Abstract
Metallothionein-I (MT-I) gene is silenced by methylation of CpG islands in mouse lymphosarcoma P1798 cells but not in the thymus, the cell type from which the tumor was derived. Bisulfite genomic sequencing revealed that all 21 CpG dinucleotides present within −216 bp to +1 bp with respect to transcription start site are methylated in the tumor cell line, but none is methylated in the thymus. The lymphosarcoma cells induced MT-I in response to heavy metals only after demethylation with 5-azacytidine (5-AsaC). The electrophoretic mobility shift assay using specific oligonucleotide probes showed that the key transcription factors regulating MT-I gene (e.g., MTF-1, Sp 1 and MLTF/USF) are active in P1798 cells. In vivo footprinting of the proximal promoter region showed that none of the metal regulatory elements (MREs) or MLTF/USF are occupied in response to heavy metals. Demethylation of the lymphosarcoma cells with 5-AzaC resulted in constitutive footprinting at MLTF/ARE, and zinc-inducible footprinting at MRE-c, MRE-d and MRE-e sites. Demethylation of just 10 − 20% of the CpG islands was sufficient to render the gene inducible by cadmium or zinc. The MT-I induction persisted in the cancer cells for several generations even after withdrawal of 5-AzaC from the culture medium.
Keywords: metallothionein, methylation, lymphosarcoma, thymus, bisulfite sequencing
Introduction
DNA methylation in mammals plays a crucial role in development, X chromosome inactivation, epigenetic silencing, aging, carcinogenesis and certain human genetic disease (Baylin, 1997; Jaenisch, 1997; Lewin, 1998). The most significant methylation occurs in the cytosines of the CpG dinucleotides, which leads to alteration in transcription of the gene (Kass et al., 1997). In normal tissues, extensive methylation of CpG islands in the promoter region is exclusively associated with transcriptional silencing of imprinted alleles and genes on the inactive X chromosome (Baylin et al., 1998) whereas the methylation profiles are often modified in neoplasia. As compared to control cells, many cancer cells have been shown to exhibit global hypomethylation of DNA compared to control cells (Feinberg et al., 1988; Lengauer et al., 1997). This finding is consistent with the oncogenic potential of cells following treatment with 5-azacytidine (5-AzaC), a demethylating agent that irreversibly inactivates the methyltransferase (Santi et al., 1984). On the other hand, hypermethylation of tumor suppressor genes resulted in their silencing or inactivation (Norris et al., 1994) that leads to oncogenesis. In many cancers, different tumor suppressor or growth regulatory genes are silenced due to methylation of their promoter sequence, whereas methylation within the coding region causes cytosine to thymine transition due to spontaneous deamination of methyl cytosines (Baylin et al., 1998; Ng et al., 1997). This study further lends support to the notion that modification in the DNA methylation machinery constitutes a common cancer-related change, and probably plays a key role in the early stages of carcinogenesis.
An interesting outcome of DNA methylation is the transcriptional silencing of whole chromosomes, transgenes, certain developmentally regulated genes and human disease genes (Knight et al., 1993; Li et al., 1993; Norris et al., 1994). Methylation in CpG island results in alteration of chromatin structure (Klein and Costa, 1997) and in some cases directly impedes binding of the positive factors to the regulatory elements (Kass et al., 1997). Recent study has, however, revealed that methylation per se cannot silence a gene (Nan et al., 1997). Some methylated DNA binding proteins (MeCP) are required in this process. Because many fully methylated genes can be transcribed at nearly normal rates in the absence of methyl-CpG binding proteins it is unlikely that the CpG methylation by itself renders these sites inaccessible to the basal transcriptional machinery or prevents interaction of the transcription factors with the promoters.
Metallothioneins are a group of small cysteine-rich, metal binding proteins that are expressed widely in yeasts, plants and animals. The mouse cells contain four isoforms of MT, designated MT-I, II, III, and IV, arranged in tandem in chromosome 8 (for recent reviews, see Quaife et al., 1994; Aschner et al., 1997). The MT-I and MT-II genes are expressed in all tissues whereas MT-III gene is expressed largely in the brain and MT-IV is expressed mainly in the stratified squamous epithelium of skin, tongue etc. The basal expression of MT-I and MT-II genes is relatively low in most tissues except in the brain and testis, but can be induced by a variety of agents that include heavy toxic metals, steroid hormones, interleukins, phorbol esters, interferons, restraint stress and other agents that produce oxygen intermediates or free oxygen radicals (Kagi, 1991; Ghoshal et al., 1998). Overexpression of MT in cells diminishes the sensitivity of the cells to the compounds that generate free oxygen radicals (Pitt et al., 1997), DNA damaging agents such as UV radiation (Chubatsu and Meneghini, 1993), nitric oxide (Schwarz et al., 1995) and certain anticancer drugs (Kelley et al., 1988). On the contrary, highly inducible MT-I and MT-II genes are silent in some lymphoid derived tumor cell lines, W-7 and S-49, but can be induced by heavy metals after treatment with 5-azacytidine (5-AzaC) (Compere and Palmiter, 1981), which suggests a role for hypermethylation in the silencing of MT gene. These studies have, however, not analysed the methylation states of MT gene or the methylation sites within the promoter in the tumor cells as well as the parental cells. MT-I and MT-II genes consist of six metal regulatory elements (MRE) in the region spanning from −216 bp to +1 bp. These elements are involved in both constitutive and induced expression of MT. MTF-1 (metal-regulatory transcription factor) binds to these elements and activates the gene (Radtke et al., 1993; Heuchel et al., 1994). In addition, the promoter and proximal promoter region contain sites for the binding of MLTF/USF (that also harbors a single ARE, antioxidant response element), C'BP-1 and C'BP-2 (Datta and Jacob, 1993, 1997; Aniskovitch and Jacob, 1997, 1998). In addition to MREs, several other cis elements, e.g., MLTF/ARE (a composite element containing overlapping E-box and antioxidant response element, MRE-c' have been identified in mouse MT-I promoter that is responsible for basal or oxidant-induced expression of the gene.
Our laboratory has been involved in the identification of key promoter elements and transactivating factors that modulate MT-I transcription (Datta and Jacob, 1993, 1997; Aniskovitch and Jacob, 1997, 1998). In the course of this study, we observed that this gene is not induced in mouse lymphosarcoma (P1798) cells, as opposed to normal induction in the parental thymus cells. The present study was undertaken to determine the potential role of promoter methylation in the repression of MT-I gene expression in the lymphosarcoma cells.
Results
Lymphosarcoma cells express MT-I or MT-II genes in response to heavy metals only after treatment with 5-azacytidine
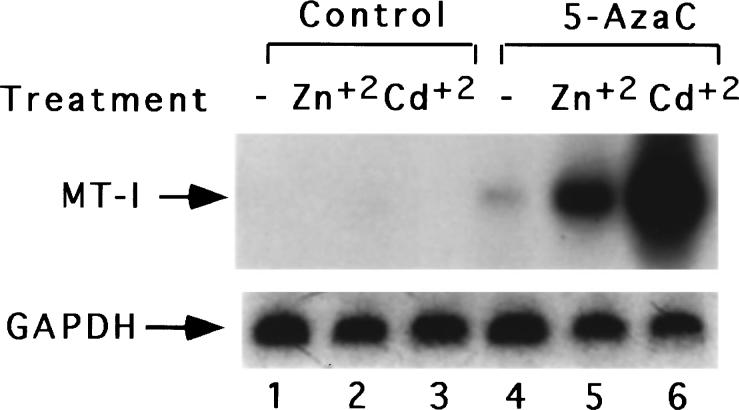
Initially, we observed that mouse lymphosarcoma cells, P1798 are extremely sensitive to heavy metals such as cadmium or zinc, as shown by the markedly reduced cell viability even in the presence of relatively low concentration of the metal. We reasoned that the relatively low viability of the cells may be due to decrease in induction of metallothionein, the protein that detoxifies the heavy metals, in these cells. To test this possibility, we analysed the mRNA level in these cells after CdSO4 or ZnSO4 treatment. Under these conditions, MT-I mRNA was not induced (Figure 1). In some tumor cell lines of lymphoid origin, e.g., S-49, W-7, MT-I gene is silent (Compere and Palmiter, 1981) and can be induced only after treatment with 5-AzaC, a DNA demethylating agent (Santi et al., 1983). It is logical to assume that the lack of induction of MT-I gene in P1798 cells, a cell line derived from thymus (Wood and Thompson, 1984), is also due to potential hypermethylation at critical site(s). To test this possibility we pre-treated these cells with the cytosine analog and measured the MT-I mRNA level following metal exposure of the 5-AzaC-treated cells. 5-AzaC is incorporated into DNA during replication followed by covalent binding of DNA methyltransferase (DNA-MTase) to the analog causing irreversible inactivation of the enzyme and formation of demethylated DNA (Santi et al., 1984). Treatment of the lymphosarcoma cells with this base analog at a concentration of 2.5 μm for 72 h resulted in induction of MT-I gene when exposed to zinc or cadmium. A representative Northern blot is shown in Figure 1. Demethylation of DNA alone resulted in twofold increase in the constitutive level of MT-I mRNA in these cells, whereas treatment of 5-AzaC-exposed cells with zinc or cadmium led to a significantly higher level of MT-I induction (Figure 1). Phosphorimager analysis showed that MT-I mRNA level increased by five and tenfold treatment with Zn2+ and Cd2+. The level of induction was highest after cadmium treatment, as observed for many cell lines and tissues (Kagi, 1991; Palmiter, 1987). This experiment was repeated at least three times with different batches of cells. MT-I cDNA probe used for Northern blot analysis in this study hybridizes to both MT-I and MT-II mRNAs. Because these two isoforms are coordinately expressed in all tissues (Yagle and Palmiter, 1985), our results suggest that both MT-I and MT-II are silent in the lymphosarcoma cells.
Figure 1.
Northern blot analysis for MT-1 mRNA in lymphosarcoma cells before and after 5-azacytidine treatment and heavy metals. RNA isolated (30 μg) from the control cells and the cells treated with 5-AzaC (2.5 μm) for 72 h, were separated by formaldehyde-agarose (1.2%) gel electrophoresis, transferred to the nylon membrane and UV-crosslinked. The membrane was then subjected to Northern blot analysis first with random-primed, (32P-labeled) mouse MT-I cDNA and subsequently with rat GAPDH cDNA. Lanes 1 − 3 indicated the RNA levels in the untreated control cells, cells treated for 3 h with ZnSO4 (50 μm) and CdSO4 (15 μm), respectively. Lanes 4 − 6 denote the RNA levels in the untreated cells, cells treated for 3 h with ZnSO4 (100 μm) and CdSO4 (30 μm) respectively following treatment with 5-AzaC
Genomic footprinting analysis shows that none of the transcriptional activators in the lymphosarcoma cells can bind to the MT-I promoter in vivo
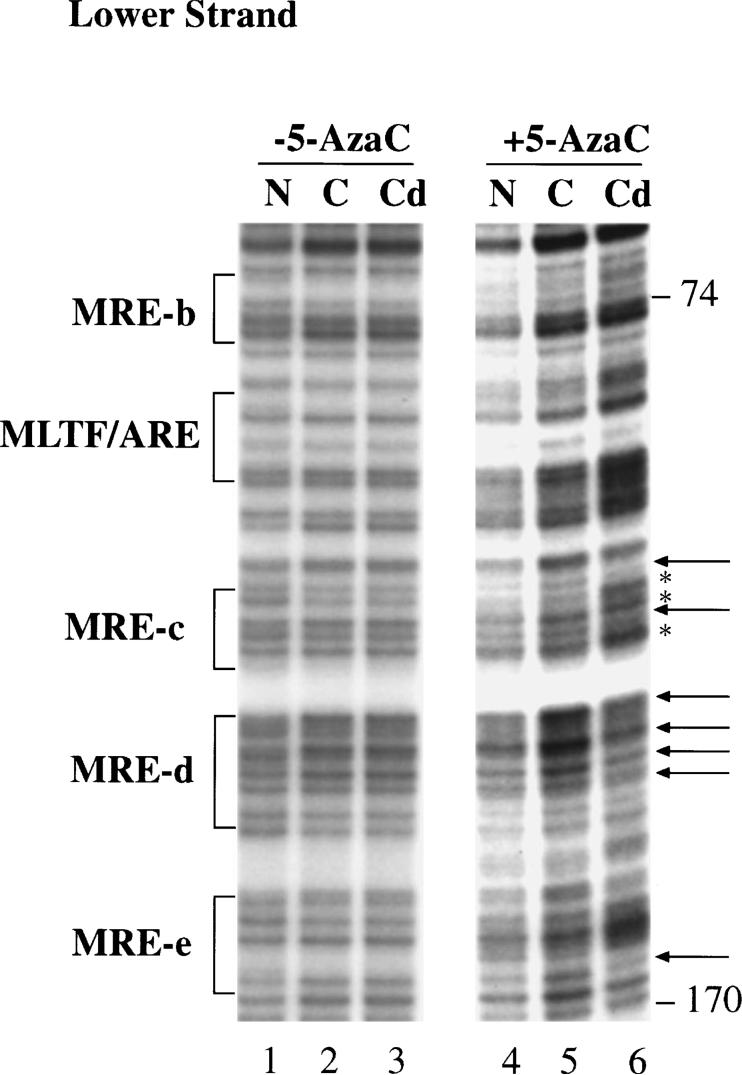
There are several plausible mechanisms by which methylation can inhibit induction of MT-I in the lymphosarcoma cells in response to heavy metals. These include: (a) direct inhibition of the binding of one or more transcription factors to their cognate elements; (b) alteration of chromatin structure; (c) positioning of inhibitory nucleosome; or (d) binding of a repressor molecule that physically blocks binding of a positive factor(s). To distinguish between these possibilities, we performed in vivo genomic footprinting (IVGF) of MT-I promoter region spanning all six metal regulatory elements (MREs) and MLTF/ARE with primers specific for the upper and lower strands. The results for the lower strands are shown in Figure 2. The upper strand-specific primers gave similar results (data not shown). We analysed footprinting in the control lymphosarcoma cells (lane 2) and cells treated with CdSO4 (15 μm) for 3 h (lane 3), and compared those with the G-ladder obtained after dimethyl sulfate treatment and piperidine cleavage of naked DNA (lane 1). To obtain better footprinting, we performed the experiment with cells treated with CdSO4 instead of ZnSO4, as Cd2+ induces MT-I to higher level than Zn2+ (see Figure 1). None of the G residues in the control or cadmium-treated cells appeared to be either protected or hypersensitive compared to those in the naked DNA (Figure 2). We, therefore, concluded that none of the transcription factors, e.g., MTF-1, MLTF, C'BP-1 or C'BP-2 show either constitutive or heavy metal-inducible binding in lymphosarcoma cells. On the contrary, IVGF analysis of DNA from 5-AzaC treated (72 h) P1798 cells after CdSO4 treatment showed heavy metal-induced footprinting at MRE-c, MRE-d, MRE-e (compare lane 6 with lane 5). The footprinting results correlates well with the Northern blot analysis (Figure 1). This observation, therefore, implies that methylation at CpG dinucleotides in MT-I promoter does not allow the essential positive factors to bind to their cognate elements in lymphosarcoma cells, unless the promoter is demethylated with 5-AzaC.
Figure 2.
In vivo footprinting analysis of MT-I promoter with 5′ lower strand-specific primers before and after 5-AzaC treatment. Lymphosarcoma cells before and after treatment with 5-AzaC were incubated with 0.1% dimethyl sulfate as described in Materials and methods. Genomic DNA was purified and cleaned with piperidine. Using LM-PCR, the lower strand of the MT-I proximal promoter was amplified and separated on 6% sequencing gel. The G-ladder was detected by autoradiography. N-Naked G-ladder, C-Control and Cd2+-15 μm CdSO4 treatment. The regulatory elements MRE-b, MRE-c, MRE-d, MRE-e and MLTF/ARE are indicated adjacent to the G-ladder. Arrows (←) indicate G-residues protected due to CdSO4 treatment and asterisks (*) indicate hypersensitive G-residues. Lanes 1 − 3 represent G-ladder in the naked DNA, control and CdSO4-treated P1798 cells respectively, whereas lanes 4 − 6 denote those from the cells treated with 5-AzaC for 72 h. Appropriate positioning of the lower arrow in MRE-c binding site is difficult due to smiling of the gel, particularly lane 6
The transcription factors that regulate basal as well as inducible expression of MT-1 gene are active in lymphosarcoma cells
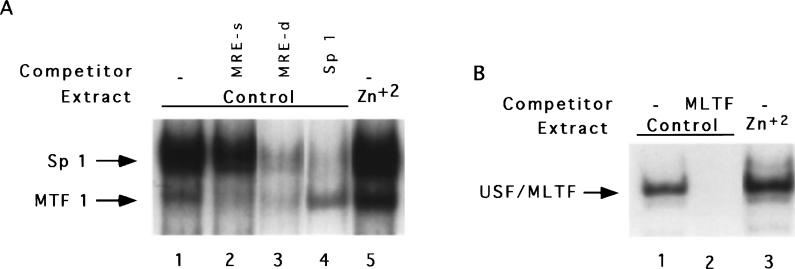
The lack of basal or inducible binding of transcriptional activators to MT-I promoter in vivo in lymphosarcoma cells may be due to absence of one or more key transcription factors required for activation of this gene, which might be induced after 5-AzaC treatment. To address this issue, we measured the DNA binding activity of a few key transcription factors in the extract from these cells. One such factor is MTF-1 (metal-responsive transcription factor) that was cloned and characterized by Walter Schaffner's group (Heuchel et al., 1994; Radtke et al., 1993). This factor is a 70 − 80 kDa protein that interacts with the metal regulatory elements (MREs), particularly MRE-d, and augments both constitutive and induced expression of MT. To test the possibility that MTF-1 is not active in lymphosarcoma cells, its DNA binding activity was assayed by EMSA with 32P-labeled MRE-d oligo in S-100 extract from these cells (Figure 3a). MRE-d oligo spans the binding sites for both MTF-1 and Sp 1 (Radtke et al., 1993). In the extract from the control cells, two complexes were formed (lane 1). The faster migrating complex was formed by MTF-1, as its formation could be competed by an excess of unlabeled mutant oligo, MRE-s (Radtke et al., 1993), to which only MTF-1 can bind (lane 2). The upper complex corresponds to that formed by Sp 1, as its formation could be competed by Sp 1 consensus oligo (lane 4). Both of these complexes could be competed by an excess of cold MRE-d oligo (lane 3). Phosphorimager analysis revealed fourfold activation of MTF-1 and twofold activation of Sp 1 in the extract prepared from the cells treated with 50 μm ZnSO4 (compare lane 5 with lane 1). Both of these transacting factors are zinc-finger proteins and activation of MTF-1 is more pronounced, as it has six zinc fingers in comparison to three zinc fingers for Sp 1. These results show that the lack of expression of MT in the lymphosarcoma cells is not due to inactivation of the transcription factors MTF-1 or Sp 1. We also measured the DNA binding activities of MLTF/USF which stimulates the basal (Carthew et al., 1987) and heavy metal-induced (Li et al., 1998) expression of MT-I in some cell lines (Figure 3b). One major complex was formed in the control lymphosarcoma cell extract (lane 1) that could be competed with an excess of E-box consensus oligo (lane 2). In the extract from the zinc-treated cells, MLTF was activated twofold (lane 3). These results indicate that although these transcriptional activators are expressed in lymphosarcoma cells and are activated also in response to heavy metals, they are unable to footprint on the MT-I promoter due to methylation at CpG islands of the promoter. Our study also shows that demethylation of 10 − 20% of mCpG is adequate for rendering the promoter accessible to positive factors.
Figure 3.
Electrophoretic mobility shift assay of MTF-1, Sp1 and MLTF/ARE in S-100 extract of lymphosarcoma cells. (a) S-100 extracts (10 μg of protein) from the control and ZnSO4 (50 μm for 3 h)-treated cells were incubated with 32P-labeled MRE-d oligo under optimal binding conditions and the DNA-protein complexes were separated on a polyacrylamide (4% acrylamide, acrylamide:bisacrylamide=38.7:1.3) gel with 0.25×TBE as the running buffer. Lanes 1 and 5 represent the complexes formed with the extract from the control and zinc-treated cells, respectively. Lanes 2 − 4 indicate the control extract incubated with 100-fold molar excess of unlabeled MRE-s, MRE-d and Sp 1 oligo respectively. (b) The DNA binding activity of MLTF/USF was measured using 32P-labeled MLTF/ARE oligo with the extract described in (a) and the complexes were resolved on a 6% acrylamide gel with Tris (40 mm) -glycine (267 mm) as running buffer. Lanes 1 and 3 indicate the complexes formed with the extracts from the control and zinc-exposed cells, respectively and lane 3 denotes the control extract incubated with 100-fold molar excess of E-box consensus oligo
MT-I promoter is not methylated in the parental cells (mouse thymus) but is methylated at all CpG sequences in mouse lymphosarcoma cells
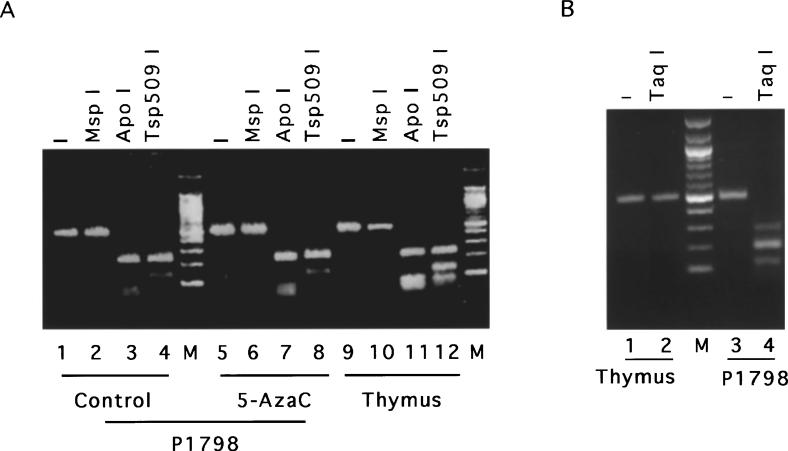
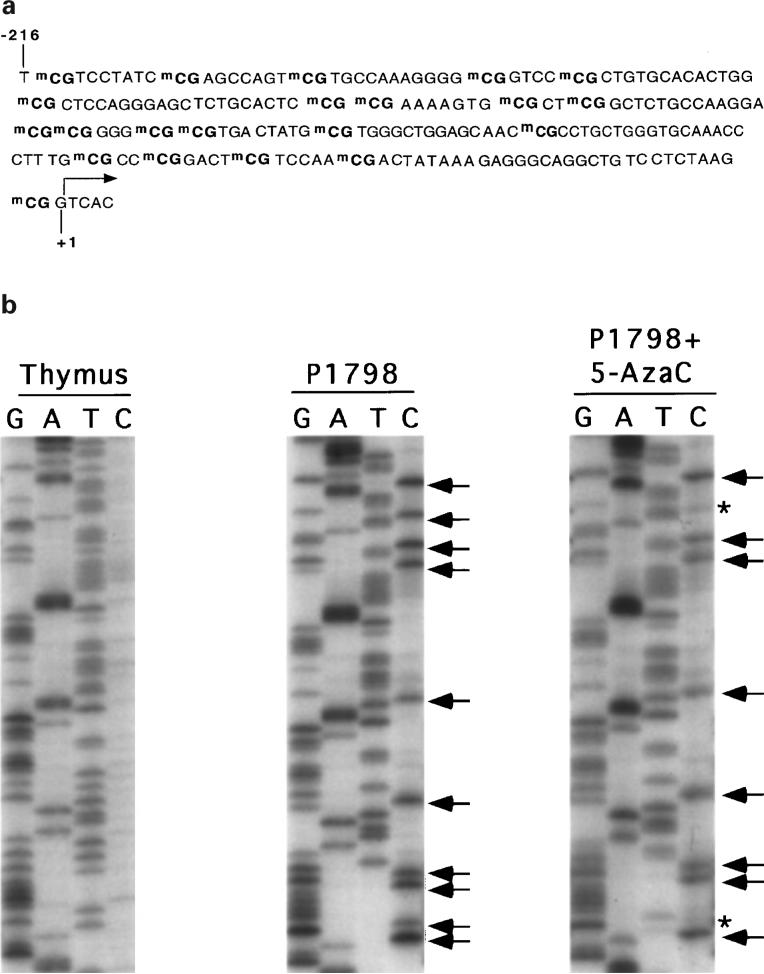
MT-I gene is inducible by heavy metals in lymphosarcoma cells only after 5-AzaC treatment despite the presence of functionally active transcription factors in the cell. As this tumor cell line was derived from the thymus that can express MT in response to various inducers (Wood and Thompson, 1984), we investigated the methylation state of the immediate promoter of the MT-I gene in P1798 cells and thymus. The mouse MT-I promoter sequence and the region proximal to the promoter (−502 bp to +1 bp) contains 32 CpG base pairs that are potential sites for methylation by DNA-MTase. With the advent of bisulfite genomic sequencing it is possible to determine precisely the methylation state of specific CpG base pairs in a DNA sequence (Clark et al., 1994; Frommer et al., 1992). Sodium bisulfite treatment converts the cytosine residues in the genomic DNA to uracil, which are amplified as thymine during subsequent PCR and sequencing (see Materials and methods). However, the methylated cytosines remain unaltered upon bisulfite treatment and read as cytosine upon sequencing. Genomic DNAs were isolated from mouse thymus, P1798 cells, and 5-AzaC treated (for 72 h) P1798 cells, and then treated with bisulfite reagent. MTI-promoter (from −502 bp to −2 bp) was amplified with gene specific primers designed to amplify the upper strand of the bisulfite-converted DNA (see Materials and methods). We designed the primers spanning the region of the promoter that does not contain any CpG sequence to avoid preferential amplification of the methylated DNA (Warnecke et al., 1997). In order to confirm that the bisulfite conversion of the genomic DNA was essentially complete, we digested the PCR-amplified DNA from each sample with ApoI (G/C ↓AATT↑ A/T) and TSp509 I (↓AATT↑). These restriction sites are absent in the control (bisulfite-untreated) MT-I promoter region from −502 bp to −2 bp and are generated only after conversion of C to T residues by bisulfite treatment and PCR amplification. As can be seen in Figure 4a, the PCR amplified DNAs from all three samples were cleaved completely by ApoI and Tsp509 I, respectively, indicating complete bisulfite conversion. To detect the presence of CpG sites, if any, after bisulfite treatment, we digested the amplified DNA with Taq I (T↓CG↑A). This restriction site is not present within the −502 bp to −2 bp region of mouse MT-I DNA, but is generated after CmCGA (internal C methylation) is converted to TCGA upon bisulfite conversion. Complete digestion of the amplified product from the lymphosarcoma DNA and lack of digestion of that from the thymus with TaqI (Figure 4b) revealed that CpG methylation occurred at some step during the development of the tumor. Recent studies have shown the existence of mCpmCpG sequences in some mammalian cells (Clark et al., 1995). To identify the occurrence of both external and internal cytosine methylation, the bisulfite-treated, amplified product from P1798 cells was digested with MspI (C↓CG↑G). The restriction site of this enzyme is retained only if both cytosines are methylated in the chromosomal DNA. MspI failed to cleave the amplified DNA from the control and 5-AzaC-treated lymphosarcoma cells as well as from the thymus (Figure 4a) indicating lack of methylation of external cytosines of CpCpG sequences in P1798 cells or thymus. Bisulfite treatment and subsequent PCR were performed with several batches of DNA. The samples that showed complete bisulfite conversion (as evidenced by restriction enzyme digestion) were subjected to PCR sequencing.
Figure 4.
Restriction enzyme site mapping of the PCR-amplified mouse MT-I promoter with strand-specific primer after bisulfite conversion of the chromosomal DNA. Chromosomal DNA was isolated from the control and 2.5 μm 5-AzaC-treated (72 h) P1798 cells as well as from mouse thymus. These samples were then treated with bisulfite and the upper strand of MT-I promoter was amplified with strand specific primers by nested PCR as described in Materials and methods. The amplified product (1 μg) was then digested with various restriction enzymes, separated on a 2% low-melting agarose gel and stained with ethidium bromide. (a) Lanes 2 − 4 represent control lymphosarcoma DNA digested with MspI, ApoI and Tsp509I, respectively. Similarly, lanes 6 − 8 denote amplified DNAs from 5-AzaC-treated cells and lanes 10 − 12 indicate thymus DNA digested with the same restriction enzymes as the control. Lanes 1, 5 and 9 represent the PCR product from the control, 5-AzaC-treated lymphosarcoma and thymus, respectively and lane M denotes the 100 bp DNA ladder (New England Biolab). (b) Lanes 1 − 4, represent the TaqI digested products of the amplified DNA from the control and 5-AzaC-treated lymphosarcoma cells, respectively
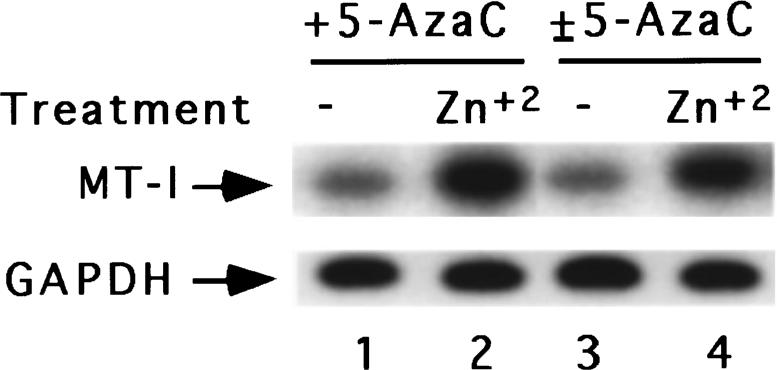
Mouse MT-I promoter spanning −502 bp to +1 bp with respect to transcription initiation site contains 32 CpG dinucleotides. To confirm whether all of the CpG islands are methylated in the control cells and to determine how many are demethylated after 5-AzaC treatment, we sequenced the PCR amplified products. Figure 5a delineates the methylated CpG dinucleotides of the MT-I promoter in P1798 cells, whereas Figure 5b depicts a part of the sequence of amplified DNA from mouse thymus and lymphosarcoma cells before and after 5-AzaC treatment. Critical analysis of the sequencing data revealed that cytosine residues of all the 21 CpG sequences in the −216 bp to +1 bp region encompassing the positive factor binding sites are methylated in P1798 cells. However, none of the CpG dinucleotides are methylated in thymus (Figure 5b). When MT-I is induced by heavy metals following pretreatment of P1798 cells with 5-AzaC, 10 − 20% of the CpG dinucleotides are demethylated depending on the extent of 5-AzaC treatment (time of exposure of cells to 5-AzaC) (data not shown). This data implicates that demethylation of all the CpG dinucleotides are not necessary for expression of the MT-I gene in these cells. The demethylated CpG dinucleotides persisted even after 5-AzaC-treated cells were grown in drug-free medium for ten doubling time (±5-AzaC) (data not shown). This data is consistent with the Northern blot results, where similar basal and heavy metal (zinc)-induced expression of MT-I were observed in the cells treated with 5-AzaC (+5-AzaC) and cells treated with 5-AzaC and grown subsequently in the drug-free medium (±5-AzaC) (Figure 6). The methylation status of MT-I promoter in ±5-AzaC cells suggests that demethylated CpGs are not immediately remethylated by the DNA methyltransferase at least within ten generation time.
Figure 5.
(a) Methylated CpG dinucleotides in mouse MT-I promoter as identified by bisulfite genomic sequencing in lymphosarcoma cells. (b) PCR sequencing of the bisulfite-converted and amplified upper strand DNA of MT-I promoter from lymphosarcoma cells before and after 5-AzaC treatment and from mouse thymus. PCR-amplified upper strand of MT-I gene was subject to PCR sequencing using femtomol DNA sequencing kit (Promega) with the same primers used in the second round of nested PCR, (a) thymus, (b) control and (c) 5-AzaC-treated lymphosarcoma cells
Figure 6.
Northern blot analysis of MT-I mRNA in cells treated with 5-AzaC and then grown in absence of 5-AzaC. Cells treated with 5-AzaC (+5-AzaC) and subsequently grown in the drug-free media (±5-AzaC) were treated with 50 μm ZnSO4 for 3 h. Total RNA isolated (30 μg) from these cells was subjected to Northern blot analysis with MT-I or GAPDH as probe as described in Figure 1. Lanes 1 and 2 represent the RNAs from untreated or zinc-treated +5-AzaC cells, whereas lanes 3 and 4 denote RNAs from untreated and zinc-treated±5-AzaC cells, respectively
Discussion
The demethylating agent, 5-AzaC has been shown to derepress MT-I gene in two thymoma cell lines, S-49 and W-7 (Compere and Palmiter, 1981). These studies did not, however, investigate whether the parental cells (thymus) are capable of inducing MT in response to heavy metals and whether the reactivation of MT-I promoter is due to direct effect of 5-AzaC on the promoter or to demethylation and consequent activation of a gene specific transcription factor. If MT-I promoter is hypermethylated, the nature of methylation sites in the active and inactive states has not been established. Further, none of the studies have addressed whether silencing of MT genes by promoter methylation is a characteristic of thymus derived tumors. Our results show that MT-I gene is silent in the lymphosarcoma cells but not in the thymus due to methylation of CpG islands within the promoter or the region proximal to the promoter. We have also observed recently that silencing of MT-I gene due to methylation is not confined to lymphoid-derived tumors (K Ghoshal and ST Jacob, unpublished data). These observations suggest that at some stage of tumor deveopment, MT-I promoter becomes susceptible to methylation by DNA methyl transferase (DNA-MTase). It is not known why only certain genes containing CpG islands are specifically methylated in the tumors.
Inducibility of MT-I gene by heavy metals after 5-AzaC treatment gave the first indication that the gene is silent in lymphosarcoma cells due to methylation. As these cells grow very slowly in presence of 5-AzaC, the cells were treated with relatively low concentration of the drug for a prolonged time (72 − 90 h). It was necessary that cells were allowed to divide at least once, as DNA replication was necessary for demethylation (Santi et al., 1984). To explore the methylation state of MT-I proximal promoter region that contains 32 CpG bps, the chromosomal DNA was subjected to genomic sequencing after bisulfite treatment and PCR. This novel technique has some limitations, which include (a) partial conversion of nonmethylated cytosines to uracils after bisulfite treatment giving overestimation of methyl CpG, and (b) preferential amplification of methylated molecules over nonmethylated ones or PCR bias (Warnecke et al., 1997). The protocol modified by us (B Yuan, unpublished data) converted, nonmethylated cytosines completely to uracils, as determined by restriction mapping and sequencing (see Materials and methods for details). In many cases, the cytosines can be partially converted to uracils during bisulfite conversion, suggesting the existence of CpCpG methylation (Harrison et al., 1998). We did not observe this type of methylation in MT-I gene in lymphosarcoma cells although there are several potential CpCpG sites, which confirms the complete conversion of C to U by this modified bisulfite method. Interestingly, demethylation of few of 32 CpG dinucleotides was sufficient to render the gene inducible to heavy metals. In fact, demethylation of just 10 − 20% CpG elements in the proximal promoter was adequate to de-repress the silent MT-I gene in the mouse lymphosarcoma cells. Even after prolonged treatment with the analog only 20 − 25% demethylation occurred in these cells and we never observed complete demethylation. The extent of CpG methylation that occurs in the promoter region of different genes that are silent due to methylation varies from tissue to tissue and also differs from gene to gene. Interestingly methylation of just one CpG sequence has been shown to be sufficient to silence Epstein Barr virus latency C promoter that codes for six viral proteins (Robertson et al., 1995). There was, however, a strong correlation of demethylation and expression level of MT-I gene. The extent of induction after heavy metal exposure increased with the extent of demethylation (data not shown). It is known that the density of methyl CpG is important for silencing the promoter (Cameron et al., 1999). Accordingly, when methylation density drops below a critical value, the gene becomes inducible.
We have also explored the potential mechanism by which methylation represses MT-I gene expression. Methylation can directly inhibit binding of transactivators to the specific promoter elements. In vitro studies have, however, shown that methylation at Sp 1 site does not inhibit its binding to its cognate site. On the other hand, methylation of cytosines at MRE-d sequence increases binding of MTF-1 several fold (Radtke et al., 1996). Only MLTF binding to MLTF/USF binding site has been shown to be inhibited by methylation (Watt and Molloy, 1990). It is likely that methylation may inhibit binding of all positive factors by altering chromatin structure or a specific methyl-CpG binding protein (MeCP) that directly prevents access of positive factors to the promoter. Alternatively, methylation can lead to recruitment of large repressor molecules like Sin3A and histone deactylase that alter chromatin structure and inhibit binding of activators. Genomic footprinting analysis did not reveal constitutive or inducible binding of factors in the region of the promoter spanning MREs, Sp 1, MRE-c' or MLTF/ARE. We could not detect any MeCP binding by in vivo DMS-footprinting, which is probably due to weak binding of MeCP to methyl CpG in vivo. Interaction of MeCPs with the methylated CpG sequences appears to be weak as in vitro DNase 1 footprinting or methylation interference assay of these factors is known to be technically cumbersome (Nan et al., 1997). This relatively weak interaction could lead to easy penetration of small molecules like DMS and methylation of guanosines in contact with MeCP. UV crosslinking studies and electrophoretic mobility shift assay with the methylated proximal promoter region of MT-I and lymphosarcoma nuclear extract can address whether a MeCP specifically binds to this region and is associated with some other polypeptides to form repressor complex on the methylated promoter. Studies along this line are in progress.
A striking observation was the continued response of P1798 cells to heavy metals despite withdrawal of 5-AzaC after initial demethylation and growth of the cells for as much as ten doubling time. These data support the notion that de novo methylase is not highly active in these lymphosarcoma cells. It is possible that the de novo methylase activity was induced transiently at the initial stage of development of the lymphosarcoma from thymus and methylated MT-I gene. Later on, the methylation state of the gene was perpetuated by maintenance methylase that uses hemimethylated DNA as substrate (Szyf, 1996). When both strands are demethylated after 5-AzaC treatment the maintenance methylase cannot methylate those CpG dinucleotides. Recently, it has been shown that multiple forms of DNA-MTase are generated in different tissues by alternative splicing (Deng and Szyf, 1998). Although the functions of these variants are not yet known, it is likely that P1798 cells express the form that catalyzes maintenance methylation but not the one that displays de novo methylase activity.
Significantly higher methylation that occurs in many tumor cell lines appears to be due to higher DNA methyl transferase activity (Szyf, 1996). The direct role of DMA-MTase in tumor propagation has been shown by overexpression of the cDNA for this protein that resulted in cell transformation, whereas the expression of the antisense RNA prevented it. It would be of interest to compare the activity and the expression level of this enzyme in the thymus and the lymphosarcoma. We also measured the expression level of several tumor suppressor genes, e.g., pRb, p53, p16 that are silent in a variety of tumors by methylation. Surprisingly, all three of them are expressed in the lymphosarcoma cells (C Dong and ST Jacob, unpublished data). In this context, one can speculate that another tumor suppressor gene that may be specific to T cell-derived tumors is suppressed in P1798 cells by methylation of its promoter. Alternatively, metallothionein by itself or in concert with another tumor suppressor could function as a suppressor of growth in these cancer cells. Studies along these lines are in progress. Nevertheless, this study has conclusively demonstrated that hypermethylation of MT-I promoter is solely responsible for its repression in the mouse lymphosarcoma cells.
Materials and methods
Cell culture, treatment with 5-azacytidine and heavy metals
Mouse lymphosarcoma P1798 cells were a gift from Dr AE Thompson, University of Texas, Galvastone, USA. These cells were grown in RPMI 1640 medium containing 25 mm HEPES (pH 7.2), 2 mm glutamine, 2% sodium bicarbonate, 0.2 μm β-mercaptoethanol and 5% fetal bovine serum. Cells at a density of 0.5 × 106 ml were treated with 2.5 μm 5-azacytidine (Sigma) for 72 − 90 h until the cells divide at least once. The control or 5-AzaC-treated cells at a density of 1 × 106/ml were treated with CdSO4 (15 μm) or ZnSO4 (50 μm) for 3 h.
Isolation of total RNA and Northern blot analysis
Total RNA was isolated from 107 cells by guanidinium thiocyanate-acid phenol method (Chomczynski and Sacci, 1987). Thirty micrograms of total RNA was separated by formaldehyde-agarose (1.2%) gel electrophoresis and transferred to nylon membrane which was then hybridized to mouse random-primed, α-32P-dCTP-labeled MT-I cDNA or rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe in rapid hybridization buffer (Amersham) following the manufacturer's protocol.
Electrophoretic mobility shift assay (EMSA) of transactivators that bind to mouse MT-1 promoter
The DNA binding activities of factors were measured in the S-100 extract prepared from the lymphosarcoma cells following published protocol (Ghoshal and Jacob, 1996). For this purpose, 10 μg of the extract was incubated with 0.1 − 0.5 ng of the 32P-labeled deoxyoligonucleotides in the buffer containing 10 mm HEPES (pH 7.9), 60 mm KCl, 5 mm MgCl2, 0.5 mm DTT, 10% glycerol, and 1 μg of poly(dI-dC). The oligonucleotides are labeled by end-filling with α-32P-dGTP and three other dNTPs with Klenow. MTF-1 and Sp 1 activities were measured with MRE-d oligo (Ghoshal et al., 1998). For competitive EMSA, the extract was preincubated with 100-fold molar excess of the unlabeled competitor for 15 min on ice before addition of the labeled oligo. The reaction mixture was incubated on ice for 30 min and the DNA-protein complex was separated by polyacrylamide (4% acrylamide) gel electrophoresis. The sequences of the upper strand of the deoxyoligonucleotides used are the following: (1) MRE-d oligo (for Sp 1 and MTF-1) 5′-GATCCAGGGAGCTCTGCACTCCGCCCGAAAAGTA; (2) MRE-s oligo (for MTF-1) 5′-GATCCAGGGAGCTCTGCACACGGCCCGAAAAAGTA; (3) MLTF/ARE (for MLTF) 5′-GATCCGCGGGGCGCGTGACTATGCGTGGGCTGGA; (4) MLTF (for MLTF) 5′-CACCCGCACGTGCCTACACC.
Bisulfite genomic sequencing of MT-I promoter in lymphosarcoma cells to determine the methylated cytosine residues
For bisulfite sequencing of MT-I promoter in lymphosarcoma cells or mouse thymus, we followed the method of Clark et al. (1994), with some modifications, to facilitate complete conversion of cytosines to uracils, methyl cytosines remaining unaltered. DNA was isolated from lymphosarcoma cells and mouse thymus and denatured (5 μg) with 0.3 m NaOH (freshly prepared) at 37°C for 30 min. Freshly prepared sodium metabisulfite solution (2.35 m) containing hydroquinone (0.04 m) were added to the denatured DNA, and cycled twenty times in a thermal cycler at 50°C for 30 min and 95°C for 2 min. The bisulfite-treated DNA was desalted using Wizard DNA Clean Up kit (Promega), desulfonated with 50 mm NaOH at 37°C for 30 min, neutralized with sodium acetate (pH 4.5) (0.2 m, final concentration) and purified using Wizard DNA Clean Up kit. An aliquot of the converted DNA was used for amplification of the MT-I promoter. The primers used to amplify the upper strand of the bisulfite-converted MT-I gene are the following: For first round of PCR (m=mouse): mMTI-S1: 5′-TAGAGTAGATGGGTTAAGGTGAGTG; mMTI-A1: 5′-ATCCCCACTTAATA-TTCTAAAAACC. For nested PCR: mMTI-S2: 5′-AGGAGTAGAGAATAATGTTGAGATGAGT; mMTI-A2: 5′-CTT-AAAAAACAACCTACCCTCTTTATAAT (annealing temperature, 60°C).
To test the efficiency of bisulfite reaction the amplified DNA (500 bp) was first digested with the restriction enzymes ApoI (G/C ↓AATT↑ A/T) and Tsp509 I (↓AATT↑) that can cleave only the converted but not the uncoverted DNA. The restriction sites for these two enzymes are generated only when C residues are converted to T residues. Completion of bisulfite conversion is confirmed by complete digestion of the amplified DNA with ApoI or Tsp509 I. Next we sequenced the PCR products from lymphosarcoma cells and thymus by fmol sequencing system (Promega) with nested PCR primers as well as the internal primer 5′-TTGGGGAAAGTATTATAGGGATATGATG for the upper strand. In the bisulfite-treated DNA only methylated cytosines will be sequenced as Cs and the converted unmethylated C residues will be sequenced as Ts. Bisulfite genomic sequencing was also performed with chromosomal DNA from the cells treated with 2.5 μm 5-AzaC for 72 h which express MT-1 in response to heavy metals.
In vivo genomic footprinting analysis of the promoter region of mouse MT-I gene of lymphosarcoma cells before and after demethylation with 5-AzaC
In vivo methylation of cellular DNA and extraction of DNA were as described by Mueller and Wold (1989). The DNA sequence of interest was amplified by ligation-mediated PCR (LM-PCR) according to the procedure of Meuller and Wold, as modified by Ping et al. (1996). Briefly, the procedure involves exposure of the lymphosarcoma cells in the culture media (RPMI) to dimethyl sulphate (1 μl/ml) for 2 min at room temperature, followed by purification of genomic DNA. The methylated DNA was then cleaved at positions of methyl guanine with piperidine (10%) at 90°C for 30 min. The purified, cleaved DNA (2 μg) was then subjected to LM-PCR to analyse the footprinting at MRE sequences and MLTF/ARE with mouse MT-I promoter specific primers as described previously (Mueller and Wold, 1989). The primers used for LM-PCR of the lower strand are the following: (1) 5′-GAGTTCTCGTAAACTCCAGAGCAGC; (2) 5′-CAGAGCAGCGATAGGCCGTAATATC; (3) 5′-GATAGGCCGTAATATCGGGGAAAGCAC.
Acknowledgments
The authors are grateful to Aubrey Thompson and Richard Palmiter for providing mouse lymphosarcoma P1798 cells and mouse MT-I minigene, respectively. This work was supported, in part, by the USPHS Grant CA61321 to ST Jacob and ACS Institutional Research Grant to K Ghoshal.
References
- Aniskovitch LP, Jacob ST. Arch. Biochem. Biophys. 1997;341:337–346. doi: 10.1006/abbi.1997.9976. [DOI] [PubMed] [Google Scholar]
- Aniskovitch LP, Jacob ST. Oncogene. 1998;16:1475–1486. doi: 10.1038/sj.onc.1201659. [DOI] [PubMed] [Google Scholar]
- Aschner M, Cherian MG, Klaassen CD, Palmiter RD, Erickson JC, Bush AI. Toxicol. Appl. Pharmacol. 1997;142:229–242. doi: 10.1006/taap.1996.8054. [DOI] [PubMed] [Google Scholar]
- Baylin SB. Science. 1997;277:1948–1949. doi: 10.1126/science.277.5334.1948. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Chodosh LA, Sharp PA. Genes Dev. 1987;1:973–980. doi: 10.1101/gad.1.9.973. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chubatsu LS, Meneghini R. Biochem. J. 1993;291:193–198. doi: 10.1042/bj2910193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Frommer M. Nat. Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compere SJ, Palmiter RD. Cell. 1981;25:233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- Datta PK, Jacob ST. Cell. Mol. Biol. Res. 1993;39:439–449. [PubMed] [Google Scholar]
- Datta PK, Jacob ST. Biochem. Biophys. Res. Commun. 1997;230:159–163. doi: 10.1006/bbrc.1996.5655. [DOI] [PubMed] [Google Scholar]
- Deng J, Szyf M. J. Biol. Chem. 1998;273:22869–22872. doi: 10.1074/jbc.273.36.22869. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Jacob ST. Biochem J. 1996;317:689–695. doi: 10.1042/bj3170689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Li Z, Jacob ST. Proc. Natl. Acad. Sci. USA. 1998;95:10390–10395. doi: 10.1073/pnas.95.18.10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Wang Y, Sheridan J, Jacob ST. J. Biol. Chem. 1998;277:27904–27910. doi: 10.1074/jbc.273.43.27904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Stirzaker C, Clark SJ. Anal. Biochem. 1998;264:129–132. doi: 10.1006/abio.1998.2833. [DOI] [PubMed] [Google Scholar]
- Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. EMBO J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Kagi JA. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- Kass SU, Pruss D, Wolffe AP. Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- Kelley SL, Basu A, Teicher BA, Hacker MP, Hamer DH, Lazo JS. Science. 1988;241:1813–1815. doi: 10.1126/science.3175622. [DOI] [PubMed] [Google Scholar]
- Klein CB, Costa M. Mutat. Res. 1997;386:163–180. doi: 10.1016/s1383-5742(96)00052-x. [DOI] [PubMed] [Google Scholar]
- Knight SJ, Flannery AV, Hirst MC, Campbell L, Christodoulou Z, Phelps SR, Pointon J, Middleton-Price HR, Barnicoat A, Pembrey ME, et al. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Cell. 1998;93:301–303. doi: 10.1016/s0092-8674(00)81154-x. [DOI] [PubMed] [Google Scholar]
- Lie E, Beard C, Forster AC, Bestor TH, Jaenisch R. Cold Spring Harb. Symp. Quant. Biol. 1993;58:297–305. doi: 10.1101/sqb.1993.058.01.035. [DOI] [PubMed] [Google Scholar]
- Li Q, Hu N, Daggett MA, Chu WA, Bittel D, Johnson JA, Andrews GK. Nucleic Acids Res. 1998;26:5182–5189. doi: 10.1093/nar/26.22.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, Wold B. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Ng MH, Chung YF, Lo KW, Wickham NW, Lee JC, Huang DP. Blood. 1997;89:2500–2506. [PubMed] [Google Scholar]
- Norris DP, Patel D, Kay GF, Penny GD, Brockdorff N, Sheardown SA, Rastan S. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Palmiter RD. EXS. 1987;52:63–80. doi: 10.1007/978-3-0348-6784-9_4. [DOI] [PubMed] [Google Scholar]
- Ping D, Jones PL, Boss JM. Immunity. 1996;4:455–469. doi: 10.1016/s1074-7613(00)80412-4. [DOI] [PubMed] [Google Scholar]
- Pitt BR, Schwarz M, Woo ES, Yee E, Wasserloos K, Tran S, Weng W, Mannix RJ, Watkins SA, Tyurina YY, Tyurin VA, Kagan VE, Lazo JS. Am. J. Physiol. 1997;273:L856–L865. doi: 10.1152/ajplung.1997.273.4.L856. [DOI] [PubMed] [Google Scholar]
- Quaife CJ, Findley SD, Erickson JC, Froelic GJ, Kelly EJ, Zambrowicz BP, Palmiter RD. Biochemistry. 1994;33:7250–7259. doi: 10.1021/bi00189a029. [DOI] [PubMed] [Google Scholar]
- Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Hug M, Georgiev O, Matsuo K, Schaffner W. Biol. Chem. Hoppe Seyler. 1996;377:47–56. doi: 10.1515/bchm3.1996.377.1.47. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Hayward SD, Ling PD, Samid D, Ambinder RF. Mol. Cell. Biol. 1995;15:6150–6159. doi: 10.1128/mcb.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi DV, Garrett CE, Barr PJ. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE. Proc. Natl. Acad. Sci. USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, Lazo JS, Yalowich JC, Allen WP, Whitmore M, Bergonia HA, Tzeng E, Billiar TR, Robbins PD, Lancaster JR, Jr, et al. Proc. Natl. Acad. Sci. USA. 1995;92:4452–4456. doi: 10.1073/pnas.92.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. Pharmacol. Ther. 1996;70:1–37. doi: 10.1016/0163-7258(96)00002-2. [DOI] [PubMed] [Google Scholar]
- Warnecke PM, Stirzacker C, Melki JR, Millar DS, Paul CL, Clark SJ. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F, Molloy PL. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1990;326:267–275. doi: 10.1098/rstb.1990.0010. [DOI] [PubMed] [Google Scholar]
- Wood KM, Thompson AE., Jr Mol. Cell. Endocrinol. 1984;37:169–180. doi: 10.1016/0303-7207(84)90049-2. [DOI] [PubMed] [Google Scholar]
- Yagle MK, Palmiter RD. Mol. Cell. Biol. 1985;5:291–294. doi: 10.1128/mcb.5.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]