Götte M, Gallwitz D. FEBS Lett. 1997;411:48–52. doi: 10.1016/s0014-5793(97)00575-9. [DOI] [PubMed] [Google Scholar]
- 18.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukada M, Gallwitz D. J Cell Sci. 1996;109:2471–2481. doi: 10.1242/jcs.109.10.2471. [DOI] [PubMed] [Google Scholar]
- 20.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 123–128. [Google Scholar]
- 21.Hicke L, Schekman R. EMBO J. 1989;8:1677–1684. doi: 10.1002/j.1460-2075.1989.tb03559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicke L, Yoshihisa T, Schekman R. Mol Biol Cell. 1992;3:667–676. doi: 10.1091/mbc.3.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis M J, Pelham H R B. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- 24.Aalto M K, Ronne H, Keranen S. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becherer K A, Rieder S E, Emr S D, Jones E W. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada Y, Nakamura N, Ohsumi Y, Hirata A. J Cell Sci. 1997;110:1299–1306. doi: 10.1242/jcs.110.11.1299. [DOI] [PubMed] [Google Scholar]
- 27.Abeliovich H, Grote E, Novick P, Ferro-Novick S. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- 28.Holthuis J C M, Nichols B J, Dhruvakumar S, Pelham H R B. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel M O, Guillaud P, Devilliers G, Rossanese O W, Glick B S, Riezman H, et al. Mol Biol Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett M K, Garcia-Arraras J E, Elferink L A, Peterson K, Fleming A M, Hazuka C D, Scheller R H. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 31.Dascher C, Ossig R, Gallwitz D, Schmitt H D. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Søgaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Söllner T. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 33.Fields S, Song O A. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 34.Gimeno R E, Espenshade P, Kaiser C A. Mol Biol Cell. 1996;7:1815–1823. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaywitz D A, Espenshade P J, Gimeno R E, Kaiser C A. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 36.Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates J R, III, Abeliovich H, Ferro-Novick S. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNew J A, Søgaard M, Lampen N M, Machida S, Ye R R, Lacomis L, Tempst P, Rothman J E, Söllner T H. J Biol Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- 38.Roberg K J, Bickel S, Rowley N, Kaiser C A. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]