Abstract
Although cementoblasts express Toll-like receptors (TLR)-2 and -4, little is known regarding the possible participation of cementoblasts in the inflammatory response. We investigated the effects of Porphyromonas gingivalis lipopolysaccharide (LPS), tetra- and penta-acylated lipid A species (designated PgLPS1435/1449 and PgLPS1690, respectively), on gene expression of osteoclastogenesis-associated molecules in murine cementoblasts. Real-time quantitative RT-PCR analysis revealed that receptor activator of NF-κB ligand (RANKL), interleukin-6, Regulated on activation, normal T-cell expressed, and secreted (RANTES), macrophage inflammatory protein-1α, and monocyte chemoattractant protein-1 were rapidly and dramatically induced upon stimulation with PgLPS1690, but only slightly induced with PgLPS1435/1449. Osteoprotegerin, which was expressed constitutively, was not altered significantly. ELISA demonstrated synthesis of corresponding proteins. PgLPS1690 significantly induced transcripts for NF-κB, and this activation was inhibited by pre-treatment with anti-TLR-2 but not with TLR-4 antibodies. These results suggest that cementoblasts participate in the recruitment of osteoclastic precursor cells by up-regulation of chemokines/cytokines.
Keywords: cementoblast, osteoclastogenesis, Toll-like receptor, lipopolysaccharide, Porphyromonas gingivalis
INTRODUCTION
Porphyromonas gingivalis has been implicated as a major pathogen in the development and progression of periodontitis (Socransky and Haffajee, 1992). This bacterium possesses a large number of potential virulence factors such as fimbriae, hemagglutinin, lipopolysaccharide (LPS), and various proteases (Holt et al., 1999). LPS is known to induce not only inflammatory responses but also bone resorption by enhancing osteoclastogenesis via osteoblast-mediated activities. When osteoblasts are stimulated with Escherichia coli LPS, the expression of receptor activator of NF-κB ligand (RANKL) is up-regulated, and the expression of osteoprotegerin (OPG), which acts as a decoy receptor for RANKL, is down-regulated (Kikuchi et al., 2001; Suda et al., 2004). Furthermore, interleukin (IL)-6, which functions in the induction of osteoclast activity and subsequent bone resorption (Kotake et al., 1996), is released by osteoblasts upon stimulation with E. coli LPS (Kwan Tat et al., 2004). Moreover, osteoblasts release a series of chemokines—such as monocyte chemoattractant protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, RANTES (Regulated on activation, normal T-cell expressed, and secreted)—that recruit osteoclast precursors and directly enhance osteoclast formation through a pathway dependent on the presence of RANKL (Jiang and Graves, 1999; Yu et al., 2004; Kim et al., 2005).
In contrast to the observation of significant destruction of bone with inflammatory conditions of the oral cavity, root resorption is rarely seen under inflammatory conditions. Cementum is a thin mineralized tissue covering the tooth root surface and an important component of the periodontal attachment apparatus. Although cementum shares many properties with bone, most notably a remarkable similarity in biochemical composition, it has yet to be established whether cementoblasts and osteoblasts have a common precursor cell. Cementum differs from bone in its histology by lacking innervation and vascularization. Furthermore, cementum has limited remodeling potential when compared with bone, where a system consisting of osteoblasts, osteocytes, bone-lining cells, and osteoclasts is required (Saygin et al., 2000; Bosshardt, 2005).
Our laboratory developed an immortalized murine cementoblast cell line (OCCM-30) that reflects genes and proteins associated with these cells in situ, such as expression of bone sialoprotein, osteocalcin (OCN), and type I collagen (D'Errico et al., 2000). Previously, we reported that, similar to osteoblasts, OCCM-30 expresses Toll-like receptor (TLR) -2 and -4, and that, further, upon exposure of OCCM-30 to P. gingivalis LPS, genes associated with cementum formation are altered (Nociti et al., 2004). P. gingivalis LPS has been reported to contain an unusual amount of lipid A heterogeneity (Darveau et al., 2004), and has been shown to have the ability to utilize TLR-2 or TLR-4 (Hajishengallis et al., 2002; Zhou et al., 2005). We have generated two different structural types of lipid A, tetra- and penta-acylated lipid A species designated PgLPS1435/1449 and PgLPS1690, respectively (Reife et al., 2006).
These observations led us to investigate whether PgLPS1690 and PgLPS1435/1449 affect osteoclastogenesis-associated gene expression of cementoblasts.
MATERIALS & METHODS
Reagents
Monoclonal antibodies (mAb) for mouse/human TLR-2 (T2.5, mouse IgG1) and mouse TLR-4/MD-2 (MTS510, Rat IgG2a), and isotype-matched control IgGs, were purchased from eBioscience (San Diego, CA, USA). Ascorbic acid (AA) and Escherichia coli LPS 0111:B4 LPS were purchased from Sigma (St. Louis, MO, USA).
Cells
An immortalized murine cementoblast cell line, OCCM-30, was established by the isolation of tooth root-surface cells from transgenic mice containing an SV40 large T-antigen under the control of the OCN promoter (D'Errico et al., 2000). Cells were maintained in DMEM (Gibco, Rockville, MD, USA) plus 10% FBS and antibiotics. All procedures were approved by the University of Washington, Committee on Use and Care of Animals, and were in compliance with state and Federal laws.
Purification of PgLPS1435/1449 and PgLPS1690
The PgLPS1435/1449 was purified from P. gingivalis ATCC 33277 by the MgCl2/ETOH procedure (Darveau et al., 2004). PgLPS1690 was obtained by the phenol water procedure (Westphal and Jann, 1965) and treated for the removal of trace amounts of endotoxin protein, and then subjected to SDS-PAGE and stained for protein by the enhanced colloidal gold procedure (Manthey and Vogel, 1994). The colloidal gold procedure revealed less than 0.1% protein contamination, based upon the amount of LPS loaded onto the gel and the relative intensity of the major protein band to known BSA standards. Gas chromatography/mass spectrometry of the fatty acids present in the PgLPS1690 preparation was performed, and the identification of all the peaks found in the preparation after transmethylation was performed by identification of the mass spectral patterns (Darveau et al., 2004). Matrix-assisted laser desorption-time-of-flight mass spectroscopy was performed (Guo et al., 1997) and revealed one major cluster of lipid A structures centered around a mass of PgLPS1690.
Reverse Transcription and Real-time Quantitative PCR
Total cellular RNA was extracted with the use of Trizol® reagent (Gibco) according to the manufacturer's instructions, and was DNase-treated (DNA-free™, Ambion Inc., Austin, TX, USA). The transcription of total RNA into cDNA was carried out with the use of a Transcriptor First Strand cDNA Synthesis Kit® (Roche Diagnostic Co., Indianapolis, IN, USA) according to the manufacturer's instructions. Primers were designed with the use of LightCycler probe design software (Roche Diagnostics GmbH, Mannheim, Germany), and primer sequences for each gene, IL-6, MCP-1, MIP-1α, RANTES, RANKL, OPG, and GAPDH were as follows (forward/reverse): IL-6 (ACTGATGCTGGTGACA/GCAAGTGCATCATCGT); MCP-1 (CTCGGGCAGCTAGAAT/GGCGTTGTGATGCAAA); MIP-1α (GGAAGATTCCACGCCAA/CCTCGATGTGGCTACT); RANTES (GAAGGAACCGCCAAGT/CCGATTTTCCCAGGACC); RANKL (CATCGGGTTCCCATAAAGTC/TTGCCCGACCAGTTTTTC); OPG (TGAATGCCGAGAGTGTAG/CTGCTCGCTCGATTTG); GAPDH (ACCACAGTCCATGCCATCAC/TCCACCACCCTGTTGCTGTA). The amplification profile was 95/0; 55/7; 72/20 [temperature (°C)/time (sec)], with a slope of 20°C/sec and 43 cycles. PCR was performed in a LightCycler system (Roche Diagnostic GmbH) with the FastStart DNA Master SYBR Green I kit (Roche Diagnostic Co.), with optimization of 3 mM MgCl2 and 0.5 μM primer. Reaction product was quantified (Roche Quantification Software, Roche Diagnostics GmbH) with GAPDH as the reference gene.
Preparation of Cell Lysates
Confluent cells cultured in 35-mm-diameter culture dishes were harvested with a cell scraper and subjected to lysis in 100 μL of cell lysis buffer (10 mM phosphate buffer [pH 7.0], 150 mM NaCl, 1% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF). After lysis, cells were incubated on ice for 30 min, followed by centrifugation at 12,000 x g for 10 min, and then the supernatants were collected and stored at -20°C. The protein concentration of cell lysates was measured by DC Protein Assay® (Bio-Rad Laboratories, Hercules, CA, USA).
Enzyme-linked Immunosorbent Assay (ELISA)
Supernatants from cell cultures were harvested by centrifugation, and kept at -20°C. The amounts of OPG, MCP-1, IL-6, RANTES, and MIP-1α in the supernatants and RANKL in the cell lysate (50 μg) were measured by means of mouse ELISA kits (R&D Systems Inc., Minneapolis, MN, USA). The assays were performed precisely as instructed by the ELISA manufacturer.
NF-κB Luciferase Assay
The NF-κB firefly luciferase reporter construct pNF-κB-TA-Luc was obtained from BD Bioscience Clontech (Palo Alto, CA, USA). The β-actin-Renilla luciferase reporter construct was generously provided by C. Wilson (University of Washington, Seattle). Cells were seeded in a 24-well plate at a density of 1 x 105 cells per well one day before transfection. The cells were transiently co-transfected with 0.4 μg of pNF-κB-TA-Luc and 0.01 μg of pβ-actin-Renilla Luc by means of LipofectAMINE2000™ (Invitrogen, San Diego, CA, USA) as described previously (Coats et al., 2003). Eighteen hours after the transfection, the transfected cells were stimulated with activators as indicated in the text. Following 5 hrs of stimulation in DMEM with 10% FBS, the cells were rinsed with PBS, subjected to lysis with 100 μL of passive lysis buffer, and analyzed with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) as instructed by the manufacturer. Data are presented as means ± standard deviations (SD) of triplicate experiments.
Statistical Analysis
Experimental values are given as means ± SD. The significance of differences between control and treatments was evaluated by one-way ANOVA.
RESULTS
Effect of P. gingivalis LPS on Gene and Protein Expression
Cells were exposed to 1 μg/mL of PgLPS1435/1449 or PgLPS1690 and then examined for changes in selected transcripts, over a 24-hour period, by quantitative RT-PCR. Expression of RANKL and chemokines/cytokines rapidly increased, with a peak point of 3 hrs noted for cells exposed to PgLPS1690, while for PgLPS1435/1449 a minimal effect on expression of these genes was noted. Induction of these genes declined rapidly, except for RANTES, which exhibited a slow decline. The expression of OPG was not altered significantly among the various treatments (Fig. 1A). Cementoblast response to E. coli (Ec) LPS was similar in terms of RANKL and OPG (Fig. 1B). ELISA analysis revealed that RANKL, MCP-1, IL-6, RANTES, and MIP1-α protein levels were rapidly (3-6 hrs) and significantly induced after stimulation with PgLPS1690, with only slight induction upon stimulation with PgLPS1435/1449 noted. OPG protein levels were induced over time in untreated cells, with no changes by either PgLPS1690 or PgLPS1435/1449 (Fig. 2). Interestingly, a rapid decrease of RANKL was observed after 6 hrs, unlike other factors examined. This was probably due to shedding of RANKL by endogenous enzymes following cell activation, as reported in T-lymphocytes (Hikita et al., 2005), and/or related to the increase in OPG secretion with time. These results suggest that both P. gingivalis LPSs have the potential to promote osteoclastogenesis, with a more dramatic impact for PgLPS1690 vs. PgLPS1435/1449.
Figure 1.
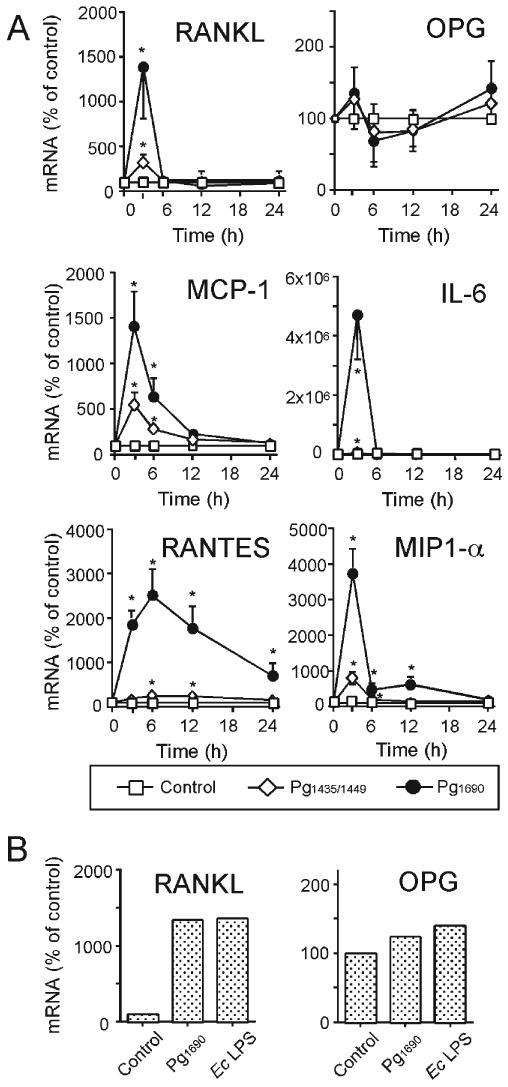
Effects of P. gingivalis and E. coli LPS on selected mRNA levels. Confluent OCCM-30 cells in 60-mm-diameter culture dishes was cultured in DMEM (5% FBS) with AA (50 μg/mL) in the presence or absence of P. gingivalis LPS (1 μg/mL) for 1, 3, 6, 12, and 24 hrs (A) or E. coli LPS (10 ng/mL) for 3 hrs (B). Total cellular RNA was extracted, and the mRNA expressions were analyzed by real-time quantitative RT-PCR. Data in (A) represent mean ± SD from 3 separate experiments. Data in (B) are representative of 2 independent experiments. Statistical significance is shown (*P < 0.05 vs. control).
Figure 2.

Effect of P. gingivalis LPS on selected protein levels. Confluent OCCM-30 in 35-mm-diameter culture dishes was cultured in DMEM (5% FBS) with AA (50 μg/mL) in the presence or absence of LPS (1 μg/mL) for 3, 6, 12, and 24 hrs. The amounts of OPG, MCP-1, IL-6 RANTES, and MIP-1α in the supernatant and RANKL in the cell lysate were analyzed by ELISA. Data represent mean ± SD from 2 separate experiments done by triplicate assay. Statistical significance is shown (*P < 0.05 vs. control).
Effects of Anti-TLR-2 and TLR-4 mAbs on NF-κB Activation
Since P. gingivalis LPS has been reported to interact with both TLR-2 and -4 (Darveau et al., 2004; Zhou et al., 2005), and cementoblasts express both receptors (Nociti et al., 2004), we focused on determining whether these receptors were involved in P. gingivalis LPS-mediated response. Cells were transiently transfected with an NF-κB-luciferase reporter construct, and blocking experiments with mAbs were performed. E. coli LPS and peptidoglycan were used as specific ligand controls for TLR-4 and TLR-2, respectively. NF-κB activation by E. coli LPS was significantly inhibited by TLR-4 mAb, which neutralized TLR-4 signaling (Kikuchi et al., 2001), and activation by peptidoglycan was completely inhibited by TLR-2 mAb. NF-κB activation by PgLPS1690 was almost completely inhibited by TLR-2 mAb ,but not by TLR-4 mAb, and, further, the combination of both mAbs showed no additional effect (Fig. 3). A similar inhibitory pattern was observed for PgLPS1435/1449-treated cells, although NF-κB activation was induced slightly. These results indicate that PgLPS1690 and PgLPS1435/1449 acted on cementoblasts through a TLR-2-dependent pathway. Next, we examined whether the changes in gene expression mediated by P. gingivalis LPS were inhibited by TLR-2 mAb. PgLPS1690-mediated induction of RANKL, IL-6, and RANTES was markedly inhibited by pre-treatment with TLR-2 mAb, but not with TLR-4 mAb. Pre-treatment with both mAbs showed no additional effect. Although the induction of these genes by PgLPS1435/1449 was weak, a similar inhibitory pattern was observed (Fig. 4).
Figure 3.

Effects of anti-TLR-2 and TLR-4 mAbs on NF-κB activation. OCCM-30 cells were co-transfected with pNK-κB-TA-Luc and pβ-actin Renilla luciferase, as described in MATERIALS & METHODS. Eighteen hours later, transfectants were pre-treated with 20 μg/mL of anti-TLR-2 and -4 mAbs, as well as with 20 μg/mL of control Abs, for 30 min, followed by stimulation with E. coli LPS (10 ng/mL), peptidoglycan (1 μg/mL), PgLPS1690 (1 μg/mL), or PgLPS1435/1449 (1 μg/mL) for 5 hrs. Cells were harvested, and luciferase activity was measured. Values were calculated as fold increase of relative luciferase units (firefly luciferase/Renilla luciferase) compared with the non-stimulated control response, which was set at 1. Representative data of 3 separate experiments are shown as means ± SD of triplicate assays. Statistical significance is shown (*P < 0.05 vs. control).
Figure 4.

Effects of anti-TLR-2 and TLR-4 mAbs on gene expression. Confluent OCCM-30 cells in 35-mm-diameter culture dishes were pre-treated with 20 μg/mL of anti-TLR-2 and -4 mAbs, as well as with 20 μg/mL of control Abs, for 30 min, followed by stimulation with PgLPS1690 (1 μg/mL) or PgLPS1435/1449 (1 μg/mL) in DMEM (5% FBS) with AA (50 μg/mL) for 3 hrs. Total cellular RNA was extracted, and mRNA expression of RANKL, IL-6, and RANTES was analyzed by real-time quantitative RT-PCR. Values were calculated as fold increase compared with calibrator cDNA, which was generated from OCCM-30 stimulated with 50 μg/mL AA for 5 days. Representative data of 3 separate experiments are shown as means ± SD of triplicate assays. Statistical significance is shown (*P < 0.05 vs. control).
DISCUSSION
We showed that osteoclastogenesis-associated chemokines/cytokines and RANKL were rapidly induced in cementoblasts upon stimulation with PgLPS1690, and that this effect was mediated through the TLR-2 signaling pathway. OPG was constitutively expressed by cementoblasts, and was not significantly altered by P. gingivalis LPS stimulation.
Previous studies with murine osteoblasts have shown that the expression of RANKL is rapidly induced upon stimulation by E. coli LPS via TLR-4 (Kikuchi et al., 2001; Suda et al., 2004). Analysis of existing data indicates that osteoblasts have the potential to release various chemokines/cytokines when stimulated with pro-inflammatory cytokines or LPS. These chemokines/cytokines have been shown to induce osteoclastogenesis directly or indirectly by the mechanism described previously in the INTRODUCTION. Our findings indicated that, when cementoblasts were exposed to inflammatory stimuli, they responded in a similar fashion to osteoblasts in terms of stimuli-mediated induction of proteins/genes associated with osteoclast activation, as well as the recruitment of osteoclastic precursors, suggesting that cementoblasts have the potential to enhance osteoclastogenesis during periodontal disease in a manner comparable with that of osteoblasts.
However, and in seeming contrast to osteoblasts, two lines of evidence indicate that cementoblasts may protect the tooth root surface from resorption during times of local LPS challenge. First, it was shown that OPG was expressed by cementoblasts constitutively, and the gene and protein levels were not altered upon stimulation with LPS. In contrast, OPG expressed by osteoblasts has been reported to be suppressed by E. coli LPS (Suda et al., 2004), rendering this cell type less effective for inhibiting bone or tooth resorption. This is unlikely to represent a TLR-2 vs. TLR-4 phenomenon, considering that E. coli and P. gingivalis LPS exhibited similar effects in terms of RANKL/OPG. Second, the rapid appearance of RANKL and slow increase in OPG by cementoblasts suggest that RANKL-dependent osteoclast formation mediated by MIP1-α and RANTES (Yu et al., 2004) may be inhibited further, preventing osteoclast formation near the tooth root surface. In addition, osteoclasts from the blood may favor migration to the well-vascularized surrounding bone vs. the avascular cementum. These results are consistent with the clinical observations that osteoclast-mediated root resorption is rarely linked to periodontal disease, even at a stage of severe disease with marked bone loss. Therefore, differences in the regulation of OPG by osteoblasts vs. cementoblasts, while speculative at this point, may be related to protection of the tooth root surface in response to LPS. Future studies, using a co-culture system of osteoclastic precursors with cementoblasts or other cell types, including osteoblasts, in the presence or absence of LPS, may enable us to verify the validity of our hypothesis.
TLR-4 is the principal signal transducer for most types of LPS, while analysis of existing data suggests that TLR-2 is a signal transducer for other bacterial components, such as peptidoglycan and lipoprotein (Takeuchi et al., 1999). However, P. gingivalis LPS is unusual, in that it has been reported to be an agonist for both TLR-2 and -4 (Hajishengallis et al., 2002; Darveau et al., 2004; Zhou et al., 2005). Here, it was shown that both the PgLPS1435/1449 and PgLPS1690 activated NF-κB in cementoblasts through the TLR-2 pathway. In another ongoing study from our group, we observed that PgLPS1690 stimulated E selectin expression on human endothelial cells, whereas PgLPS1435/1449 did not, and, furthermore, that it blocked E selectin expression in response to PgLPS1690 (Reife et al., 2006). These findings demonstrate that the different P. gingivalis LPS lipid A structural types activate different cementoblast activation pathways.
In conclusion, we report for the first time that cementoblasts potentially participate in the recruitment of osteoclastic precursors by inducing chemokines/cytokines during periodontitis. In addition, constant expression of OPG in cementoblasts may play a critical role in protecting the tooth from osteoclast activity under physiological conditions such as stress associated with mastication, as well as under pathological conditions.
Acknowledgments
This work was supported by NIH grant R01DE09532.
References
- Bosshardt DD. Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J Dent Res. 2005;84:390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- Coats SR, Reife RA, Bainbridge BW, Pham TT, Darveau RP. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at Toll-like receptor 4 in human endothelial cells. Infect Immun. 2003;71:6799–6807. doi: 10.1128/IAI.71.12.6799-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Errico JA, Berry JE, Ouyang H, Strayhorn CL, Windle JJ, Somerman MJ. Employing a transgenic animal model to obtain cementoblasts in vitro. J Periodontol. 2000;71:63–72. doi: 10.1902/jop.2000.71.1.63. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both Toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–5051. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, et al. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Martin M, Schifferle RE, Genco RJ. Counteracting interactions between lipopolysaccharide molecules with differential activation of Toll-like receptors. Infect Immun. 2002;70:6658–6664. doi: 10.1128/IAI.70.12.6658-6664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita A, Kadono Y, Chikuda H, Fukuda A, Wakeyama H, Yasuda H, et al. Identification of an alternatively spliced variant of Ca2+-promoted Ras inactivator as a possible regulator of RANKL shedding. J Biol Chem. 2005;280:41700–41706. doi: 10.1074/jbc.M507000200. [DOI] [PubMed] [Google Scholar]
- Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Graves DT. Periodontal pathogens stimulate CC-chemokine production by mononuclear and bone-derived cells. J Periodontol. 1999;70:1472–1478. doi: 10.1902/jop.1999.70.12.1472. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Matsuguchi T, Tsuboi N, Mitani A, Tanaka S, Matsuoka M, et al. Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J Immunol. 2001;166:3574–3579. doi: 10.4049/jimmunol.166.5.3574. [DOI] [PubMed] [Google Scholar]
- Kim MS, Day CJ, Morrison NA. MCP-1 is induced by receptor activator of nuclear factor-{kappa}B ligand, promotes human osteoclast fusion, and rescues granulocyte macrophage colony-stimulating factor suppression of osteoclast formation. J Biol Chem. 2005;280:16163–16169. doi: 10.1074/jbc.M412713200. [DOI] [PubMed] [Google Scholar]
- Kotake S, Sato K, Kim KJ, Takahashi N, Udagawa N, Nakamura I, et al. Interleukin-6 and soluble interleukin-6 receptors in the synovial fluids from rheumatoid arthritis patients are responsible for osteoclast-like cell formation. J Bone Miner Res. 1996;11:88–95. doi: 10.1002/jbmr.5650110113. [DOI] [PubMed] [Google Scholar]
- Kwan Tat S, Padrines M, Theoleyre S, Heymann D, Fortun Y. IL-6, RANKL, TNF-alpha/IL-1: interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev. 2004;15:49–60. doi: 10.1016/j.cytogfr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Manthey CL, Vogel SN. Elimination of trace endotoxin protein from rough chemotype LPS. J Endotoxin Res. 1994;1:84–91. [Google Scholar]
- Nociti FH, Jr, Foster BL, Barros SP, Darveau RP, Somerman MJ. Cementoblast gene expression is regulated by Porphyromonas gingivalis lipopolysaccharide partially via Toll-like receptor-4/MD-2. J Dent Res. 2004;83:602–607. doi: 10.1177/154405910408300804. [DOI] [PubMed] [Google Scholar]
- Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, et al. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 2006;8:857–868. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- Saygin NE, Giannobile WV, Somerman MJ. Molecular and cell biology of cementum. Periodontol 2000. 2000;24:73–98. doi: 10.1034/j.1600-0757.2000.2240105.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- Suda K, Udagawa N, Sato N, Takami M, Itoh K, Woo JT, et al. Suppression of osteoprotegerin expression by prostaglandin E2 is crucially involved in lipopolysaccharide-induced osteoclast formation. J Immunol. 2004;172:2504–2510. doi: 10.4049/jimmunol.172.4.2504. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Westphal O, Jann K. Bacterial lipopolysaccharide: extraction with phenol-water and further applications of the procedure. In: Whistler RL, editor. Methods in carbohydrate chemistry. New York: Academic Press Inc.; 1965. p. 83. [Google Scholar]
- Yu X, Huang Y, Collin-Osdoby P, Osdoby P. CCR1 chemokines promote the chemotactic recruitment, RANKL development, and motility of osteoclasts and are induced by inflammatory cytokines in osteoblasts. J Bone Miner Res. 2004;19:2065–2077. doi: 10.1359/JBMR.040910. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Desta T, Fenton M, Graves DT, Amar S. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect Immun. 2005;73:935–943. doi: 10.1128/IAI.73.2.935-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


