Abstract
The Drosophila gene bicoid functions as the anterior body pattern organizer of Drosophila. Embryos lacking maternally expressed bicoid fail to develop anterior segments including head and thorax. In wild-type eggs, bicoid mRNA is localized in the anterior pole region and the bicoid protein forms an anterior-to-posterior concentration gradient. bicoid activity is required for transcriptional activation of zygotic segmentation genes and the translational suppression of uniformly distributed maternal caudal mRNA in the anterior region of the embryo. caudal genes as well as other homeobox genes or members of the Drosophila segmentation gene cascade have been found to be conserved in animal evolution. In contrast, bicoid homologs have been identified only in close relatives of the schizophoran fly Drosophila. This poses the question of how the bicoid gene evolved and adopted its unique function in organizing anterior–posterior polarity. We have cloned bicoid from a basal cyclorrhaphan fly, Megaselia abdita (Phoridae, Aschiza), and show that the gene originated from a recent duplication of the direct homolog of the vertebrate gene Hox3, termed zerknüllt, which specifies extraembryonic tissues in insects.
Drosophila body pattern formation is initiated in response to asymmetrically distributed proteins, emanating from prelocalized mRNA in the pole regions of the egg (1–3). The factor required for the establishment of the anterior body part, including head and thorax, is encoded by the homeobox gene bicoid (3). bicoid, which is located within the homeotic gene complex (Hox-C) next to zerknüllt (Fig. 1A; reviewed in refs. 4–7), is expressed maternally in response to a general transcription factor, encoded by the gene serendipity, in the nurse cell/oocyte syncytium (8). The bicoid mRNA is transferred into the oocyte and becomes localized in the anterior pole region of the egg. After egg deposition and translation of the transcript, the bicoid protein (Bicoid), and to a lesser degree the mRNA, spread posteriorly, thereby generating a concentration gradient of the protein (3).
Figure 1.
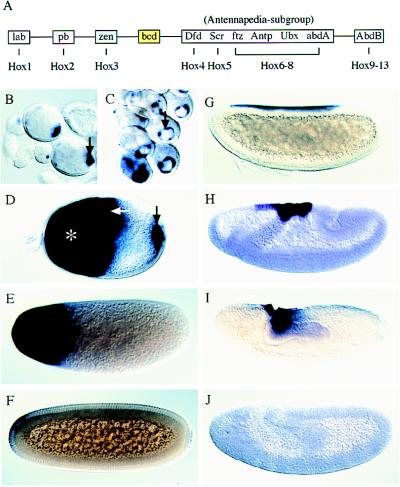
Hox gene cluster and expression patterns of Ma-bcd and Ma-zen in Megaselia. (A) The Hox-C of Drosophila and homologous genes in vertebrates (Hox1–13) (4–7). Dfd (Deformed), Scr (Sex combs reduced), ftz (fushi tarazu), Antp (Antennapedia), Ubx (Ultrabithorax), and abdA (abdominal A) form a subgroup of Antennapedia related genes. lab, labial; pb, proboscipedia; zen, zerknüllt; bcd, bicoid; AbdB, Abdominal B. (B to F) In situ hybridization showing the transcript distribution of Ma-bcd in the oocyte (B–D; arrows), nurse cells (D; asterisk), and embryos before (E) and during (F) cellularization. (G–J) Patterns of Ma-zen transcripts before (G), during (H), and after (I, J) gastrulation. In Drosophila, zerknüllt is expressed also in the pole cells (22).
The Bicoid gradient controls two regulatory aspects of gene expression in the early embryo. Firstly, it acts as a threshold-dependent transcriptional activator of zygotic segmentation genes, which are required to metamerize the anterior region of the embryo and to specify the segments and pattern elements (3). Secondly, Bicoid prevents the translation of uniformly distributed maternal caudal mRNA in the anterior region of the early embryo and thereby causes a second homeodomain transcription factor gradient, that of Caudal, in the opposite direction to Bicoid (2). The combined activities of the two transcription factors are necessary to activate the zygotic segmentation gene cascade in the precellular blastoderm embryo (2). Whereas caudal genes have been identified in a large variety of species and consistently show posterior-to-anterior protein concentration gradients (refs. 9 and 10 and references therein), bicoid genes have not been found in species other than Drosophila or some related schizophoran flies (11, 12). Here we present evidence for the evolutionary origin of bicoid by a gene duplication event involving the insect Hox3 homolog zerknüllt.
MATERIALS AND METHODS
Cloning of Homologs.
PCR clones of Ma-bcd and Ma-zen were obtained by using the primer pairs ATGMGTMGTCCDMGDMGNAC/GCKGCKRTTYTTRAACCA (6) and CARCTBGTDGARCTIGARAAYGARTT/TTYTTRWAYTTCATICKICKRTTYTG, respectively, on genomic DNA. Genomic sequence was obtained from partially MboI digested DNA cloned in the phage vector Lambda FixII (Stratagene). For the Ma-bcd cDNA 3′ and 5′ rapid amplification of cDNA ends (RACE) experiments were performed. cDNA was prepared from 3 μg of poly(A)+ RNA according to the instructions of the Marathon cDNA amplification kit (CLONTECH). The ORF of Ma-zen was deduced from 2.1 kb of genomic sequence by using splice site and promotor prediction software from the Berkley Drosophila Genome Project Sequence Analysis Tools (www.fruitfly.org/reg_tools) package. The sequence includes two promotor consensus sequences and a polyadenylation signal in the 3′ untranslated region. The Ma-bcd and Ma-zen sequences are deposited in the GenBank database (accession nos. AJ133024, AJ133025).
Whole-Mount in Situ Hybridization.
Hybridization to Megaselia embryos was done with DNA probes at 48°C as described for Drosophila (13) with the following modifications. To burst the vitelline layer, −80°C cold methanol was used in the devitellinization step, and the embryos were heated in methanol for 1 min to +70°C. The temperature shock was repeated two to three times during the following methanol washes.
Phylogenetic Analysis.
Sequence similarities in percentage were calculated with the PAM250 residue weight table by using megalign/dnastar software. Molecular phylogenetic trees were constructed from protein sequences to avoid possible distortion by codon usage differences. The Parsimony method and the Neighbor Joining method on a Dayhoff distance matrix, implemented by using phylip 3.572c, were applied (14). Topology robustness was assessed by 500 bootstrap resamplings. Statistical likelihood of alternative user-defined trees was assessed by a resampling of estimated likelihood method implemented with protml in molphy 2.2 (15).
RESULTS AND DISCUSSION
Cloning and Expression Patterns of Megaselia bicoid.
In a search for bicoid homologs in lower dipteran species, we used degenerated PCR primers to amplify DNA fragments containing the homeodomain-encoding region from a primitive cyclorrhaphan fly, Megaselia abdita. The cyclorrhaphan flies are divided into two subordinate groups: the Aschiza and the Schizophora. The phorid Megaselia is an aschizan fly (16). Therefore, it is different from the monophyletic group of schizophoran flies that includes Drosophila and the few other species where bicoid could be identified so far (11, 12). We isolated genomic DNA encompassing the transcription unit and performed 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments to clone the cDNA. The cDNA and corresponding genomic DNA were sequenced to establish the identity and structure of the bicoid homologous gene, termed Ma-bcd, and the transcript distribution during oogenesis and embryogenesis was visualized by in situ hybridization.
Ma-bcd transcripts accumulate first in the oocyte, where a transient ring-shaped pattern is observed (Fig. 1 B and C). Later, during oogenesis, transcripts are expressed in the nurse cells; they accumulate in the anterior region of the oocyte and transiently at its posterior pole (Fig. 1D). In the early embryo, Ma-bcd transcripts spread from the anterior pole forming an enlarging anterior cap until the onset of cellularization (Fig. 1E). Subsequently, transcripts disappear rapidly (Fig. 1F). No zygotic expression was observed during embryogenesis (not shown). These findings establish identical expression patterns of bicoid and Ma-bcd in Drosophila (3, 17) and Megaselia.
The Homeodomains of Ma-bcd and zerknüllt Are Closely Related.
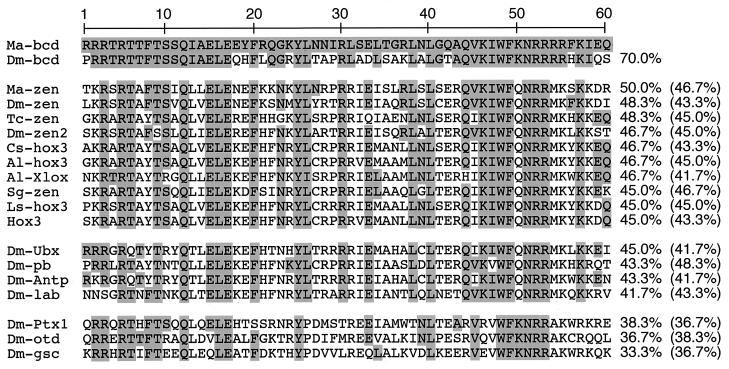
A comparison of the Ma-bcd homeodomain and homeodomain proteins of Drosophila indicates that aside from bicoid, Ma-bcd is most similar to zerknüllt (48.3%), whereas the similarity to homeodomains encoded by the other members of the Drosophila Hox-C is less pronounced (45.0%–36.7%) (Fig. 2 and data not shown). The homeodomain of Ma-bcd is related only distantly to the homeodomains encoded by orthodenticle and the Drosophila homologs of goosecoid and Ptx1 (38.3%–33.3%), which have been classified in the past as bicoid-related genes (Fig. 2). These proteins share common DNA-binding properties that depend on the diagnostic lysine in position 50 of the homeodomain (18–21). Notably, also the zerknüllt homologs of other insects and their orthologs in various animal classes, the Hox3 genes of chordates, ribbonworm, and spider, show a higher degree of similarity to the Ma-bcd homeodomain than do the bicoid-related genes (Fig. 2). These observations suggest that, in spite of the considerable sequence divergence exhibited by the Drosophila genes, bicoid and zerknüllt are closely related.
Figure 2.
Homeodomain alignment and percent sequence similarity relative to Ma-bcd and Dm-bcd (in brackets). Numbers refer to amino acid position. Abbreviations: Ma-bcd, bicoid of Megaselia (this work); Dm-bcd, bicoid of Drosophila (27); Ma-zen, zerknüllt of Megaselia (this work); Dm-zen, zerknüllt of Drosophila (22);Tc-zen, zerknüllt of beetle (7); Cs-hox3, Hox3 of spider (24); Al-hox3 (28) and Al-Xlox (29), Hox3 and Xlox of cephalochordate, respectively; Dm-zen2, zerknüllt-2 of Drosophila (22); Sg-zen, zerknüllt of grasshopper (7); Ls-hox3, Hox3 of ribbonworm (30); Hox3, consensus reported for the Hox3 paralogy group (7). Other abbreviations refer to Hox genes (cf. legend to Fig. 1; 31–34), Ptx1, orthodenticle and goosecoid of Drosophila (18–21). Identical amino acids (reference to Ma-bcd) are underlaid.
Ma-bcd and Ma-zen Are Sister Genes.
To address the hypothesis that bicoid and zerknüllt are the closest relatives among Hox genes, we cloned the Megaselia zerknüllt gene (Ma-zen) and asked whether Ma-zen provides a link between bicoid and the Hox3 genes of the vertebrate Hox-Cs.
We isolated Ma-zen by a PCR approach and isolated the genomic DNA encompassing the transcription unit. Whole-mount in situ hybridization of Megaselia embryos reveals that Ma-zen transcripts are expressed only zygotically. They form a restricted pattern at the dorsal side of the blastoderm embryo, covering the area of the amnioserosa precursor cells, and disappear in the extended germ-band stage (Fig. 1 G–J). Thus, Ma-zen is expressed like zerknüllt in Drosophila. This suggests that Ma-bcd and Ma-zen have separate functions in Megaselia, similar to the functions of their homologs in Drosophila (3, 22).
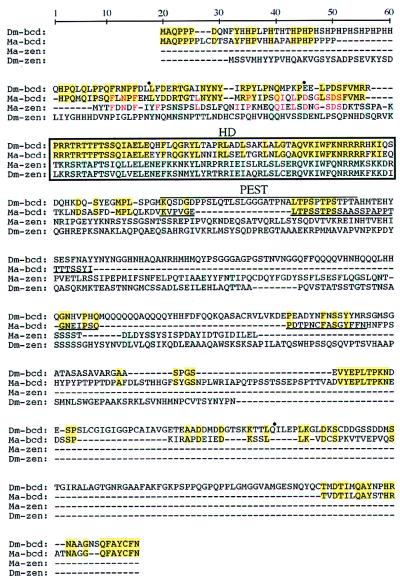
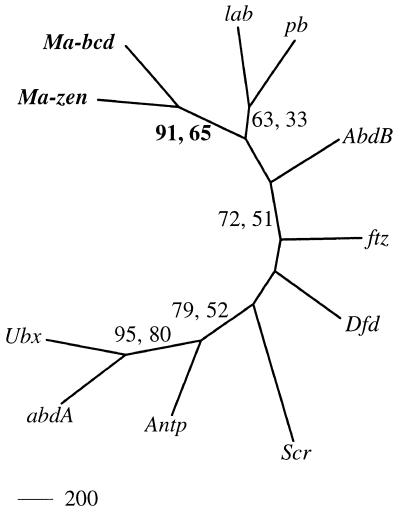
Sequence comparison of the predicted Ma-bcd and Ma-zen proteins clearly establishes a sister relationship of the two proteins (Figs. 2–4). Evidence for this is based on the following findings. The homeodomain of Ma-bcd shows a higher sequence similarity to Ma-zen (50.0%) than to any other nonorthologous homeodomain (Fig. 2). In addition, molecular phylogenetic trees involving the homeodomains of the Hox-C genes of Drospohila resolve with high confidence when the Ma-bcd and Ma-zen instead of the bicoid and zerknüllt sequences were used for the analysis (Fig. 3). It is important to note that the Drosophila homeodomains included in the tree in Fig. 3 evolve very slowly (except fushi tarazu) and can be assumed to be identical or almost identical in amino acid sequence in Megaselia and Drosophila (ref. 23; U.S.-O., unpublished results). To further test the statistical significance of this finding, we estimated likelihood values for the tree shown in Fig. 3 and for 11 modified trees where the position of Ma-bcd was changed. The tree shown in Fig. 3 is supported over the modified trees with a bootstrap value of 94.8%. Finally, the alignment of the Ma-bcd and Ma-zen proteins shows conservation of sequences not only in the homeodomain but also N-terminal to it (Fig. 4). The conserved sequences in front of the homeodomain are not evident from the comparison of bicoid and zerknüllt of Drosophila. Thus, the sister-gene relationship of bicoid and zerknüllt revealed by the Megaselia genes remains hidden when the obviously more diverged sequences of the Drosophila genes are compared.
Figure 4.
Proteins encoded by bicoid and zerknüllt of Megaselia (Ma-bcd, Ma-zen) and Drosophila (Dm-bcd, Dm-zen) (27, 22). Dashes indicate sequence gaps for optimal alignment. Amino acid identities of the bicoid proteins (yellow boxes) or the zerknüllt proteins (blue boxes) and between Ma-bcd and Ma-zen in front of the homeodomain (red letters) are highlighted. Dots above Dm-bcd mark intron positions; only the first two are conserved in Ma-bcd (64 bases and 11.5 kb). Homeodomains (HD) are boxed, PEST domains (35) are overlined (Dm-bcd) and underlined (Ma-bcd).
Figure 3.
Evolutionary relationship of Ma-bcd, Ma-zen, and Hox-C genes of Drosophila, as deduced by neighbor-joining analysis. Numbers refer to bootstrap percentages obtained from neighbor-joining (first value) and maximum parsimony analysis. Trees including the more diverged bicoid and zerknüllt genes of Drosophila remain unresolved with respect to the monophyletic cluster of bicoid and zerknüllt orthologs (bootstrap value below 50%; data not shown). For abbreviations, see legend to Figs. 1 and 2.
bicoid Is a Derived Hox Class 3 Gene.
The newly established sister-gene relationship implies that bicoid genes, like the zerknüllt genes (7, 24, 25), are direct homologs of the Hox3 genes in the Hox-C of noninsect animal classes. Thus, bicoid is a Hox gene in the phylogenetic sense, and the location of bicoid in the Hox-C of Drosophila (Fig. 1A) is an ancestral trait. The consistent failure to isolate bicoid from insects other than flies, which has been attempted in various laboratories (4, 26) including our own, suggests a recent origin of the bicoid gene. The fact that Ma-bcd is more similar to zerknüllt genes of higher insects than to other Hox3 homologs (Fig. 2) is consistent with the assumption that bicoid originated recently during insect radiation.
bicoid is expressed in the anterior egg region, where it exerts its role in patterning the anterior body of the larval fly. In contrast, zerknüllt and its orthologs function in extraembryonic anlagen. Although the extraembryonic anlage in flies, the amnioserosa, is located at the dorsal side of the blastoderm fate map (22), extraembryonic anlagen in other insects, such as the beetle Tribolium, are formed in an anterior egg position (7). This suggests that initially the sister genes bicoid and zerknüllt may have been coexpressed in the anterior egg region. The subsequent recruitment of bicoid in patterning the embryo, instead of determining the dorsally shifted extraembryonic anlagen, changed the selection conditions for the gene. Ensuing adaptations must have resulted in a new set of target genes (1, 2), as reflected by the characteristic lysine in Bicoid′s homeodomain position 50 that specifies DNA recognition (Fig. 2). The newly acquired functions of Bicoid entrained a significant change in the developmental mechanism of axis specification and furnish an outstanding model of molecular evolution in a patterning process.
Acknowledgments
We thank Heike Taubert for DNA work, Gordon Dowe for sequencing, Klaus Sander (Albert Ludwigs Universität, Freiburg, Germany) for the Megaselia culture and for advice on keeping the animals, Michael Akam (University Museum of Zoology, Cambridge, U.K.) and his former lab member Francesco Falciani for discussions, Hinrich Böger (Max-Planck-Institut für biophysikalische Chemie, Göttingen, Germany), and Ehab Abouheif (State University of New York, Stony Brook, NY) for helpful comments on the manuscript, and our colleagues Ralf Pflanz and Emanuel Busch for help with computers. The work was supported by the Max-Planck-Gesellschaft (H. J.). M.S. was funded by a grant from the Deutsche Forschungsgemeinschaft to U.S.-O.
ABBREVIATION
- Hox-C
homeotic gene complex
Footnotes
References
- 1.St Johnston D, Nüsslein-Volhard C. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 2.Rivera-Pomar R, Jäckle H. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 3.Driever W. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 301–324. [Google Scholar]
- 4.Akam M, Averof M, Castelli-Gair J, Dawes R, Falciani F, Ferrier D. Development (Cambridge, U.K.) Supplement; 1994. 209–215. [PubMed] [Google Scholar]
- 5.von Allmen G, Hogga I, Spierer A, Karch F, Bender W, Gyurkovics H, Lewis E. Nature (London) 1996;380:116. doi: 10.1038/380116a0. [DOI] [PubMed] [Google Scholar]
- 6.Randazzo F M, Seeger M A, Huss C A, Sweeney M A, Cecil J K, Kaufman T C. Genetics. 1993;133:319–330. doi: 10.1093/genetics/134.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falciani F, Hausdorf B, Schröder R, Akam M, Tautz D, Denell R, Brown S. Proc Natl Acad Sci USA. 1996;93:8479–8484. doi: 10.1073/pnas.93.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payre F, Crozatier M, Vincent A. Genes Dev. 1994;8:2718–2728. doi: 10.1101/gad.8.22.2718. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Xu P-X, Suzuki Y. Development (Cambridge, UK) 1994;120:277–285. doi: 10.1242/dev.120.2.277. [DOI] [PubMed] [Google Scholar]
- 10.Epstein M, Pillemer G, Yelin R, Yisraeli J K, Fainsod A. Development (Cambridge, UK) 1997;124:3805–3814. doi: 10.1242/dev.124.19.3805. [DOI] [PubMed] [Google Scholar]
- 11.Sommer R, Tautz D. Development (Cambridge, UK) 1991;113:419–430. doi: 10.1242/dev.113.2.419. [DOI] [PubMed] [Google Scholar]
- 12.Schröder R, Sander K. Roux′s Arch Dev Biol. 1993;203:34–43. doi: 10.1007/BF00539888. [DOI] [PubMed] [Google Scholar]
- 13.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. phylip (Phylogeny Inference Package) Version 3.572c. University of Washington, Seattle, WA: Department of Genetics; 1995. [Google Scholar]
- 15.Adachi J, Hasegawa M. molphy. Phylogeny Inference Package; 1992. Version 2.2. (Institute of Statistical Mathematics, Tokyo, Japan). [Google Scholar]
- 16.McAlpine J F. In: Manual of Nearctic Diptera. McAlpine J F, editor. Vol. 3. Hull, Quebec, Canada: Canadian Government Publishing Centre; 1989. pp. 1397–1520. [Google Scholar]
- 17.Karlin-Mcginness M, Serano T L, Cohen R S. Dev Genet. 1996;19:238–248. doi: 10.1002/(SICI)1520-6408(1996)19:3<238::AID-DVG7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 18.Finkelstein R, Perrimon N. Nature (London) 1990;346:485–488. doi: 10.1038/346485a0. [DOI] [PubMed] [Google Scholar]
- 19.Goriely A, Stella M, Coffinier C, Kessler D, Mailhos C, Dessain S, Desplan C. Development (Cambridge, UK) 1996;122:1641–1650. doi: 10.1242/dev.122.5.1641. [DOI] [PubMed] [Google Scholar]
- 20.Hahn M, Jäckle H. EMBO J. 1996;15:3077–3084. [PMC free article] [PubMed] [Google Scholar]
- 21.Vorbrüggen G, Constien R, Zilian O, Wimmer E A, Dowe G, Taubert H, Noll M, Jäckle H. Mech Dev. 1997;68:139–147. doi: 10.1016/s0925-4773(97)00139-1. [DOI] [PubMed] [Google Scholar]
- 22.Rushlow C, Levine M. Genes Dev. 1990;1:1268–1279. doi: 10.1101/gad.1.10.1268. [DOI] [PubMed] [Google Scholar]
- 23.Bürglin, T. R. in Biodiversity and Evolution, eds. Arai, R., Kato, M., Doi, Y. (The National Science Museum Foundation, Tokyo, Japan), pp. 291–336.
- 24.Damen W G M, Tautz D. Dev Genes Evol. 1998;208:586–590. doi: 10.1007/s004270050218. [DOI] [PubMed] [Google Scholar]
- 25.Telford M J, Thomas R H. Dev Genes Evol. 1998;208:591–594. doi: 10.1007/s004270050219. [DOI] [PubMed] [Google Scholar]
- 26.Sander K. Semin Cell Dev Biol. 1996;7:573–582. [Google Scholar]
- 27.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Fernandez J, Holland P W H. Nature (London) 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- 29.Brooke N M, Garcia-Fernandez J, Holland P W H. Nature (London) 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- 30.Kmita-Cunisse M, Loosli F, Bièrne J, Gehring W J. Proc Natl Acad Sci USA. 1998;95:3030–3035. doi: 10.1073/pnas.95.6.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornfeld K, Saint R B, Beachy P A, Harte P J, Peattie D A, Hogness D S. Genes Dev. 1989;3:243–258. doi: 10.1101/gad.3.2.243. [DOI] [PubMed] [Google Scholar]
- 32.Pultz M A, Diederich R J, Cribbs D L, Kaufman T C. Genes Dev. 1988;2:901–920. doi: 10.1101/gad.2.7.901. [DOI] [PubMed] [Google Scholar]
- 33.Schneuwly S, Kuroiwa A, Baumgartner P, Gehring W J. EMBO J. 1986;5:733–739. doi: 10.1002/j.1460-2075.1986.tb04275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mlodzik M, Fjose A, Gehring W J. EMBO J. 1996;7:2569–2578. doi: 10.1002/j.1460-2075.1988.tb03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rechsteiner M, Rogers S W. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]