Abstract
The p38 mitogen-activated protein kinase is activated by treatment of cells with cytokines and by exposure to environmental stress. The effects of these stimuli on p38 MAP kinase are mediated by the MAP kinase kinases (MKKs) MKK3, MKK4, and MKK6. We have examined the function of the p38 MAP kinase signaling pathway by investigating the effect of targeted disruption of the Mkk3 gene. Here we report that Mkk3 gene disruption caused a selective defect in the response of fibroblasts to the proinflammatory cytokine tumor necrosis factor, including reduced p38 MAP kinase activation and cytokine expression. These data demonstrate that the MKK3 protein kinase is a critical component of a tumor necrosis factor-stimulated signaling pathway that causes increased expression of inflammatory cytokines.
Mitogen-activated protein kinase signal transduction pathways have been implicated in multiple physiological and pathophysiological processes, including growth, differentiation, survival, and death (1–3). In mammals, three groups of MAP kinases have been identified: the extracellular signal-regulated protein kinases (ERK), the c-Jun NH2-terminal kinases (JNK), and the p38 MAP kinases. These MAP kinases are activated by conserved protein kinase signaling modules, which include a MAP kinase kinase kinase (MKKK) and a dual-specificity MAP kinase kinase (MKK). The MKKK phosphorylates and activates the MKK, which, in turn, activates the MAP kinase by dual phosphorylation on threonine and tyrosine residues within a Thr-Xaa-Tyr motif located in protein kinase subdomain VIII (2, 3). Separate protein kinase signaling modules are employed to activate different groups of MAP kinases (4). These signaling modules are coupled to different environmental stimuli. For example, the ERK MAP kinases are activated by a Ras-dependent pathway in response to many growth factors and hormones (3). In contrast, the JNK and p38 MAP kinases are activated by environmental stresses, such as UV radiation, osmotic shock, heat shock, protein synthesis inhibitors, and lipopolysaccharide (1, 2). The JNK and p38 MAP kinases also are activated by treatment of cells with proinflammatory cytokines, including interleukin 1 (IL-1) and tumor necrosis factor (TNF) (1, 2).
Functional analysis of the role of the p38 MAP kinase signaling pathway has been facilitated by the identification of pyridinyl imidazole derivatives that inhibit p38 MAP kinase (5, 6). This inhibition is mediated by specific interactions between the drug molecule and the ATP-binding site on p38 MAP kinase (7–9). Interestingly, these drugs exert antiinflammatory effects because they inhibit the expression of cytokines, including IL-1, IL-6, and TNF (5, 6). Based on this observation, it has been proposed that the p38 MAP kinase signaling pathway is a physiologically important mediator of increased cytokine biosynthesis in response to the exposure of cells to stress (5, 6). The p38 MAP kinase therefore represents a possible target for the design of novel antiinflammatory drugs.
The biochemical mechanisms that account for the effects of the p38 MAP kinase signaling pathway remain unclear. However, recent studies have led to the identification of substrates for p38 MAP kinase that may be physiologically relevant. These include the transcription factors ATF2 (10, 11), CHOP (12), ELK-1 (13, 14), MEF2C (15), and SAP-1 (13, 14), which are phosphorylated and activated by p38 MAP kinase. In addition, the p38 MAP kinase pathway has been reported to regulate NF-κB by a mechanism that has not yet been defined (16, 17). The p38 MAP kinase also has been reported to phosphorylate and activate several other protein kinases, including MNK1, MNK2, MAPKAPK2, MAPKAPK3, MSK1, and PRAK (18–24). It is likely that these protein kinases may function as effectors for some actions of the p38 MAP kinase pathway. For example, MAPKAPK2 and MSK1 may mediate the effects of p38 MAP kinase signaling on the phosphorylation and activation of the transcription factors ATF1 and CREB (23, 25, 26); MAPKAPK2 may mediate the effects of p38 MAP kinase signaling on the phosphorylation of tyrosine hydroxylase and the small heat shock protein hsp27 (20, 22, 27), and the MNK1 and MNK2 protein kinases may contribute to the effects of the p38 MAP kinase signaling pathway on translation through phosphorylation of eIF-4E (19).
Many studies of the p38 MAP kinase signaling pathway have relied extensively on the use of pyridinyl imidazole drugs to inhibit the function of p38 MAP kinase in vivo. An alternative approach to studying the p38 MAP kinase signaling pathway would be desirable because the drugs employed could inhibit other protein kinases under some experimental conditions (13, 28–31). The need for an alternative approach to complement pharmacological studies is highlighted further by the recent finding that pyridinyl imidazole drugs, including SB203580, directly inhibit thromboxane synthase, cyclooxygenase-1, and cyclooxygenase-2 (32). This observation indicates that some of the actions of p38 MAP kinase inhibitors may be secondary to changes in prostaglandin metabolism that occur independently of p38 MAP kinase inhibition.
The genetically tractable model organism Drosophila is likely to be useful for studies of the functional role of the p38 MAP kinase signaling pathway (33). Similar genetic analysis in mammals is difficult. However, targeted gene disruption strategies can be employed to study the function of signaling pathways in mice. Because there are four genes that encode p38 MAP kinase in mammals (28, 29, 34–39), which may have partially redundant functions, we have investigated the effect of targeted disruption of the genes that encode MKK that activate p38 MAP kinase. Three genes have been identified: Mkk3 (4), Mkk4 (4, 40), and Mkk6 (11, 41–43). The MKK3 and MKK6 protein kinases are specific for p38 MAP kinase, whereas the MKK4 protein kinase can activate both p38 MAP kinase and JNK. Disruption of the Mkk4 gene causes marked defects in JNK activation and early embryonic death (44–46). Mkk4 (−/−) cells therefore do not provide a good model for studies of p38 MAP kinase signaling. However, it was likely that the disruption of a gene that encodes a specific activator of p38 MAP kinase might cause a selective defect in p38 MAP kinase signaling. Here we report the effect of targeted disruption of the Mkk3 gene on the p38 MAP kinase signaling pathway in murine embryo fibroblasts.
EXPERIMENTAL PROCEDURES
Targeted Disruption of the Mkk3 Gene.
DNA clones corresponding to the Mkk3 locus were cloned from a λ Fix II phage library prepared from genomic DNA isolated from mouse strain 129/Sv (Stratagene). Positive clones were characterized by Southern blot analysis and DNA sequencing (Applied Biosystems). The targeting vector was constructed by inserting a 4.4-kb BglII-BglII fragment from the 5′ end of the Mkk3 genomic clone, a 1.6-kb PGK-neo cassette, a 0.92-kb EcoRV-HincII fragment derived from the 3′ end of the Mkk3 genomic clone, and a 3.4-kb fragment containing two copies of the Herpes simplex thymidine kinase gene into pBluescript KS vector (Stratagene). The targeting vector was linearized with NotI and electroporated into W9.5 embryonal stem cells. Genomic DNA from transfectants resistant to G418 (0.2 mg/ml) (Life Technologies, Gaithersburg, MD) and gancyclovir (2 μM) (Syntex, Palo Alto, CA) was isolated and screened by Southern blot analysis. Heterozygous Mkk3 (−/+) cells were injected into C57BL/6 blastocysts to create chimeric mice, which were crossed to obtain germ-line transmission of the disrupted Mkk3 allele. A detailed description of the targeted disruption of the Mkk3 gene in mice will be published elsewhere (55).
Preparation of Murine Embryo Fibroblasts.
Embryonic day 14 Mkk3 (−/−) embryos were employed to prepare murine embryo fibroblasts (MEF). The cells were grown in DMEM supplemented with 10% heat-inactivated FBS/2 mM glutamine/100 units/ml of penicillin/100 μg/ml of streptomycin (Life Technologies) at 37°C in a humidified atmosphere of 5% CO2. Control experiments demonstrated that the wild-type (+/+) and Mkk3 (−/−) MEF proliferated at similar rates (data not shown). The cells were treated with 10 μM SB203580 (Calbiochem), 10 ng/ml TNFα (Genzyme), 10 ng/ml IL-1β (Genzyme), 300 mM sorbitol, and 40 J/m2 UV radiation (UV-C) (10).
Protein Kinase Assays.
JNK and p38 MAP kinase activity was measured with an immunocomplex kinase assay by using recombinant c-Jun and ATF2 as substrates (10).
Cytokine Assays.
IL-1β (Genzyme) and IL-6 (PharMingen) were measured by ELISA with a kit, using procedures recommended by the manufacturer. Ribonuclease protection assays of total RNA isolated from MEF were performed by using the Riboquant kit (PharMingen).
Immunoblot Analysis.
Protein immunoblot analysis of cell lysates (30 μg) was performed by probing with a polyclonal antibody to p38 MAP kinase (Santa Cruz Biotechnology), a mAb to JNK (PharMingen), and rabbit polyclonal antibodies to MKK3, MKK4, and MKK6. The MKK antibodies were prepared by immunizing rabbits with purified recombinant human protein kinases expressed in bacteria (4, 11). Immunocomplexes were detected by enhanced chemiluminescence (Amersham).
RESULTS
The MKK3 protein kinase was identified previously as a specific activator of the p38 MAP kinase (4). We isolated the Mkk3 gene by screening a mouse genomic library with the human Mkk3 cDNA as a probe. Sequence analysis demonstrated that the gene consists of 12 exons (Fig. 1). Disruption of the murine Mkk3 gene in W9.5 embryonic stem cells was accomplished using standard techniques by replacement of Mkk3 sequences with a NeoR cassette by homologous recombination (Fig. 1). Disruption of one Mkk3 allele was confirmed by Southern blot analysis of genomic DNA. Chimeric mice were created by injecting six independent clones of heterozygous Mkk3 (−/+) stem cells into C57BL/6 blastocysts. Two clones resulted in germ-line transmission of the disrupted Mkk3 allele. Heterozygotes were intercrossed to obtain homozygous Mkk3 (−/−) mice, which were identified by Southern blot analysis of genomic DNA. The Mkk3 (−/−) mice were viable with no obvious morphological defects. The absence of a profound defect in the Mkk3 (−/−) animals may reflect a redundant function of the MKK3 protein kinase in the presence of the p38 MAP kinase activators MKK4 (4) and MKK6 (11, 41–43).
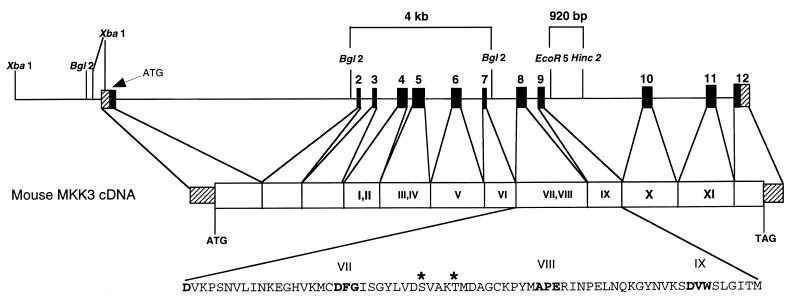
Figure 1.
Targeted disruption of the Mkk3 gene. The structure of the murine Mkk3 gene is illustrated schematically. This gene consists of 12 exons. The coding and noncoding regions of the exons are presented as solid and striped, respectively. The restriction fragments used to construct the targeting vector are indicated (4-kb BglII-BglII fragment and 0.92 kb EcoR5-HincII fragment). The disrupted Mkk3 allele substitutes a NeoR cassette for exons 8 and 9, which encode the essential protein kinase subdomains VII, VIII, and IX.
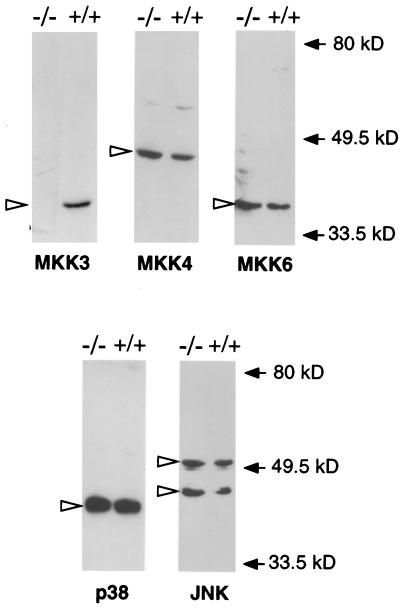
Biochemical analysis of the effect of Mkk3 gene disruption was facilitated by the preparation of primary MEF. Protein immunoblot analysis of extracts prepared from wild-type (+/+) and Mkk3 (−/−) MEF demonstrated that, in contrast to wild-type cells, the Mkk3 (−/−) cells do not express the MKK3 protein kinase (Fig. 2). However, both wild-type (+/+) and Mkk3 (−/−) cells express similar levels of the stress-activated MKKs MKK4 and MKK6 (Fig. 2). These data demonstrate that Mkk3 gene disruption caused a selective defect in the expression of the protein kinase MKK3 in the absence of changes in the expression of other stress-activated MKKs. In addition, the Mkk3 gene disruption caused no change in the expression of the stress-activated MAP kinases p38 and JNK (Fig. 2).
Figure 2.
Cells isolated from Mkk3 (−/−) mice do not express the MKK3 protein kinase. Wild-type (+/+) and Mkk3 (−/−) MEF were prepared, and extracts (30 μg) were examined by protein immunoblot analysis by using antibodies to MKK3, MKK4, MKK6, JNK, and p38 MAP kinase. Open arrows indicate the migration of the protein kinases. Solid arrows indicate the migration of molecular mass standards (kDa).
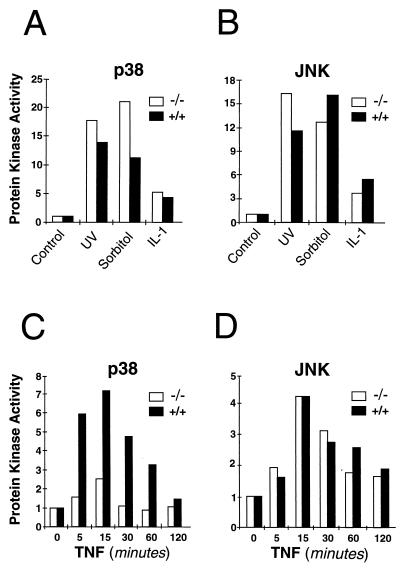
The loss of MKK3 protein kinase expression was predicted to cause defects in the p38 MAP kinase signaling pathway. Studies therefore were performed to examine the regulation of p38 MAP kinase in wild-type (+/+) and Mkk3 (−/−) MEF. The basal activity of p38 MAP kinase was reduced slightly (approximately 40% lower) in Mkk3 (−/−) MEF compared with wild-type (+/+) MEF (data not shown). Exposure to environmental stress (UV radiation and osmotic shock) and the inflammatory cytokine IL-1 caused activation of p38 MAP kinase in both wild-type (+/+) and Mkk3 (−/−) MEF (Fig. 3A). These stimuli also caused similar activation of the stress-activated JNK MAP kinase in wild-type (+/+) and Mkk3 (−/−) cells (Fig. 3B). In contrast, studies of the effect of TNFα demonstrated that the Mkk3 (−/−) MEF were selectively defective in the activation of p38 MAP kinase (Fig. 3C).
Figure 3.
Effect of Mkk3 gene disruption on stress-activated MAP kinase activity. (A and B) Wild-type (+/+) and Mkk3 (−/−) MEF were untreated or treated with UV radiation (UV-C) (40 J/m2), osmotic shock (300 mM sorbitol, 30 min), or IL-1 (10 ng/ml, 15 min). (C and D) Wild-type (+/+) and Mkk3 (−/−) MEF were untreated or treated with TNFα (10 ng/ml). The cells were harvested at the indicated times. The p38 MAP kinase (A and C) and JNK (B and D) activity was measured by using an immunocomplex kinase assay with the substrates ATF2 and c-Jun, respectively. Radioactivity incorporated into each substrate was quantitated after SDS/PAGE by PhosphorImager analysis. The data are presented as the relative protein kinase activity.
The defect in TNFα-stimulated p38 MAP kinase activity observed in Mkk3 (−/−) MEF (Fig. 3C) might reflect a general deficiency of these cells in their response to TNFα signaling. However, ribonuclease protection assays demonstrated that the expression of the p55 and p75 TNF receptors was equal in wild-type (+/+) and Mkk3 (−/−) MEF (data not shown). This observation suggested that the Mkk3 (−/−) MEF may have a selective defect in their response to TNF. To test this hypothesis, we examined the effect of TNFα on the stress-activated MAP kinase JNK. These data demonstrated that TNFα caused similar JNK activation in wild-type (+/+) and Mkk3 (−/−) MEF (Fig. 3D). These observations demonstrated that the defect in p38 MAP kinase activation in Mkk3 (−/−) MEF represents a selective impairment of TNFα signaling to the p38 MAP kinase signaling pathway. We conclude that the MKK3 protein kinase is required for efficient TNFα-stimulated p38 MAP kinase activation in MEF.
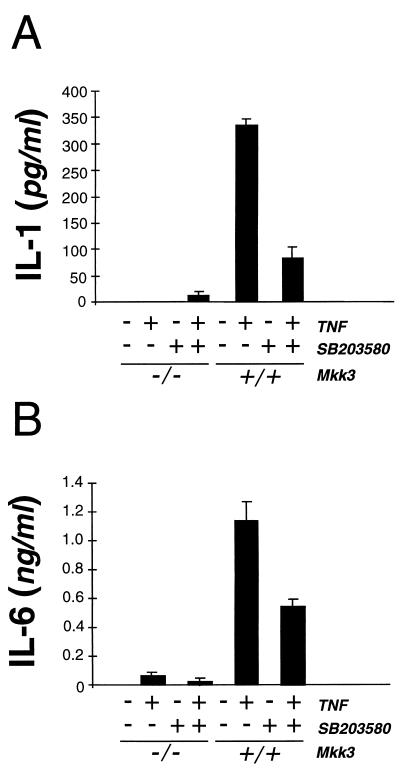
The stress-activated p38 MAP kinase signaling pathway has been implicated as a regulatory mechanism that controls the production of cytokines by cells exposed to stress. Previous studies have established that p38 MAP kinase may mediate the effect of TNFα to induce the expression of IL-1 (6) and IL-6 (47). We therefore examined cytokine expression by wild-type (+/+) and Mkk3 (−/−) MEF treated with TNFα. Treatment of wild-type (+/+) MEF with TNFα increased the production of IL-1 and IL-6 (Fig. 4). The p38 MAP kinase inhibitor SB203580 (48) partially inhibited TNFα-stimulated IL-1 and IL-6 synthesis (Fig. 4). In contrast, disruption of the Mkk3 gene markedly decreased the TNFα-stimulated production of IL-1 and IL-6 by MEF (Fig. 3). These data demonstrate that Mkk3 gene disruption caused a defect in TNFα-stimulated IL-1 and IL-6 expression, indicating that the MKK3 protein kinase is an essential component of the TNF signaling pathway that regulates cytokine expression.
Figure 4.
Effect of Mkk3 gene disruption on IL-1 and IL-6 secretion. Wild-type (+/+) and Mkk3 (−/−) MEF were untreated or treated with SB203850 (10 μM) for 2 hr before treatment with TNFα (10 ng/ml; 24 hr) and measurement of cytokine expression by ELISA. The amount of IL-1β (A) and IL-6 (B) is presented as the mean ± SD of triplicate observations. Similar data were obtained in three separate experiments.
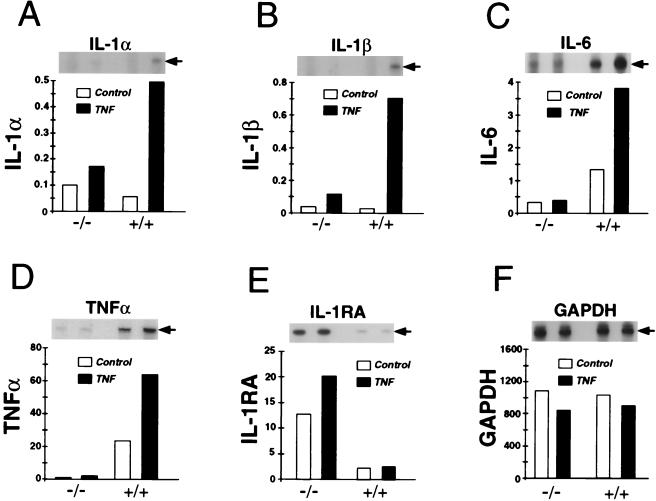
The induction of cytokine expression caused by TNFα may be mediated through the regulation of several steps in the cytokine biosynthetic pathway, including increased mRNA expression and increased mRNA translation (5, 6, 47). The defect in cytokine expression by Mkk3 (−/−) MEF therefore may result from alterations in either of these processes. Therefore, to examine the mechanism that accounted for the defective production of cytokines by Mkk3 (−/−) MEF, we examined the ability of TNFα to induce cytokine mRNA expression by using a ribonuclease protection assay. Control experiments demonstrated that the amount glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA detected in wild-type (+/+) and Mkk3 (−/−) MEF was similar and that treatment with TNFα did not cause marked changes in the amount of GAPDH mRNA (Fig. 5). In contrast, treatment of wild-type (+/+) MEF with TNFα increased the amount of IL-1, IL-6, and TNFα mRNA (Fig. 5). The Mkk3 (−/−) MEF were defective in the TNFα-stimulated accumulation of IL-1, IL-6, and TNFα mRNA (Fig. 5). This defect in cytokine mRNA expression was selective because TNFα did induce the expression of TCA3 chemokine mRNA to a similar extent in wild-type (+/+) MEF (14-fold) and Mkk3 (−/−) MEF (15-fold) (data not shown). These data demonstrate that the Mkk3 (−/−) MEF were selectively defective in TNFα-induced expression of IL-1, IL-6, and TNFα mRNA.
Figure 5.
Effect of Mkk3 gene disruption on IL-1 and IL-6 gene expression. Wild-type (+/+) and Mkk3 (−/−) MEF were untreated or treated with TNFα (10 ng/ml) for 6 hr. The amount of IL-1α (A), IL-1β (B), IL-6 (C), TNFα (D), IL-1 receptor antagonist (IL-1RA) (E), and glyceraldehyde-3-phosphate dehydrogenase (F) mRNA was measured by ribonuclease protection assay. The protected RNA was detected by autoradiography (Inset) after denaturing PAGE and was quantitated by PhosphorImager analysis (relative PhosphorImager units). For clarity, the time employed for autoradiography was different for each type of mRNA. Similar data were obtained in three separate experiments.
No defects in the expression of cytokine mRNA were detected in control cultures of Mkk3 (−/−) MEF treated without TNFα (Fig. 5). However, increased expression of the IL-1 receptor antagonist (IL-1RA) was observed in Mkk3 (−/−) MEF compared with wild-type cells (Fig. 5E). This observation suggests that MKK3 functions genetically to down-regulate IL-1RA expression. The biochemical basis for this effect of Mkk3 gene disruption is unclear, but may be related to the altered basal p38 MAP kinase activity in Mkk3 (−/−) MEF. Further studies are required to test this hypothesis.
Together, these data indicate that the defective cytokine expression by Mkk3 (−/−) MEF was accounted for, at least in part, by the failure of TNFα to induce the expression of cytokine mRNA (Fig. 5).
DISCUSSION
It has been proposed previously that the p38 MAP kinase signaling pathway is an important mediator of inflammatory processes. Several lines of experimental evidence support this contention. First, endotoxic lipopolysaccharide and proinflammatory cytokines cause marked activation of p38 MAP kinase (10). Second, specific inhibitors of p38 MAP kinase (pyridinyl imidazole drugs) inhibit the secretion of inflammatory cytokines (6). Third, transgenic animal models demonstrate that the p38 MAP kinase pathway regulates transcription of the interferon γ gene by T helper 1 cells (49). The results of the present study provide further support to the hypothesis that the p38 MAP kinase signaling pathway contributes to inflammatory processes. Disruption of the Mkk3 gene, which encodes an activator of p38 MAP kinase, caused a selective defect in TNFα-stimulated p38 MAP kinase activation (Fig. 3) and inflammatory cytokine expression (Figs. 4 and 5). Together, these observations strongly implicate the p38 MAP kinase pathway as a mediator of inflammatory signaling.
The conclusion that p38 MAP kinase is a mediator of inflammation in mammals contrasts with observations that have been reported concerning the role of p38 MAP kinase in the innate immune response of Drosophila. Two genes that encode p38 MAP kinase and one gene that encodes a p38 MAP kinase activator have been identified in Drosophila by molecular cloning (33). Inhibition of the Drosophila p38 MAP kinase signaling pathway caused increased expression of antibacterial and antifungal genes. This observation suggested that p38 MAP kinase, which is activated during the innate immune response in the fly, normally functions to down-regulate this response (33). Further studies are required to define whether this apparent difference between mammals and insects reflects an evolutionary change in the function of the p38 MAP kinase or whether the different observations indicate that the role of p38 MAP kinase can be altered in highly differentiated tissues.
Mkk3 Gene Disruption Causes a Selective Defect in p38 MAP Kinase Signaling.
The targeted disruption of the Mkk3 gene caused only a selective defect in the p38 MAP kinase signal transduction pathway (Figs. 3–5). It is likely that a more profound defect in p38 MAP kinase signaling was not observed because the function of the MKK3 protein kinase is partially redundant to the MKK4 and MKK6 protein kinases. Indeed, many studies have documented that MKK6 is a strong activator of p38 MAP kinase (11, 41–43). For example, MKK6 is the major activator of p38 MAP kinase in cells exposed to osmotic stress (50). Interestingly, Mkk3 (−/−) MEF were selectively defective in their response to TNFα. Whereas JNK activation was comparable to that observed in wild-type MEF, the activation of p38 MAP kinase was suppressed (Fig. 3). Thus, the MKK3 protein kinase is required for full activation of p38 MAP kinase in response to the treatment of MEF with TNFα. The observation that TNFα does not activate MKK4 (2) can account for the lack of complementation of the MKK3 deficiency by MKK4. In contrast, MKK6 can be activated by TNFα (50). The low level of p38 MAP kinase activation observed in TNFα-treated Mkk3 (−/−) MEF therefore may be mediated by MKK6. Further studies of other cell types derived from the Mkk3 (−/−) mice are warranted to document more fully this partial complementation by MKK6.
The MKK3 Protein Kinase Is Required for TNFα-Regulated Cytokine Expression.
Studies using pyridinyl imidazole drugs, which inhibit p38 MAP kinase, indicate that these compounds may regulate the translation of cytokine mRNAs (5, 51–53). The mechanism by which the p38 MAP kinase pathway regulates cytokine mRNA translation requires further study, but recent progress has been achieved through the identification of cytokine-responsive translational regulatory elements (52) and the identification of a group of eIF-4E protein kinases that are activated by p38 MAP kinase (18, 19). Although the effect of Mkk3 gene disruption to block TNFα-stimulated expression of IL-1, IL-6, and TNFα mRNA indicates that the regulation of cytokine mRNA expression is a primary target of the MKK3 protein kinase signaling pathway in MEF (Fig. 5), these data do not exclude the possibility that this signaling pathway may also regulate later steps in the cytokine biosynthetic pathway, including translation. It is also possible that the relative importance of regulated mRNA expression and translational regulation for TNF-stimulated cytokine expression may differ between cell types.
Previous studies have established that the p38 MAP kinase signaling pathway mediates activation of several transcription factors, including ATF1 (25, 26), ATF2 (10, 11), CHOP (12), CREB (25, 26), ELK-1 (13, 14), MEF2C (15), NF-κB (16, 17), and SAP-1 (13, 14). Defects in the activation of one or more of these transcription factors in Mkk3 (−/−) MEF may contribute to the reduced TNFα-stimulated expression of cytokine mRNA in these cells. Indeed, treatment with pyridinyl imidazole drugs inhibits the induction of IL-6 mRNA in TNFα-stimulated fibroblasts (47). The biochemical mechanism that accounts for the p38 MAP kinase-regulated expression of IL-6 mRNA may be caused by increased stability of the IL-6 mRNA (54) or by increased transcription of the IL-6 gene mediated, in part, by NF-κB (16). Further studies are required to identify the specific defects that are present in Mkk3 (−/−) MEF.
CONCLUSIONS
The MKK3 protein kinase is required in MEF for full activation of the p38 MAP kinase signal transduction pathway by TNFα. Disruption of the Mkk3 gene blocked the effect of TNFα to increase the expression of the cytokines IL-1 and IL-6 in MEF. These data provide direct genetic evidence that the p38 MAP kinase signaling pathway contributes to the regulation of cytokine expression in mammalian cells after exposure to stress.
Acknowledgments
We thank T. Barrett for DNA sequence analysis; J. Bao, D. Butkis, L. Evangelisti, C. Hughes, and J. Musco for technical assistance; and K. Gemme for administrative assistance. This work was supported by a grant from the National Cancer Institute (R.J.D.). R.A.F. and R.J.D. are Investigators of the Howard Hughes Medical Institute.
ABBREVIATIONS
- IL
interleukin
- JNK
c-Jun NH2-terminal kinase
- MAP kinase
mitogen-activated protein kinase
- MKK
MAP kinase kinase
- MKKK
MKK kinase
- TNF
tumor necrosis factor
- MEF
murine embryo fibroblasts
References
- 1.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 2.Ip Y T, Davis R J. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 3.Robinson M J, Cobb M H. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 4.Dérijard B, Raingeaud J, Barrett T, Wu I-H, Han J, Ulevitch R J, Davis R J. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 5.Lee J C, Young P R. J Leukocyte Biol. 1996;59:152–157. doi: 10.1002/jlb.59.2.152. [DOI] [PubMed] [Google Scholar]
- 6.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, et al. Nature (London) 1994;327:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 7.Wilson K P, McCaffrey P G, Hsiao K, Pazhanisamy S, Galullo V, Bemis G W, Fitzgibbon M J, Caron P R, Murcko M A, Su M S. Chem Biol. 1997;4:423–431. doi: 10.1016/s1074-5521(97)90194-0. [DOI] [PubMed] [Google Scholar]
- 8.Young P R, McLaughlin M M, Kumar S, Kassis S, Doyle M L, McNulty D, Gallagher T F, Fisher S, McDonnell P C, Carr S A, et al. J Biol Chem. 1997;272:12116–12121. doi: 10.1074/jbc.272.18.12116. [DOI] [PubMed] [Google Scholar]
- 9.Tong L, Pav S, White D M, Rogers S, Crane K M, Cywin C L, Brown M L, Pargellis C A. Nat Struct Biol. 1997;4:311–316. doi: 10.1038/nsb0497-311. [DOI] [PubMed] [Google Scholar]
- 10.Raingeaud J, Gupta S, Rogers J, Dickens M, Han J, Ulevitch R J, Davis R J. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 11.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R J. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X Z, Ron R. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 13.Whitmarsh A J, Yang S-H, Su M S-S, Sharrocks A D, Davis R J. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price M A, Cruzalegui F H, Treisman R. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 16.Vanden Berghe W, Plaisance S, Boone E, De Bosscher K, Schmitz M L, Fiers W, Haegeman G. J Biol Chem. 1998;273:3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 17.Zechner D, Craig R, Hanford D S, McDonough P M, Sabbadini R A, Glembotski C C. J Biol Chem. 1998;273:8232–8239. doi: 10.1074/jbc.273.14.8232. [DOI] [PubMed] [Google Scholar]
- 18.Fukunaga R, Hunter T. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freshney N W, Rawlinson L, Guesdon F, Jones E, Cowley S, Hsuan J, Saklatvala J. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 21.McLaughlin M M, Kumar S, McDonnell P C, Van Horn S, Lee J C, Livi G P, Young P R. J Biol Chem. 1996;271:8488–8492. doi: 10.1074/jbc.271.14.8488. [DOI] [PubMed] [Google Scholar]
- 22.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 23.New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood L J, Kato Y, Parry G C, Han J. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, Gaestel M, Rapp U R. Mol Cell Biol. 1996;16:6687–6697. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb M J. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 26.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H J, Herrlich P. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas G M, Haavik J, Cohen P. Eur J Biochem. 1997;247:1180–1189. doi: 10.1111/j.1432-1033.1997.01180.x. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 29.Enslen H, Raingeaud J, Davis R J. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 30.Cohen P, Goedert M. Chem Biol. 1998;5:R161–R164. doi: 10.1016/s1074-5521(98)90068-0. [DOI] [PubMed] [Google Scholar]
- 31.Eyers P A, Craxton M, Morrice N, Cohen P, Goedert M. Chem Biol. 1998;5:321–328. doi: 10.1016/s1074-5521(98)90170-3. [DOI] [PubMed] [Google Scholar]
- 32.Borsch-Haubold A G, Pasquet S, Watson S P. J Biol Chem. 1998;273:28766–28772. doi: 10.1074/jbc.273.44.28766. [DOI] [PubMed] [Google Scholar]
- 33.Han Z S, Enslen H, Hu X, Meng X, Wu I-H, Barrett T, Davis R J, Ip Y T. Mol Cell Biol. 1998;18:3527–3539. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z J, Jiang Y, Ulevitch R J, Han J. Biochem Biophys Res Commun. 1996;228:334–340. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 35.Cuenda A, Cohen P, Buee-Scherrer V, Goedert M. EMBO J. 1997;16:295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goedert M, Cuenda A, Craxton M, Jakes R, Cohen P. EMBO J. 1997;16:3663–3671. doi: 10.1093/emboj/16.12.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein B, Yang M X, Young D B, Janknecht R, Hunter T, Murray B W, Barbosa M S. J Biol Chem. 1997;272:19509–19517. doi: 10.1074/jbc.272.31.19509. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y, Gram H, Zhao M, New L, Gu J, Feng L, Di Padova F, Ulevitch R J, Han J. J Biol Chem. 1997;272:30122–30128. doi: 10.1074/jbc.272.48.30122. [DOI] [PubMed] [Google Scholar]
- 39.Wang X S, Diener K, Manthey C L, Wang S, Rosenzweig B, Bray J, Delaney J, Cole C N, Chan-Hui P Y, Mantlo N, et al. J Biol Chem. 1997;272:23668–23674. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- 40.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Lee J-D, Jiang Y, Li Z, Feng L, Ulevitch R J. J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 42.Stein B, Brady H, Yang M X, Young D B, Barbosa M S. J Biol Chem. 1996;271:11427–11433. doi: 10.1074/jbc.271.19.11427. [DOI] [PubMed] [Google Scholar]
- 43.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, et al. J Biol Chem. 1996;271:13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 44.Yang D, Tournier C, Wysk M, Lu H-T, Xu J, Davis R J, Flavell R A. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishina H, Fischer K D, Radvanyl L, Shahinian A, Hakem R, Ruble E A, Bernstein A, Mak T W, Woodgett J R, Penninger J M. Nature (London) 1997;385:350–353. doi: 10.1038/385350a0. [DOI] [PubMed] [Google Scholar]
- 46.Ganiatsas S, Kwee L, Fujiwara Y, Perkins A, Ikeda T, Labow M A, Zon L I. Proc Natl Acad Sci USA. 1998;95:6881–6886. doi: 10.1073/pnas.95.12.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beyaert R, Cuenda A, Vanden Berghe W, Plaisance S, Lee J C, Haegeman G, Cohen P, Fiers W. EMBO J. 1996;15:1914–1923. [PMC free article] [PubMed] [Google Scholar]
- 48.Cuenda A, Rouse J, Doza Y N, Meier R, Cohen P, Gallagher T F, Young P R, Lee J C. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 49.Rincón M, Enslen H, Raingeaud J, Recht M, Zapton T, Su M S-S, Penix L A, Davis R J, Flavell R A. EMBO J. 1998;17:2817–2829. doi: 10.1093/emboj/17.10.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriguchi T, Toyoshima F, Gotoh Y, Iwamatsu A, Irie K, Mori E, Kuroyanagi N, Hagiwara M, Matsumoto K, Nishida E. J Biol Chem. 1996;271:26981–26988. doi: 10.1074/jbc.271.43.26981. [DOI] [PubMed] [Google Scholar]
- 51.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, et al. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 52.Kruys V, Beutler B, Huez G. Enzyme. 1990;44:193–202. doi: 10.1159/000468757. [DOI] [PubMed] [Google Scholar]
- 53.Prichett W, Hand A, Sheilds J, Dunnington D. J Inflamm. 1995;45:97–105. [PubMed] [Google Scholar]
- 54.Miyazawa K, Mori A, Miyata H, Akahane M, Ajisawa Y, Okudaira H. J Biol Chem. 1998;273:24832–24838. doi: 10.1074/jbc.273.38.24832. [DOI] [PubMed] [Google Scholar]
- 55.Lu, H.-T., Yang, D. D., Wysk, M., Gatti, E., Mellman, I., Davis, R. J. & Flavell, R. A. (1999) EMBO J., in press. [DOI] [PMC free article] [PubMed]