Abstract
Previous studies showed that thymidylate synthase (TS), as an RNA binding protein, regulates its own synthesis by impairing the translation of TS mRNA. In this report, we present evidence that p53 expression is affected in a similar manner by TS. For these studies, we used a TS-depleted human colon cancer HCT-C cell that had been transfected with either the human TS cDNA or the Escherichia coli TS gene. The level of p53 protein in transfected cells overexpressing human TS was significantly reduced when compared with its corresponding parent HCT-C cells. This suppression of p53 expression was the direct result of decreased translational efficiency of p53 mRNA. Similar results were obtained upon transfection of HCT-C cells with pcDNA 3.1 (+) containing the E. coli TS gene. These findings provide evidence that TS, from diverse species, specifically regulates p53 expression at the translational level. In addition, TS-overexpressing cells with suppressed levels of p53 are significantly impaired in their ability to arrest in G1 phase in response to exposure to a DNA-damaging agent such as γ-irradiation. These studies provide support for the in vivo biological relevance of the interaction between TS and p53 mRNA and identify a molecular pathway for controlling p53 expression.
Keywords: translational regulation, gene expression
Thymidylate synthase (TS) is a folate-dependent enzyme that catalyzes the reductive methylation of dUMP by 5,10-methylenetetrahydrofolate to form dTMP and dihydrofolate (1, 2). Because the TS-catalyzed enzymatic reaction provides the sole intracellular de novo source of thymidylate, an essential precursor for DNA biosynthesis, this enzyme has been an important target for cancer chemotherapy for nearly 40 years (3–6).
In addition to its critical role in enzyme catalysis, TS functions as an RNA binding protein (7–10). Studies from this laboratory have demonstrated that translation of human TS mRNA is regulated by its own protein product via a negative autoregulatory mechanism whereby the binding of TS protein to at least two distinct sequences on its own TS mRNA results in translational repression (7, 8).
More recently, studies have documented that TS, in addition to directly interacting with its own TS mRNA, is capable of binding to several cellular RNA species (11–13). An immunoprecipitation–RNA–random (r)PCR method was developed to isolate cellular RNA sequences that formed ribonucleoprotein (RNP) complexes with TS in human colon cancer H630 cells (13). With this approach, nine different cellular mRNAs including those corresponding to the myc family and the p53 tumor suppressor gene were identified. Previous studies had shown that TS binds in vitro to the C-terminal coding region of c-myc mRNA. Further investigation revealed that this RNA–protein interaction results in translational repression of c-myc mRNA (12). These studies suggested that TS may be involved in the coordinate regulation of expression and/or function of a host of cellular genes. Although recent work has identified a direct interaction between human TS protein and p53 mRNA (13), the in vivo biological significance of this RNA–protein interaction remains to be characterized.
The p53 tumor suppressor plays an essential role for preserving the integrity of the genome and for maintaining regulation of cell cycle progression (14–17). Several investigators have shown that the levels of p53 are acutely increased in both normal and malignant cells in response to DNA-damaging agents (18–21). Although definitive mechanistic studies remain to be performed, the induced expression of p53 after DNA damage appears to be regulated at least in part, by translational and post-translational regulatory processes. The importance of translational regulatory mechanisms underlying the expression of p53 has been recently supported by studies that suggest that the expression of murine p53 is controlled by a negative autoregulatory feedback process (22, 23). Mosner et al. (23) demonstrated that the p53 protein end product binds to the 5′ untranslated region of its corresponding p53 mRNA and, in so doing, effectively represses translation. A more recent report by Fu et al. (24) confirmed that the expression of human p53 was also controlled at the translational level, presumably, through an autoregulatory feedback loop. However, in contrast to the murine system where p53 protein interacts with the 5′ untranslated region of its corresponding mRNA sequence, human p53 protein binds to a sequence contained within the 3′ untranslated region of the human message (24, 25).
Because the biosynthesis of p53 is regulated, in part, at the translational level and a TS–RNP complex composed of TS protein and a sequence corresponding to human p53 mRNA was identified in human colon cancer cells, we decided to further investigate the biological role of the interaction between TS protein and p53 mRNA. For these studies, we transfected the respective full-length human TS and Escherichia coli TS cDNAs under the control of a constitutive cytomegalovirus promoter into HCT-C cells that express wild-type p53 and nonfunctional TS protein. In this report, we show that the overexpression of human and E. coli TS in the HCT-C cell line results in marked suppression of synthesis of p53 protein with no associated change in p53 mRNA levels. Our findings demonstrate that TS protein binds directly to the p53 mRNA resulting in translational repression and identifies a mechanism by which p53 expression is controlled.
MATERIALS AND METHODS
Cell Culture.
The human colon cancer HCT-8 and HCT-C cell lines were a gift from S. Berger (University of South Carolina, Columbia) and Y. Rustum (Roswell Park Memorial Cancer Institute, Buffalo, NY). The HCT-C line is a mutant subline of the HCT-8 parental cell line, and it expresses a Ser → Leu mutation at amino acid position 216 of the human TS protein (26). Enzyme kinetic analysis has shown that this mutant TS protein is nearly completely inactive (27). Cells were maintained in RPMI 1640 medium containing 10% dialyzed fetal bovine serum and supplemented with 10 μM thymidine.
Construction of a Human TS Expressing Vector pcDNA3.1-His Tag (rHTS).
The vector pET28(+)rHTS (28) containing the His-Tag region 5′ to the human TS cDNA (rHTS) gene was gel-purified and digested with XbaI and HindIII, and the desired restriction fragment was gel-purified by using the Qiaquick gel extraction kit (Qiagen, Chatsworth, CA). The 5′ overhang ends were repaired to generate blunt ends by using the Klenow Filling-In protocol (United States Biochemicals). This blunt-ended fragment was then ligated into EcoRV-digested pcDNA3.1 (+) and transformed into INVαF′ competent cells for screening and plasmid preparation. Correct orientation of the insert was determined by NdeI restriction analysis and DNA sequencing.
Construction of an E. coli TS Expressing Vector pcDNA3.1-TS.
An NheI site was introduced into the −10 region of the pBS(KS+)-thyA TS vector by using the Stratagene Quick Change site-directed mutagenesis kit. The following oligomer and its complement 5′-GTCTGGGCATATCGTCGCTAGCCCACAGCAAC were used to introduce an NheI restriction site into the −10 region of the thyA-TS gene (29). The resulting PCR product was transformed into XLI-Blue for purposes of screening and plasmid preparation. A thyA-TS containing fragment was digested from the latter vector with NheI and HindIII and purified by agarose gel electrophoresis followed by the Qiaquick extraction procedure (Qiagen). The isolated DNA fragment was ligated into NheI/HindIII-digested pcDNA3.1(+).
Stable Transfection of pcDNA3.1-TS and pcDNA3.1-His Tag(rHTS) into HCT-C Cell Lines.
Parent HCT-C cells were transfected with the pcDNA3.1-His Tag(rHTS) plasmid containing the full-length human TS cDNA. Transfection was performed by using Lipofectin according to the manufacturer’s protocol (GIBCO/BRL). After 3 weeks of growth, colonies were selected and subsequently expanded (30). The stable transfected cell line was then designated HCT-C:His-TS+. The transfection procedure of the E. coli TS plasmid into parent HCT-C cells was identical to the method described above, and the stable transfected cell line was designated HCT-C:TS+.
Western Immunoblot Analysis.
Cells were harvested and processed as described (31). Equal amounts of protein (100 μg) from each cell line were resolved by SDS/PAGE on 10% gels by the method of Laemmli (32). Proteins were probed with mouse anti-TS 106 monoclonal antibody (1:1,000 dilution), mouse anti-His-Tag monoclonal antibody (1:1,000 dilution), rabbit anti-E. coli TS antibody (1:1,000 dilution), anti-p53 mouse monoclonal antibody (Ab-2, Oncogene Science; 1:150 dilution), or anti-α-tubulin mouse monoclonal antibody (Amersham; 1:4,000 dilution) followed by incubation with a horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Proteins were visualized with a chemiluminescence detection system using the Super Signal substrate (Pierce).
Whole-Cell Extraction and Immunoprecipitation of TS–RNP Complexes.
Whole-cell extracts were prepared and immunoprecipitation of RNP complexes was performed as described (11, 33). Immunoprecipitation of TS–RNP complexes was carried out with either an anti-His-Tag monoclonal or an anti-E. coli polyclonal antibody.
Reverse Transcription-Coupled PCR (RT-PCR) Analysis.
Immunoprecipitated RNA was subjected to reverse transcription as described (11, 12). The cDNA was then used as template for PCR amplification. The reagents used were those outlined by the Perkin–Elmer protocol (Perkin–Elmer). The primer sequences are as follows: p53 (sense), 5′-TTGGATCCATGTTTTGCCAACTGGCC-3′; p53 (antisense), 5′-TTGAATTCAGGCTCCCCTTTCTTGCG-3′; TS (sense), 5′-ACCGAGCTCCCGAGACTTTTTGGACAGCCT-3′; TS (antisense), 5′-ACCAAGCTTAAGAATCCTGAGCTTTGGGAA-3′; dihydrofolate reductase (DHFR) (sense), 5′-ACCCTCGAGCAAGAACGGGGACCT-3′; DHFR (antisense), 5′-ACCAAGCTTCATTCTTCTCATATA-3; β-actin (sense), 5′-GCGGGAAATCGTGCGTGCGTGACATT-3′; β-actin (antisense), 5′-GATGGAGTTGAAGGTAGTTTCGTG-3′.
Samples were incubated at 95°C for 45 sec, 60°C for 1 min, and 72°C for 2 min for 35 cycles. Reaction samples were then incubated for an additional 7 min at 72°C and cooled to 4°C. PCR products were resolved on a 1% nondenaturing agarose gel.
Isolation of Total Cellular RNA, Northern Blot, and RNase Protection Assay.
Total cellular RNA was extracted from cells by the method of Chomczynski and Sacchi (34). A radiolabeled antisense p53 and antisense β-actin RNA probe were synthesized by in vitro transcription using the pTRI-p53, pTRI-β-actin transcription template, and T7 RNA polymerase (Ambion, Austin, TX). Equal amounts of total RNA (20 μg) from each sample were used for the RNase protection assay as described in the Ambion protocol.
Measurement of p53 Biosynthesis by Immunoprecipitation Analysis.
Cells were labeled with [35S]methionine and processed as described (31). To determine the half-life of the p53 protein, radiolabel was removed after a 30-min incubation, and fresh methionine-containing medium was added to the cell cultures. Cells were harvested and processed at 0, 10, 20, 30, and 60 min. Immunoprecipitation of p53 protein was performed with an anti-p53 monoclonal antibody (Ab-2, Oncogene Science) and protein A-agarose (GIBCO/BRL) by the method of Harford (35). Samples were analyzed by autoradiography after electrophoresis on a 10% SDS-polyacrylamide gel. The relative levels of p53 protein were then determined by densitometric scanning.
RNA Gel Mobility Shift Assay.
RNA gel mobility shift assays were performed as described (7, 8).
Cell Cycle Analysis.
Samples were prepared for flow cytometry as described (36). In experiments examining the effect of γ-irradiation on cell cycle distribution, human colon cancer cells were exposed for 2 min to 4-Gy γ-irradiation, and cells were then harvested 24 hr later. Cell cycle analysis was performed by using a Becton-Dickinson fluorescence-activated FACStarPLUS cell analyzer and sobr model analysis program provided by the manufacturer.
RESULTS
Characterization of HCT-8 and HCT-C Cells.
The HCT-8 cell line was selected as our model system to investigate the interaction between TS and p53 mRNA. This was based on previous work that HCT-8 cells express wild-type p53 (37). A mutant subline HCT-C was established by Hoganson et al. (26) in which the TS protein had been rendered functionally inactive by a missense mutation at amino acid 216. This specific point mutation results in the near complete inactivation of TS enzyme activity. TS catalytic assay were used to confirm that HCT-C cells expressed significantly lower TS enzyme activity (0.33 pmol per min per mg of protein) than that observed in their corresponding parent HCT-8 cells (3.79 pmol per min per mg of protein). Western immunoblot analysis revealed nearly undetectable levels of TS protein in the HCT-C line when compared with the parent HCT-8 cell line despite the use of an ultrasensitive chemilluminescence detection method (Fig. 1). This finding is in contrast to previous studies by Berger et al. (27) who reported that a different anti-TS monoclonal antibody could readily detect the mutant TS protein. However, when an anti-TS polyclonal antibody was used, the levels of TS protein in HCT-8 and HCT-C cells were identical (data not shown). Thus, these findings suggest that the mutant TS protein expressed in HCT-C cells may adopt a conformation that is not easily detected by the anti-TS monoclonal antibody used in our studies.
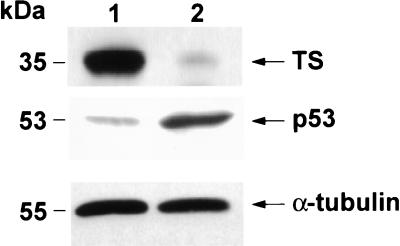
Figure 1.
Characterization of HCT-8 and HCT-C cells. Cytosolic extracts from HCT-8 (lane 1) and HCT-C (lane 2) cells were prepared and Western immunoblot analyses were performed.
In contrast, the expression of p53 protein in HCT-C cells was significantly higher than that observed in parent HCT-8 cells. Sequencing analysis of the entire coding region of the p53 cDNA confirmed the presence of a wild-type sequence in both the HCT-8 and HCT-C cell lines. Moreover, in vitro translation of the p53 cRNA derived from each cell line gave rise to an identical protein product with the predicted molecular mass of 53 kDa (data not shown).
Characterization of HCT-C and HCT-C:His-TS+.
Stable transfection of HCT-C cells with the full-length human TS changed their growth requirements as the HCT-C:His-TS+ cells were able to grow in thymidine-deficient RPMI 1640 medium. This was in sharp contrast to HCT-C cells, which required the presence of 10 μM thymidine for growth. However, the actual growth rate of the transfected cells was not significantly different from parent HCT-C cells, as the doubling time for each cell line was on the order of 22–24 hr. The TS-transfected cells expressed significantly higher TS enzyme activity, 8.03 pmol per min per mg of protein, than their corresponding HCT-C parent cells and markedly increased levels of human TS protein as visualized by Western blot analysis (Fig. 2).
Figure 2.
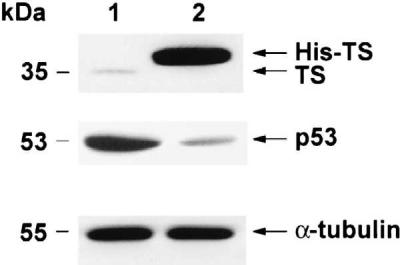
Characterization of HCT-C and HCT-C:His-TS+ cells. Cytosolic extracts from HCT-C (lane 1) and HCT-C:TS+ (lane 2) cells were prepared. Equal amounts of protein were loaded onto each lane.
To investigate whether a similar relationship existed between TS and p53 expression, a Western immunoblot analysis was performed to characterize the level of expression of p53 protein in parent HCT-C and transfected HCT-C:His-TS+ cells. As shown in Fig. 2, the expression of p53 protein was significantly reduced, by nearly 5-fold, in HCT-C:His-TS+ cells overexpressing human TS (Fig. 2, lane 2). To provide support for the specificity of this effect of TS on p53 expression, the levels of α-tubulin were identical in parent HCT-C (Fig. 2, lane 1) and TS-transfected cells (Fig. 2, lane 2) by Western immunoblot analysis. In addition, the expression of other proteins including β-actin, mdm-2, and DHFR remained unchanged upon transfection with human TS (data not shown).
RNase Protection Assay.
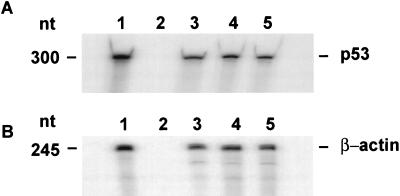
To identify the level at which the decreased expression of p53 was controlled, an RNase protection assay was performed to compare the levels of p53 mRNA in HCT-8, HCT-C, and HCT-C:His-TS+ cells. The level of expression of p53 mRNA was identical in parent HCT-8 (Fig. 3A, lane 3), mutant HCT-C (Fig. 3A, lane 4), and TS-transfected HCT-C:His-TS+ cells (Fig. 3A, lane 5). A control RNase protection experiment revealed no differences in the level of expression of β-actin mRNA in these cell lines (Fig. 3B).
Figure 3.
RNase protection assay of p53 mRNA and β-actin levels in HCT-8 cells. (A) p53 mRNA. Equal amounts (20 μg) of total RNA from HCT-8 (lane 3), HCT-C (lane 4), and HCT-C:His-TS+ (lane 5) cells were incubated with a radiolabeled p53 antisense RNA probe. In vitro-transcribed p53 mRNA (lane 1) and yeast tRNA (lane 2) were incubated with the same radiolabeled p53 antisense RNA probe and processed as above. (B) β-actin mRNA. Equal amounts of total RNA from HCT-8 (lane 3), HCT-C (lane 4), and HCT-C:His-TS+ (lane 5) cells were incubated with a radiolabeled β-actin antisense RNA. In vitro transcribed β-actin mRNA (lane 1) and 50 μg of yeast tRNA (lane 2) were incubated with the same radiolabeled β-actin antisense RNA probe and processed as above.
Effect of TS on p53 Synthesis and Stability.
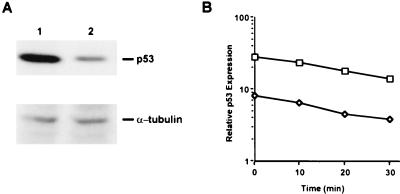
The effect of overexpression of TS on p53 biosynthesis was investigated by a pulse-labeling immunoprecipitation analysis. The level of p53 synthesis was significantly higher (5-fold) in HCT-C cells (Fig. 4A, lane 1) when compared with transfected HCT-C:His-TS+ cells (Fig. 4A, lane 2) overexpressing human TS. To confirm specificity and to show that the decrease in labeling of p53 in HCT-C:His-TS+ did not result from an artifact in loading or from inhibition of global protein synthesis, a control immunoprecipitation experiment was performed with an anti-α-tubulin monoclonal antibody. The levels of newly synthesized α-tubulin protein were identical in parent HCT-C (Fig. 4A, lane 1) and TS-transfected HCT-C:His-TS+ cells (Fig. 4A, lane 2).
Figure 4.
(A) Measurement of p53 biosynthesis in HCT-C (lane 1) and HCT-C:TS+ (lane 2) cells. (B) Determination of the half-life of p53 protein in HCT-C (□) and HCT-C:TS+ (◊). Each point represents the mean ± SEM of at least three experiments.
The half-life of p53 protein in control HCT-C and TS-transfected HCT-C:His-TS+ cells was next characterized. As seen in Fig. 4B, the half-life of p53 in parent HCT-C and HCT-C:His-TS+ cells was 15–20 min in each case, a finding that is consistent with published results (19, 20). Because no alteration in the stability of the p53 protein in the TS-transfected cells was observed when compared with parent HCT-C cells, this finding indicates that the markedly decreased level of p53 protein is not due to an enhanced degradation of p53 protein but is rather the result of an absolute decrease in biosynthesis of p53.
Immunoprecipitation of TS–RNP Complexes in HCT-C:TS+ Cells.
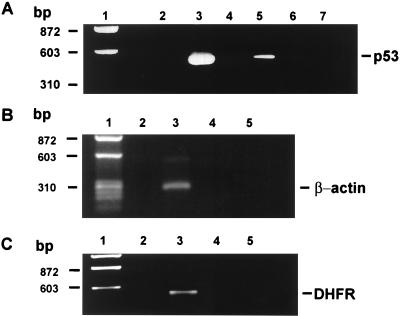
The studies presented, thus far, suggest a direct interaction between human TS protein and human p53 mRNA in TS-transfected cells. However, it was important to confirm the presence of such an in vivo RNA–protein interaction. In previous studies, an immunoprecipitation/RT-PCR method was used to identify a TS–RNP complex in cultured human colon cancer cells that was composed of TS and its own TS mRNA (11). This same strategy was used in the present study to isolate a TS–RNP complex containing the transfected human His-Tag TS protein and p53 mRNA in HCT-C:His-TS+ cells. TS–RNP complexes were immunoprecipitated with a specific anti-His-Tag monoclonal antibody, and the nucleic acid fraction specifically bound to TS was isolated and subjected to RT-PCR amplification using p53-specific primers. A 489-nt DNA fragment corresponding to nucleotides 531–1020 of p53 mRNA was amplified from HCT-C:His-TS+ cells (Fig. 5A, lane 5) but not from its corresponding HCT-C cells (Fig. 5A, lane 4). This band resolved at the same position as a DNA product obtained from a PCR using the identical set of p53-specific primers and the 1587-nt p53 cDNA as DNA template (Fig. 5A, lane 3). To confirm specificity, either no antibody (Fig. 5A, lane 6) or an unrelated antibody, anti-α-tubulin, (Fig. 5A, lane 7) was included in the immunoprecipitation reaction. With these control antibodies, no amplification of the 489-nt DNA product was observed.
Figure 5.
(A) RT-PCR analysis of p53 RNA immunoprecipitated from human colon cancer cells. The 1587-nt p53 cDNA was used as DNA template in a control PCR amplification reaction using p53-specific primers (lane 3). Lane 2 represents a control reaction with p53-specific primers and no exogenously added nucleic acid (lane 2). Whole-cell extracts from HCT-C (lane 4) and HCT-C:His-TS+ (lane 5) cells were immunoprecipitated with anti-His-Tag TS monoclonal antibody. Whole-cell extracts from HCT-C:His-TS+ cells were immunoprecipitated with no antibody (lane 6) or with an anti-α-tubulin monoclonal antibody (lane 7). (B) RT-PCR analysis of β-actin RNA immunoprecipitated from cells. The single-strand cDNA synthesized from total cellular RNA was used as the DNA template in a control PCR amplification reaction using β-actin-specific primers (lane 3). Lane 2 represents a control reaction with only β-actin-specific primers and no exogenously added nucleic acid (lane 2). Whole-cell extracts from HCT-C (lane 4) and HCT-C:His-TS+ (lane 5) cells were immunoprecipitated with anti-His-Tag TS monoclonal antibody, and the isolated nucleic acid fraction was then RT-PCR amplified by using the same β-actin-specific primers as in the control reaction. (C) RT-PCR analysis of DHFR RNA immunoprecipitated from cells. The single-strand cDNA was used as the DNA template in a control PCR amplification reaction using DHFR-specific primers (lane 3). Lane 2 represents a control reaction with only DHFR-specific primers and no exogenously added nucleic acid (lane 2). Whole-cell extracts from HCT-C (lane 4) and HCT-C:His-TS+ (lane 5) cells were immunoprecipitated with anti-His-Tag TS monoclonal antibody, and the isolated nucleic acid fraction was then RT-PCR-amplified by using the same DHFR-specific primers as in the control reaction.
To determine whether the TS–RNP complexes contained RNAs other than p53, we used the same RT-PCR method, but included primer sets specific for unrelated genes such as β-actin (Fig. 5B) and DHFR (Fig. 5C). Neither of these cellular RNAs formed an RNP complex with the human His-Tag TS protein. Control experiments revealed that each of these primer sets were able to amplify their respective genes from total cellular RNA isolated from HCT-C:His-TS+ cells by PCR (Fig. 5 B, lane 3, and C, lane 3). This finding provides additional support for the specificity of the TS protein–p53 RNA in vivo interaction.
RNA Gel Shift Analysis of Binding of Human Recombinant His-Tag TS Protein to Human p53 mRNA.
An RNA gel mobility shift assay was used to confirm that human recombinant His-Tag TS protein could directly interact with human p53 mRNA. Recombinant human His-Tag TS protein bound with high affinity to human p53 mRNA (Kd = 1.2 nM; data not shown). This binding affinity is on the same order of magnitude as that previously observed for the human recombinant TS protein–p53 mRNA interaction (Kd = 1.5 nM) (13).
Characterization of HCT-C and HCT-C:TS+ Cells.
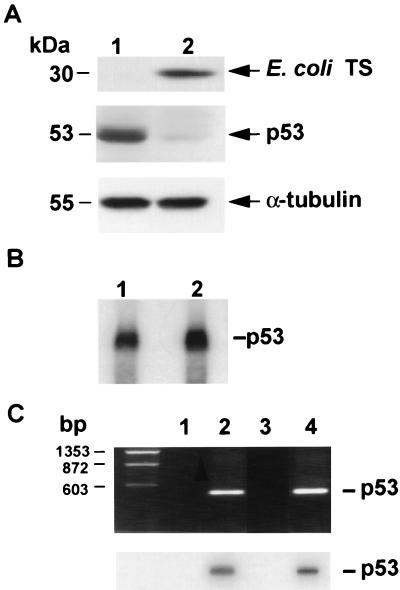
The studies presented, thus far, suggest that human TS directly regulates the expression of p53 at the translational level. Although TS represents one of the most highly conserved proteins identified to date, one issue we wished to investigate was whether a different species of TS, such as E. coli, could regulate the expression of p53 in a similar manner. For these studies, mutant HCT-C cells were transfected with the pcDNA3.1 plasmid containing the full-length E. coli TS cDNA sequence to give the HCT-TS+ cell line. HCT-C:TS+ cells and their parent HCT-C cells have a similar growth rate with the doubling time of 23 and 24 hr, respectively. HCT-C: TS+ cells (Fig. 6A, lane 2) expressed significantly higher levels of TS than parent HCT-C cells (Fig. 6A, lane 1). However, the expression of p53 protein was reduced by more than 10-fold in HCT-C:TS+ cells overexpressing E. coli TS. The levels of α-tubulin were found to be identical in parent (Fig. 6A, lane 1) and TS-transfected cells (Fig. 6A, lane 2). RNA Northern blot analysis revealed nearly identical levels of p53 mRNA in HCT-C (Fig. 6B, lane 1) and transfected HCT-C:TS+ cells (Fig. 6B, lane 2). Pulse-labeling immunoprecipitation studies were performed and confirmed that the half-life of the p53 protein remained unaffected by the increased levels of E. coli TS in the transfected cells (data not shown). Thus, these results demonstrate that E. coli TS protein can translationally regulate the expression of p53 in a manner identical to that observed in the human species.
Figure 6.
(A) Western immunoblot analysis of p53 in HCT-C cells. Cytosolic extracts from HCT-C (lane 1) and HCT-C:TS+ (lane 2) cells were prepared. TS protein was detected by immunoblot analysis by using an anti-E. coli TS polyclonal antibody. Filter membranes were also probed with an anti-p53 monoclonal antibody and then reprobed with an anti-α-tubulin monoclonal antibody to control for loading and integrity of protein. (B) RNA Northern blot analysis of p53 mRNA levels in HCT-C and HCT-C:TS+ cells. (C) RT-PCR analysis of p53 RNA immunoprecipitated from each cells. The 1587-nt p53 cDNA was used as DNA template in a control PCR amplification reaction using p53-specific primers (Upper, lane 2). Lane 1 represents a control reaction with only p53-specific primers and no exogenously added nucleic acid. Whole-cell extracts from HCT-C (lane 3) and HCT-C:TS+ (lane 4) cells were immunoprecipitated with anti-E. coli TS monoclonal antibody. The PCR products were then transferred onto a nitrocellulose membrane. Hybridization of the filter membrane with a radiolabeled p53 RNA antisense probe confirmed that these PCR-amplified products were specific for p53 (Lower).
An immunoprecipitation/RT-PCR method was performed to confirm the presence of the E. coli TS–p53 mRNA complexes in transfected HCT-C:TS+ cells. The p53 sequence was isolated from HCT-C:TS+ cells transfected with and expressing E. coli TS protein (Fig. 6C Upper, lane 4). In contrast, the same p53 RNA sequence was not RT-PCR-amplified from HCT-C cells. Hybridization of the filter membrane with a radiolabeled p53 RNA antisense probe confirmed that these PCR-amplified products were specific for p53 (Fig. 6C Lower, lane 4). These findings provide further support for the specificity of the in vivo TS protein–p53 mRNA interaction.
Cell Cycle Analysis.
Given the well-documented role of p53 in the G1-phase cell cycle checkpoint, we next investigated the effect of suppression of p53 expression on this critical cellular function. For these experiments, the parent HCT-8, the mutant HCT-C, and the human TS-transfected HCT-C:His-TS+ cells were investigated. There were no alterations in the normal cell cycle distribution between these three cell lines, as determined by flow cytometric analysis. The extent to which these three cell lines were able to arrest at the G1/S-phase boundary of the cell cycle after exposure to a DNA-damaging agent was evaluated by exposure of each cell line to the same dose of γ-irradiation (4 Gy). The G1/S-phase block is displayed by comparing the ratio of the percentage of cells in the G1 phase relative to S phase in untreated and treated cells. Mutant HCT-C cells, expressing detectable levels of wild-type p53, exhibited a pronounced 4.5- to 5-fold increase in the G1/S-phase ratio after γ-irradiation (Fig. 7). In contrast, parent HCT-8 cells and TS-transfected HCT-C:His-TS+ cells, both expressing nearly undetectable levels of p53 relative to HCT-C cells, were severely impaired in their ability to arrest in G1 phase after γ-irradiation (Fig. 7).
Figure 7.
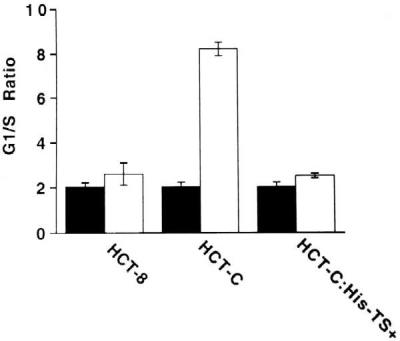
Measurement of G1/S-phase arrest after treatment with γ-irradiation in HCT-8, HCT-C, and HCT-C:His-TS+ cells. G1/S-phase ratios for human colon cancer cells were determined in the absence (solid bars) and presence (open bars) of γ-irradiation (4 Gy), as indicated. Cell cycle distribution was quantitated by flow cytometry.
DISCUSSION
In this study, we used as our experimental model system a human colon cancer HCT-8 cell line and a mutant HCT-C subline in which the TS protein had been rendered marginally active and largely immunologically undetectable. The HCT-8 cell line expresses wild-type p53. Transfection of the mutant HCT-C cell line with the pcDNA3.1 plasmid containing the entire human TS cDNA gave rise to a HCT-C:His-TS+ cell line that expressed significantly higher levels of TS enzyme activity (24.3-fold) and markedly elevated (25-fold) levels of immunoreactive TS protein. However, the expression of p53 protein was significantly reduced, by 5-fold, in cells transfected with and expressing higher levels of the human TS protein. Although our analysis of the effect of TS on the expression of cellular proteins was restricted to only a limited number of other proteins including α-tubulin, β-actin, DHFR, and mdm2, our results suggest that one of the intracellular downstream targets of human TS is p53 biosynthesis.
To specifically address the concern of decreased expression of mutant TS via Western immunoblot in HCT-C cells, we performed a Western immunoblot analysis with an anti-TS monoclonal antibody or an anti-TS polyclonal antibody by using cellular extracts from HCT-8 and HCT-C cells. Our results show that the levels of TS protein are identical in both cell lines when the anti-TS polyclonal antibody was used (data not shown). However, when the anti-TS monoclonal antibody was used as described in Fig. 1, the immunostaining of TS in HCT-C cells was significantly weaker when compared with TS in parental HCT-8 cells. In addition, previous studies (27) have shown that wild-type human TS and various mutant TS proteins including the Ser → Leu mutant isolated from HCT-C cells are recognized to the same extent with an anti-TS monoclonal antibody that is different than the one used in our study. These results suggest that the near absent staining of TS protein in the HCT-C subline is most likely caused by the inability of our anti-TS monoclonal antibody to recognize certain key epitopes in this mutant TS protein and that it is not due to an alteration in protein stability.
A series of experiments were subsequently performed to characterize the molecular level at which human TS was able to suppress the expression of p53. Our studies have shown that (i) the level of p53 mRNA is identical in HCT-C and TS-transfected HCT-C:TS+ cells, (ii) the half-life of p53 protein in HCT-C and TS-transfected HCT-C:TS+ cells is nearly identical—being on the order of 20 min for each cell line, (iii) the level of newly synthesized p53 is significantly higher (5-fold) in HCT-C cells when compared with TS-transfected HCT-C:TS+ cells, (iv) immunoprecipitable TS–RNP complexes containing human His-Tag protein and p53 mRNA are present in TS-transfected cells but not in nontransfected cells, and (v) there is direct binding of human recombinant His-Tag TS protein to human p53 mRNA. Thus, these results indicate that the decreased level of p53 protein expression in TS-transfected cells is due to an absolute decrease in the translational efficiency of p53 mRNA, and these findings are consistent with a scenario in which the direct binding of human TS protein to p53 mRNA results in translational repression.
TS has been purified and well-characterized from various species including human, E. coli, bacteriophage T4, yeast, several viruses, parasites, mouse, and rat (2, 5, 38). Careful analysis of the predicted primary amino acid sequences of TS isolated from nearly 30 different species reveals that it is one of the most highly conserved proteins identified to date (5, 38). Specifically, there is nearly complete sequence homology at both the folate-binding and nucleotide-binding regions of these proteins. With this in mind, we characterized the effect of E. coli TS on p53 expression in HCT-C cells, and virtually identical results were observed with the human and E. coli TS species. This finding suggests that the domain(s) required for binding to p53 mRNA and for regulating p53 expression are highly conserved between these two species. Previous studies suggest that the folate-binding domain, as well as the nucleotide active site cysteine sulfhydryl, may play important elements in this process (9, 10). However, it remains unclear as to whether these two sites directly interact with RNA or whether they maintain the protein in a certain conformational state that allows the actual domains access for RNA binding. Our recent work has identified a 35-amino acid domain in the folate-binding region that directly interacts with human TS mRNA (39). Of interest, this sequence is nearly completely conserved between the human and E. coli TSs.
Although the human TS protein in mutant HCT-C cells contains only a single amino acid mutation at position 216, our studies suggest that this mutant TS protein has lost its capacity to interact with p53 mRNA. Recent work from Berger’s group (27) has suggested that the conformation of this mutant protein may be different than that assumed by wild type TS. This finding would be consistent with our observation that the anti-TS 106 monoclonal antibody was unable to detect this mutant protein on Western immunoblots. Thus, it is conceivable that the conformation of this mutant protein may be altered in such a way that the actual RNA binding domain on TS is no longer accessible.
There are now several well-characterized mechanisms that mediate the regulation of p53 expression and function. In normal cells and under physiologic conditions, it appears that the expression of p53 protein is maintained at relatively low levels (16–18, 40). However, on exposure of cells to DNA-damaging agents such as UV irradiation and/or various cytotoxic agents, p53 levels are markedly increased. The experimental evidence, to date, suggests that translational as well as post-translational events are involved in this process (18–20, 22, 40). With regard to malignant tumors, wild-type p53 function is inactivated by several different mechanisms. For example, the adenovirus E1b and simian virus 40 tumor antigen proteins directly inactivate p53 transcriptional activity (15), whereas binding of the human papilloma viral protein E6 to p53 results in its rapid degradation (41). As an additional example, the mdm2 oncoprotein, found to be expressed in a number of human tumors, physically interacts with p53, and this protein–protein interaction results in inhibition of p53 transcriptional activity (40, 42). In addition to these examples of post-translational regulation, there now is growing evidence that the expression of wild-type p53 is controlled through a negative autoregulatory feedback pathway in which p53 protein binds to its own p53 mRNA, an interaction that results in translational inhibition similar to that described for TS (22–25). This process of translational autoregulation may represent an important mechanism by which the expression of p53 can be tightly and efficiently controlled. The studies described herein, provide further evidence for the role of translational regulation in determining the intracellular levels of p53 protein, but in the present case, we have identified a molecular pathway in which the translation of p53 mRNA is controlled by the binding of TS to the p53 mRNA.
With regard to the potential biological relevance of the TS protein–p53 mRNA interaction, we have shown that the suppression of p53 synthesis is observed not only in HCT-C:His-TS+ cells overexpressing TS after gene transduction but also in parent HCT-8 cells expressing basal levels of TS. This finding suggests that the levels of TS are sufficiently high even in nontransfected cells to repress p53 mRNA translation and subsequent synthesis of p53 protein. Thus, the interaction between TS protein and p53 mRNA appears to be taking place under both selected and physiologic conditions. Our preliminary results demonstrate that parent HCT-8 cells and cells transfected with and overexpressing TS are each impaired to a significant degree in their respective G1-phase checkpoint function after exposure to a DNA-damaging agent, γ-irradiation. A disruption in G1-phase arrest has been shown to have two main consequences. One is that the suppression of p53 by TS can abrogate G1-phase arrest and the subsequent process of DNA repair, resulting in the generation of genetically unstable DNA. The second is that on exposure to a DNA-damaging agent, such cells are unable to undergo the process of apoptosis, resulting in a drug-resistant phenotype. In contrast, in cells expressing a mutant TS protein, as is observed in mutant HCT-C cells, the TS protein–p53 mRNA interaction is not allowed to occur. Such a situation would lead to the efficient translation of p53 mRNA and subsequent synthesis of p53 protein, resulting in maintenance of normal cell cycle control and checkpoint function.
Further work is required to more carefully elucidate the in vivo biological consequences of the TS protein–p53 mRNA interaction. Studies are underway to determine whether malignant cells overexpressing TS display a more aggressive and/or drug-resistant phenotype. Our preliminary studies suggest that this impaired G1-phase checkpoint function may result in the development of cellular drug resistance. This finding is consistent with previous studies that have demonstrated that endogenous p53 status may effectively predict in vitro chemosensitivity (43–45). Because it has been shown that malignant tumors express relatively higher levels of TS than their normal tissue counterparts presumably due to enhanced metabolic requirements for DNA synthesis by the tumor, our results may provide a rational mechanism for the inherent resistance of tumors expressing wild-type p53 to TS inhibitor compounds and to other anticancer agents.
In conclusion, the studies presented herein provide direct evidence that TS, whether it be human or E. coli, specifically regulates the in vivo expression of p53 at the translational level. This work expands our current understanding of the various molecular mechanisms that control the expression of p53 and provides further evidence for the role of TS as an important regulator of cellular gene expression. Additional experiments are required to more carefully elucidate the specific molecular elements underlying the interaction between TS and p53 mRNA. It will also be important, given the critical role of p53 in the control of the G1- and G2-phase cell cycle checkpoints and apoptosis, to investigate in further detail, the effect of the TS protein-p53 mRNA interaction on these important downstream events. However, this study suggests a pathway by which the expression of p53 can be suppressed in vivo, and it provides further evidence for the fundamental role of translational regulation in the control of cellular gene expression.
Acknowledgments
We thank Drs. Yung-chi Cheng, Bruce Dolnick, John Schmitz, Leslie Parsels, Xiukun Lin, and Tian-Men Chen for helpful discussions and for review of the manuscript and Ms. Edna McCarthy for editorial assistance in the preparation of this manuscript. This work was supported, in part, by grants from the National Cancer Institute (CA44355 to F.M., CA16359 and CA75712 to E.C., and the Department of Veterans Affairs (VA Merit Review Award to E.C.).
ABBREVIATIONS
- TS
thymidylate synthase
- RNP
ribonucleoprotein
- RT-PCR
reverse transcription-coupled PCR
- DHFR
dihydrofolate reductase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Friedkin M, Kornberg A. Chemical Basis of Heredity. Baltimore: John Hopkins Press; 1957. pp. 609–614. [Google Scholar]
- 2.Carreras C, Santi D V. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 3.Cohen S S, Flaks J G, Barner H D, Lichtenstein J. Proc Natl Acad Sci USA. 1958;44:1004–1012. doi: 10.1073/pnas.44.10.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danenberg P V. Biochem Biophys Acta. 1977;473:73–79. doi: 10.1016/0304-419x(77)90001-4. [DOI] [PubMed] [Google Scholar]
- 5.Hardy L W, Finer-Moore J S, Montfort W R, Jones M O, Santi D V, Stroud R M. Science. 1987;235:448–455. doi: 10.1126/science.3099389. [DOI] [PubMed] [Google Scholar]
- 6.Appelt K, Baquet R J, Bartlett C A, Booth C L. J Med Chem. 1993;34:1925–1934. doi: 10.1021/jm00111a001. [DOI] [PubMed] [Google Scholar]
- 7.Chu E, Koeller D M, Casey J L, Drake J C, Chabner B A, Elwood P C, Zinn S, Allegra C J. Proc Natl Acad Sci USA. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu E, Voeller D, Koeller D M, Drake J C, Takimoto C H, Maley G, F, Maley F, Allegra C J. Proc Natl Acad Sci USA. 1993;90:517–521. doi: 10.1073/pnas.90.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voeller D M, Changchien L M, Maley G F, Maley F, Takechi T, et al. Nucleic Acids Res. 1995;23:869–875. doi: 10.1093/nar/23.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu E, Voeller D M, Morrison P F, Jones K L, Takechi T, Maley G F, Maley F, Allegra C J. J Biol Chem. 1994;269:20289–20293. [PubMed] [Google Scholar]
- 11.Chu E, Voeller D M, Jones K L, Takechi T, Maley G F, Maley F, Segal S, Allegra C J. Mol Cell Biol. 1994;14:207–213. doi: 10.1128/mcb.14.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu E, Takechi T, Jones K L, Voeller D M, Copur S M, Maley G F, Maley F, Segal S, Allegra C J. Mol Cell Biol. 1995;15:179–185. doi: 10.1128/mcb.15.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu E, Cogliati T, Copur S M, Borre A, Voeller D M, Allegra C J, Segal S. Nucleic Acids Res. 1996;24:3222–3228. doi: 10.1093/nar/24.16.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine A J, Momand J, Finlay C A. Nature (London) 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 15.Levine A J. Annu Rev Biochem. 1993;62:623–651. doi: 10.1146/annurev.bi.62.070193.003203. [DOI] [PubMed] [Google Scholar]
- 16.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 17.Vogelstein B, Kinzler K W. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 18.Maltzman W L, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 20.Zhan Q, Carrier F, Fornace A J., Jr Mol Cell Biol. 1993;13:4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewen M E, Miller S J. Biochem Biophys Acta. 1996;1242:181–184. doi: 10.1016/0304-419x(95)00010-d. [DOI] [PubMed] [Google Scholar]
- 23.Mosner, J., Mummenbrauer, T., Bauer, C., Sczakiel, F. G. & Deppert, W. EMBO J.14, 4442–4449. [DOI] [PMC free article] [PubMed]
- 24.Fu L, Minden M D, Benchimol S. EMBO J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 25.Fu L, Benchimol S. EMBO J. 1997;13:4117–4125. doi: 10.1093/emboj/16.13.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoganson, D. K., Williams, A. & Berger, S. H. (1999) Biochem. Pharmacol., in press. [DOI] [PubMed]
- 27.Williams A W, Dunlap R B, Berger S H. Biochemistry. 1998;37:7096–7102. doi: 10.1021/bi972562+. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen-Lane J, Maley G F, Chu E, Maley F. Protein Exp Purif. 1997;10:256–262. doi: 10.1006/prep.1997.0750. [DOI] [PubMed] [Google Scholar]
- 29.Belfort M, Maley G, Pedersen-Lane J, Maley F. Proc Natl Acad Sci USA. 1983;80:4914–4918. doi: 10.1073/pnas.80.16.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufman R J, Sharp P A. Mol Cell Biol. 1982;2:1304–1319. doi: 10.1128/mcb.2.11.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu E, Drake J C, Koeller D M, Zinn S, Jamis-Dow C A, Yeh G C, Allegra C J. Mol Pharmacol. 1991;39:136–143. [PubMed] [Google Scholar]
- 32.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Steitz J. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- 34.Sacchi N, Chomczynski P. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 35.Harford J. Nature (London) 1984;311:493–495. [Google Scholar]
- 36.Fried J, Perez A G, Clarkson V D. J Histochem Cytochem. 1978;26:921–926. doi: 10.1177/26.11.82573. [DOI] [PubMed] [Google Scholar]
- 37.Matsui S I, Arredondo M A, Wrzosek C, Rustum Y M. Cancer Res. 1996;56:4715–4723. [PubMed] [Google Scholar]
- 38.Perry K M, Fauman E B, Finer-Moore J S, Montfort W R, Maley G F, Maley F, Stroud R M. Proteins. 1990;8:315–333. doi: 10.1002/prot.340080406. [DOI] [PubMed] [Google Scholar]
- 39.Chu E, Allegra C J. Bioessays. 1996;18:191–198. doi: 10.1002/bies.950180306. [DOI] [PubMed] [Google Scholar]
- 40.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 42.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 43.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 44.Lowe S W, Ruley H E, Jacks T, Houseman D E. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 45.Wu G S, El-Deiry W S. Nat Med. 1996;2:255–256. doi: 10.1038/nm0396-255a. [DOI] [PubMed] [Google Scholar]