Abstract
MicroRNAs are natural, single-stranded, small RNA molecules that regulate gene expression by binding to target mRNAs and suppress its translation or initiate its degradation. In contrast to the identification and validation of many miRNA genes is the lack of experimental evidence identifying their corresponding mRNA targets. The most fundamental challenge in miRNA biology is to define the rules of miRNA target recognition. This is critical since the biological role of individual miRNAs will be dictated by the mRNAs that they regulate. Therefore, only as target mRNAs are validated will it be possible to establish commonalities that will enable more precise predictions of miRNA/mRNA interactions. Currently there is no clear agreement as to what experimental procedures should be followed to demonstrate that a given mRNA is a target of a specific miRNA. Therefore, this review outlines several methods by which to validate miRNA targets. Additionally, we propose that multiple criteria should be met before miRNA target validation should be considered “confirmed.”
Keywords: microRNA, bioinformatics, reporter genes, mRNA targets
Introduction
MicroRNAs (miRNAs) are a family of small nonprotein coding RNAs that have emerged as important regulators of gene expression [1-3]. Currently 474 human miRNAs have been characterized [4]; however, recent reports estimate that the human genome harbors ~1000 miRNA genes [5,6]. MiRNAs are expressed as long hairpin-forming precursor RNAs that get cleaved into partially double-stranded RNAs that are further processed into mature miRNAs (~22 nucleotides) [reviewed in 2]. Mature miRNAs recognize their target mRNAs by base-pairing interactions between nucleotides 2-8 of the miRNA (the seed region) and complementary nucleotides in the 3′-untranslated region (3′-UTR) of mRNAs. MiRNAs inhibit gene expression by targeting mRNAs for translational repression or cleavage [7-9]. Each miRNA has hundreds of evolutionarily conserved targets and several times that number of non-conserved targets [5]. It is currently estimated that 30% of all human genes may be regulated by miRNAs [10].
Identification of miRNA target genes has been a great challenge. Computational algorithms have been the major driving force in predicting miRNA targets [11-14]. These approaches are mainly focused on programming alignment to identify complementary elements in the 3′-UTR with the seed sequence of the miRNA and the phylogenetic conservation of the complementary sequences in the 3′-UTRs of orthologous genes. However, evidence suggests that perfect seed pairing may not necessarily be a reliable predictor for miRNA interactions [15], which may explain why some predicted target sites are nonfunctional. Hence, with few exceptions, most physiologic, and clinically relevant, targets for miRNAs remain to be identified or verified experimentally.
Currently there is no clear consensus as to what criteria should be followed to determine miRNA targets and to confirm their biological efficacy. Therefore, one goal of this review is to establish a set of guidelines that investigators can follow to validate that a given miRNA regulates a specific mRNA target. This review will also discuss a number of experimental procedures that can be utilized to confirm miRNA targets. Based on our previous experience with miRNAs [21,22], we propose a working scheme (Fig. 1) to be followed in order to reduce the number of bioinformatically predicted miRNA binding sites and to expedite the biological validation of the sites of interest. We also advocate that after miRNA/mRNA interactions have been demonstrated, at least three additional criteria should be met before confirmation of a given mRNA target is complete as shown in Figure 1. Although all of these criteria might not be met under all conditions, it is advisable that as many be achieved as possible.
Figure 1.
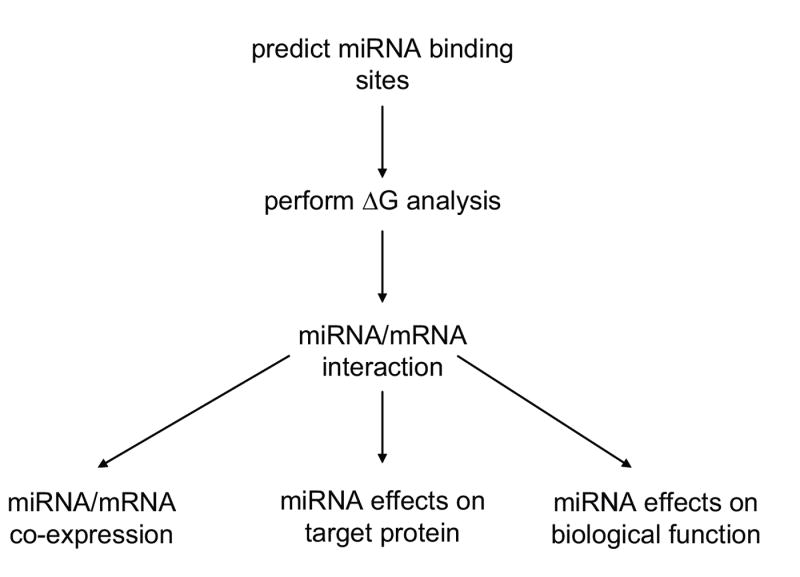
Proposed flow diagram for microRNA validation.
Description of Methods
Bioinformatic Prediction of miRNA Targets
To begin to investigate a predetermined gene as a target of miRNA regulation, individual gene sequences (i.e. 3′-UTR mRNA sequences) may be analyzed by various computational algorithms which utilize distinct parameters to predict the probability of a functional miRNA binding site within a given mRNA target. Due to performance values (i.e. sensitivity and specificity in target prediction) [30], we suggest that the following three bioinformatic algorithms be utilized to predict miRNA target sites: miRanda (http://microrna.sanger.ac.uk) [4,16]; TargetScan (http://www.targetscan.org) [10,17,18]; and, PicTar (http://pictar.bio.nyu.edu) [19,20]. All three of these computational programs allow the investigator to enter a specific “Gene Symbol” and the algorithm will compute all predicted miRNA target sites within that gene. Additionally, these algorithms will determine all of the putative mRNA targets of a given miRNA.
It is important to reiterate that multiple algorithms be utilized since these programs will often predict distinct miRNA binding sites. For example, when the human angiotensin II type 1 receptor (hAT1R) gene (i.e. gene symbol, AGTR1) was analyzed for putative miRNA binding sites, miRanda predicted 27 miRNA binding sites within the hAT1R 3′-UTR (Table 1) [21,22]. TargetScan predicted that over 37 miRNA binding sites were harbored within the hAT1R. Importantly, only eight of the predicted miRNA binding sites overlap between these two bioinformatic algorithms (Table 1). In contrast to these results, when the hAT1R gene was analyzed by the PicTar algorithm, no miRNA binding sites were predicted due to the rigid requirement of conservation of these sites across species (i.e. human, chimp, mouse, rat, dog). Of note, although used as part of the algorithm scheme in all these programs, the conservation of miRNA target sites is not a requirement for a functional miRNA. With the large discrepancies between the results obtained from these computational algorithms, how is it possible to determine which predicted miRNA binding site should be investigated further? We recommend that at least two of the three algorithms must predict the same miRNA binding site(s) before additional validation experiments are conducted. Therefore, in our example above, we would only pursue miR-155, miR-181, miR-527, miR-559, miR-562, miR-586, miR-589, and miR-624 as potential regulators of hAT1R gene expression (these data are summarized in Table 1).
Table 1.
Bioinformatic analyses of the miRNA binding sites that are predicted to target the human AT1R gene
| miRanda | TargetScan | PicTar | ||
|---|---|---|---|---|
| miR-9 | miR-527 | miR-1/206 | miR-410 | No targets |
| miR-21 | miR-559 | miR-7 | miR-431 | |
| miR-29b | miR-562 | miR-33 | miR-493-5p | |
| miR-31 | miR-586 | miR-34/449 | miR-505 | |
| miR-124a | miR-589 | miR-93/291-3p/294/295/302/372/373/520 | miR-511 | |
| miR-132 | miR-595 | miR-96 | miR-512 | |
| miR-148a | miR-613 | miR-103/107 | miR-516-5p | |
| miR-155 | miR-624 | miR-134 | miR-527 | |
| miR-181 | miR-644 | miR-148/152 | miR-559 | |
| miR-200a | miR-648 | miR-155 | miR-562 | |
| miR-350 | miR-699 | miR-181 | miR-573 | |
| miR-489 | miR-757 | miR-182 | miR-586 | |
| miR-505 | miR-770-5p | miR-205 | miR-589 | |
| miR-524 | miR-223 | miR-616 | ||
| miR-299-3p | miR-624 | |||
| miR-375 | miR-629 | |||
| miR-381 | miR-641 | |||
| miR-409-3p | miR-802 |
miRNA binding sites that are predicted by multiple algorithms are shown in boxes.
ΔG Analysis (miRNA Accessibility)
To further reduce the number of putative miRNAs that may repress the expression of a given mRNA target we recommend further in silico analysis since many miRNA targets predicted by seed sequence matching fail validation tests in vivo [15]. There is increasing acceptance that contextual features may also govern miRNA/mRNA interactions. For example, much of a given mRNA sequence is highly structured and only certain single-stranded regions may be accessible for binding with miRNAs. Thus, complex RNA secondary structures may prevent miRNA/mRNA interactions. Recently Zhao et al. [23,24] demonstrated that a common feature of most validated targets is that miRNAs preferentially target 3′-UTR sites that do not have complex secondary structures and are located in accessible regions of the RNA based on favorable thermodynamics. Since RNA accessibility may be a critical feature of miRNA target recognition, we suggest that the free energy (ΔG) of the 70 nucleotides flanking the 5′ and 3′ sides of the predicted miRNA binding sites be determined using mFold [25] as described by Zhao et al. [23,24]. When the ΔG was calculated using nucleotide sequence surrounding the eight predicted hAT1R miRNA binding sites (Table 1), all but miR-589 binding site had a higher ΔG than randomly expected (ΔG = -13.4 kcal/mol) [22] (Table 2) suggesting that the other seven sites may be accessible to miRNAs. Taken together, the bioinformatic data suggests that miR-155, miR-181, miR-527, miR-559, miR-562, miR-586, and miR-624, and possibly miR-589, may play a biologically relevant role in regulating the expression of the hAT1R and should be further pursued.
Table 2.
Predicted ΔG (-kcal/mol) of the 70 nucleotides flanking the 5′ and 3′ regions of the potential miRNA target sites.
| Predicted Overlapping
Human AT1R 3′-UTR miRNA binding sites |
5′70 bp (ΔG) | 3′70 bp (ΔG) |
|---|---|---|
| miR-155 | -10.4 | - 9.0 |
| miR-181 | - 7.8 | -11.0 |
| miR-527 | - 6.6 | -11.2 |
| miR-559 | - 3.7 | - 6.5 |
| miR-562 | -10.4 | - 6.6 |
| miR-586 | - 8.1 | - 6.4 |
| miR-589 | -10.9 | -14.3 |
| miR-624 | -11.4 | -10.6 |
miRNA/mRNA Interactions
Once bioinformatic analyses have been performed and the predicted accessible miRNA binding sites have been determined, the functional importance of a predicted miRNA/mRNA interaction can be validated (Criterion 1). Since the algorithm search may predict a large number of putative miRNA binding sites on a specific mRNA target, a rapid and reproducible assay is needed to quickly eliminate any interaction sites that are not functional. Therefore, we recommend that a reporter system be utilized. The rationale for using this assay is that the binding of a given miRNA to its specific mRNA target site will repress reporter protein production thereby reducing activity/expression that can be measured and compared to a control. The experimental approach is to clone the 3′-UTR of the target gene of interest immediately downstream of the luciferase (Photinus or Renilla) or green fluorescent protein (GFP) open reading frame sequence contained in the reporter plasmid. It is important that the entire 3′-UTR be included since a truncated version of this sequence may provide inappropriate accessibility to a given miRNA. Additionally, by subcloning the entire 3′-UTR of the target gene of interest, a single reporter construct can be utilized to investigate all of the algorithm-predicted miRNA/mRNA binding sites. The recombinant plasmid and a miRNA of interest are then transiently transfected into a host cell, preferably one that does not endogenously express this miRNA, and luciferase activity or fluorescence is measured 24-48 hours after transfection. Alternatively, just the reporter construct can be transfected into cells which express the relevant miRNAs, along with vectors which express mutant versions of the miRNA binding sites. In this experimental system the wild-type reporters should have less activity than their respective mutants. Importantly, this method avoids the need to over-express a given miRNA mimic.
Important control experiments that need to be performed include the use of plasmids which do not contain the 3′-UTR fragment [21], the 3′-UTR insert in the reverse orientation [21], and/or the 3′-UTR insert which does not harbor the miRNA binding site of interest [21]. A control luciferase reporter plasmid (distinct from the plasmid generated above) must also be cotransfected so that the data can be normalized for experimental variation in transfection efficiencies. It is also important to optimize miRNA transfection conditions for every cell type utilized. This can easily be confirmed utilizing fluorescent-labeled miRNAs [21]. In addition, specific and scrambled double-stranded RNA control molecules can be purchased from multiple sources (Ambion, Dharmacon, etc.). These reagents mimic the Dicer cleavage product and are subsequently processed into mature miRNAs (i.e. miRNA mimics).
To demonstrate the utility of this assay, luciferase reporter constructs harboring the hAT1R 3′-UTR in the right and wrong orientation (designated pMIR/883/5′→3′ or pMIR/883/3′→5′) were transfected into host CHO cells with individual miRNA mimics and luciferase activity was determined. Importantly, only the miR-155 mimic was able to reduce luciferase activity (Fig. 2) suggesting that this small RNA was able to interact with the miR-155 binding site harbored within the hAT1R 3′-UTR. The specificity of this interaction was demonstrated by the wrong orientation control and the miR-155 binding site deletion constructs (Fig. 2). Interestingly, even though miR-124a was predicted to regulate hAT1R expression (Table 1, miRanda), the luciferase experiments clearly demonstrated that this miRNA did not target hAT1R mRNA. One possible explanation for this negative result may be due to the lack of seed sequence complementarity between miR-124a and the hAT1R 3′-UTR recognition site [21]. A miR-365 mimic was used as a negative control since none of the algorithms predicted such a binding site in the hAT1R 3′-UTR (Table 1).
Figure 2.
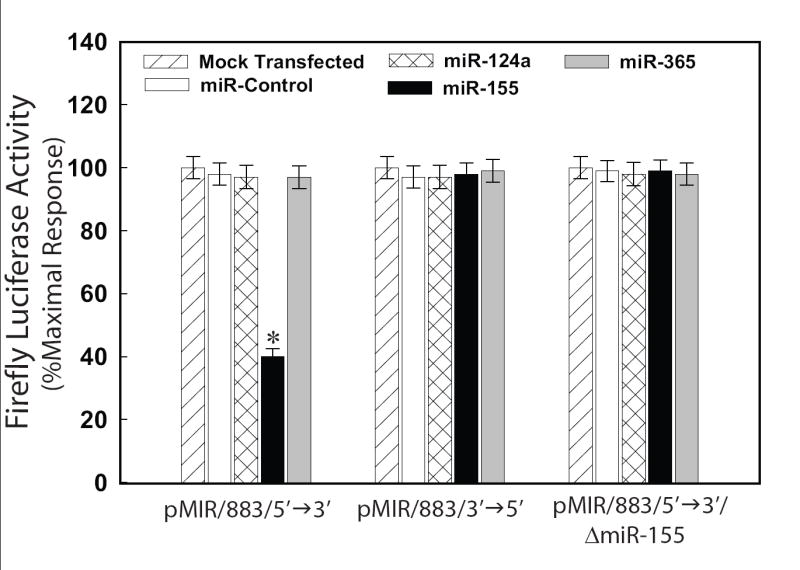
miR-155 can specifically inhibit luciferase reporter activity. CHO cells were cotransfected with pMIR/883/5′→3′, pMIR/883/3′→5′, pMIR/883/5′→3′/ΔmiR-155, pRL-CMV and 50 nM of a given miRNA mimic. Forty-eight hours after transfection luciferase activities were measured. F-luc activity was normalized to r-luc expression and the mean activities ± S.E. from five independent experiments are shown. (*p<0.01 versus CHO cells transfected with miR-control mimic)
Although the ability of miRNAs to repress translation of chimeric reporter genes is a useful screening tool, it remains a surrogate for testing the effects of miRNAs on their putative targets. Additionally, reporter assays can result in misleading assessment of targets since transfection of supra-physiological concentrations of a miRNA may create a situation in which two molecules with complementary surfaces may engage in non-physiological interactions. Finally, the expression of the 3′-UTR and the tested miRNA in a heterologous context may also lead to non-physiological interactions because of an incorrect cofactor environment. Therefore, the validity of each miRNA binding site ascertained by reporter assays must be more directly tested by further experimentation. One possible experimental procedure that could be utilized to validate miRNA/mRNA interactions, without the use of reporter assays, may be the miRNA “pull-down” strategy recently published by Easow et al. [37] and Beitzinger et al. [38]. These investigators demonstrated that by using antibodies against members of the Argonaute (Ago) protein family, a group of proteins known to bind to miRNAs, and to partially complementary sequences in the 3′-UTR of specific target mRNAs, they could co-immunoprecipitate Ago-bound mRNAs. Therefore, this approach provides a means to identify functional miRNA targets based on their physical interaction in vivo. Since “predetermined” target genes are being characterized, this procedure could be utilized to validate whether a given mRNA of interest is interacting with miRNAs in vivo.
miRNA and Target mRNA Co-expression
Clearly the miRNA and its target mRNA must be co-expressed in order for the miRNA to repress the expression of its biological target (Criterion 2). Co-expression is typically demonstrated by simply performing Northern blot analysis or quantitative real-time PCR (qPCR) using total RNA isolated from a specific cell type, and probes or primers specific for a given miRNA and mRNA target. We recommend that qPCR experiments be performed utilizing TaqMan assays (Applied Biosystems, Foster City, CA) specific for a given miRNA and mRNA target due to the ease and reproducibility of these assays. Since the bioinformatic data obtained above suggested that the hAT1R 3′-UTR harbored a putative miR-155 binding, we attempted to demonstrate that miR-155 and the hAT1R were co-expressed in specific cell types. It is well-established that AT1Rs are expressed on endothelial cells, vascular smooth muscle cells (VSMCs) and fibroblasts [26,27]. Therefore, total RNA isolated from primary VSMCs was utilized as template to investigate whether miR-155 and AT1R mRNA were co-expressed in VSMCs. The data shown in Figures 3A and 3B clearly demonstrated that miR-155 and hAT1R mRNA were both expressed in VSMCs suggesting that, if the miR-155 target site was accessible in vivo, the hAT1R levels may be regulated by miR-155.
Figure 3.

Mature miR-155 and the hAT1R are co-expressed in VSMC. The TaqMan® miR-155 and TaqMan® AT1R gene expression assay kits were utilized to quantify the expression of miR-155 and hAT1R levels in VSM and CHO cells. The expression of mature miR-155 or hAT1R relative to 18S rRNA was determined using SYBR green real-time quantitative PCR assay as described [21].
Another experimental technique that can be utilized to demonstrate co-expression of a specific miRNA and target mRNA is in situ hybridization. Importantly, these types of experiments can be utilized to demonstrate that miRNAs are expressed in a tissue- or cell-specific manner from physiologically relevant samples [22,28]. The enhanced efficiency, stability and discriminatory power of “locked nucleic acid-” (LNA) modified oligonucleotide probes make them an ideal tool in detecting mature miRNAs [22,28]. Digoxigenin (DIG)-labeled LNA antisense miRNA-specific probes are synthesized (e.g. Exiqon, Vedbaek, Denmark) and hybridization is performed utilizing fixed and mounted tissues at 37°C overnight followed by a low stringency wash [29]. The probe-target complex is visualized utilizing a digoxigenin antibody conjugated to alkaline phosphatase acting on the chromogen nitroblue tetrazidium and bromochloroindolyl phosphate. In situ hybridization experiments clearly demonstrated that miR-155 was expressed in endothelial cells and VSMCs (Fig. 4A), thus supporting our conclusions that miR-155 was co-expressed with the hAT1R mRNA (Fig. 3). Despite the power of this methodology, direct localization by in situ hybridization should be used as an adjunctive technique with other supporting experiments since data interpretation can be problematic due to the relatively narrow window between signal and background. Alternative in situ-based methods including reverse transcription (RT) in situ PCR for the direct localization of precursor and mature miRNAs may result in more reliable and reproducible in situ assays.
Figure 4.
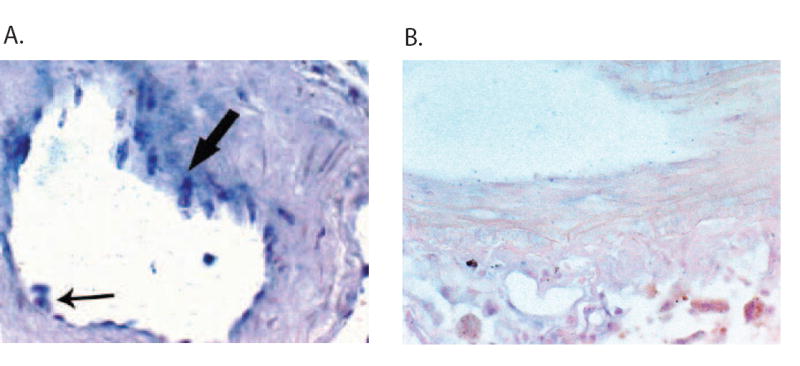
miR-155 is expressed in endothelial and VSM cells. A. Representative example of the distribution of mature miR-155 after in situ hybridization analysis with an LNA miR-155-specific antisense probe. In the arteriole, the endothelial (small arrow) and VSM (large arrow) cells abundantly express miR-155. B. No signal was evident in the arteriole when the serial section was probed with a scrambled probe.
miRNA Effect on Target Protein
If a given mRNA is a true target of a specific miRNA, then modulation of miRNA concentration should correspond to a predictable change in the amount of protein encoded by the target mRNA (Criterion 3). Therefore, a typical approach to validate the functional importance of a miRNA/mRNA target pair is a transient over-expression (“gain-of-function”) of a given miRNA mimic in a cell type known to express the putative target protein and subsequent Western analysis using a specific antibody against that protein [24]. Alternatively, ELISA or immuno-cytochemistry experiments could also be utilized to quantify differences in protein expression. However, if antibodies are not available, then other validation assays (i.e. binding, enzymatic, etc.) can be employed. For example, our laboratory routinely utilizes radioreceptor ligand binding assays to quantitate differences in hAT1R expression [21,22,31]. Since luciferase reporter experiments suggested that miR-155 could target hAT1R mRNAs (Fig. 2), over-expression of a miR-155 mimic should result in the attenuation of hAT1R expression. AT1R binding assays demonstrated that only VSMCs transfected with a miR-155 mimic showed a significant reduction (i.e. “knock-down”) in the endogenous expression of hAT1Rs when compared with controls (Fig. 5A). Since miRNAs modulate gene expression by both translational repression and mRNA cleavage [7-9], the effect of a miRNA mimic should also be assayed at the mRNA level.
Figure 5.
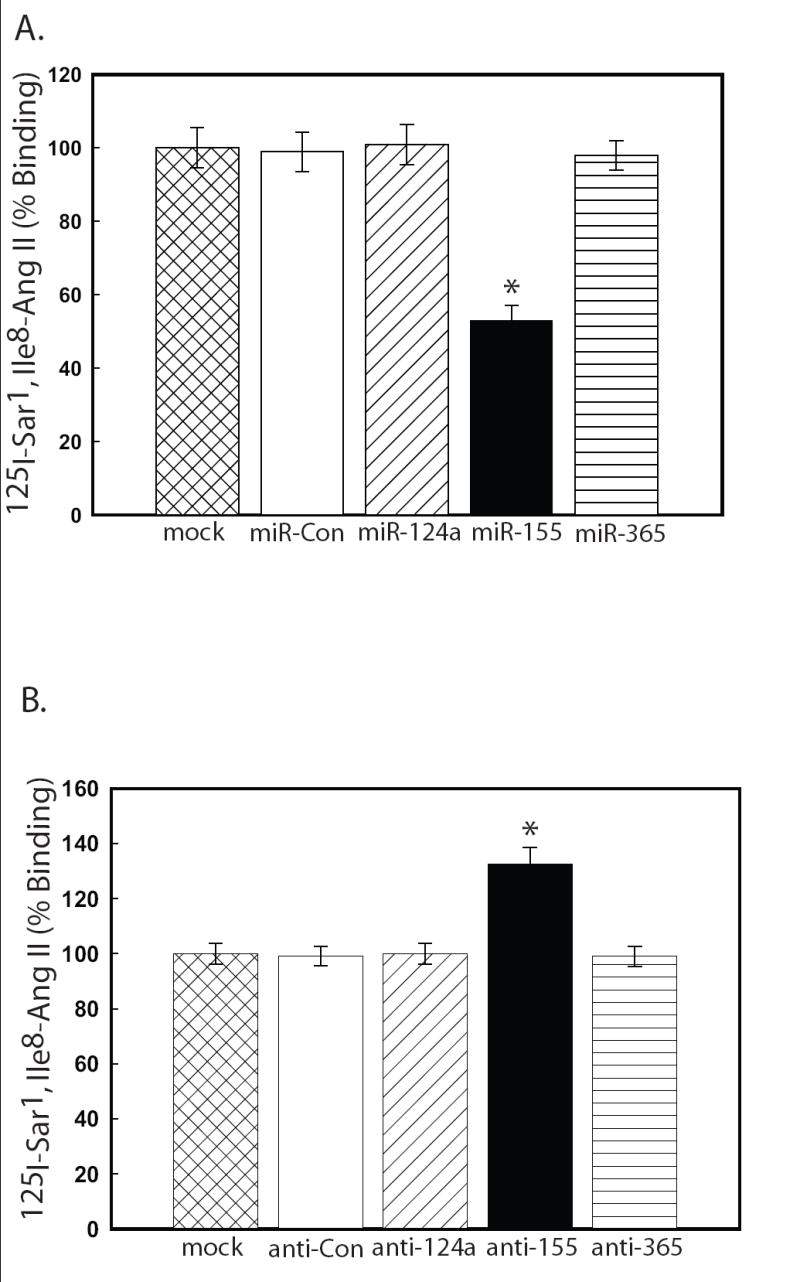
miR-155 regulates the endogenous hAT1R expression in VSMCs. VSMCs were either mock transfected or transfected with the miRNA mimics (A) or antisense miRNAs (B) as indicated. 48 hrs after transfection, the cells were utilized for AT1R radioreceptor binding assays as described [21]. The data have been normalized for protein and transfection differences and represent specific binding. The values are shown as percent of maximal specific binding of mock transfected hPFBs and represent the mean ± S.E. from four independent experiments (*p<0.001 vs. mock transfected cells).
Knock-down studies that are based entirely on over-expression of a given miRNA must be interpreted with caution since mis-expression of miRNAs could target genes that would otherwise not be affected in a physiologic context (i.e. “off-target” effects). Therefore, a complementary approach is to use “loss-of-function” studies in which a specific endogenous mature miRNA function can be inhibited using antisense oligoribonucleotides (ASO) [32-34]. These single-stranded RNAs are chemically modified (i.e. 2′-O′-methoxyethyl phosphorothioate or LNA) to improve stability and potency, and specific miRNA inhibitors can be purchased from various sources (Ambion, Dharmacon, Exiqon, etc.). Since miRNA inhibitors suppress the function of endogenous miRNAs, if the miRNA/mRNA interaction is authentic, then the expression of the target gene should be increased.
To demonstrate this phenomenon, VSMCs were transfected with various ASOs and hAT1R expression was assessed. AT1R binding assays demonstrated that only VSMCs transfected with anti-miR-155 showed a significant increase in the expression of endogenous hAT1Rs when compared with controls (Fig. 5B).
The choice of cells utilized for performing gain-of-function and loss-of-function studies is important as each cell line has varying levels of endogenous miRNA and target gene expression. If possible, it is important to select a cell culture system that expresses the appropriate level of endogenous miRNA and target gene so that the effects of the miRNA mimics and inhibitor on protein levels or activities can be clearly detected. For example, since miRNA mimics decrease target gene expression, they are best used in cells that express low levels of endogenous miRNAs and correspondingly high target mRNA expression. Therefore, under these conditions, when a given miRNA mimic is over-expressed, a decrease in the target protein should be easily detectable.
In contrast, if experiments were performed in cells with high endogenous miRNA levels and correspondingly low target expression, the effects of miRNA mimic over-expression on the target gene may not be detectable. Instead, these cells are best for investigating the effects of miRNA inhibitors (i.e. ASOs) since the resulting increase in gene expression will be more pronounced and easily quantified in these cells compared with cells that highly express the target gene.
Although the methods described above are commonly used to validate miRNA/mRNA target interactions, several additional techniques have recently been described. For example, Care et al. [35] utilized a “decoy target” strategy to overexpress a 3′-UTR with tandem sequences complementary to mouse miR-133 fused downstream of the EGFP reporter gene. The complementary sequences act as a decoy sequestering endogenous miR-133. Thus, EGFP expression becomes a surrogate marker of miR-133 expression levels. Zhu et al. [36] investigated miRNA targets by utilizing two dimensional differential-in-gel electrophoresis (2D-DIGE) and mass spectrometry analysis on cell extracts of cells transfected with or without antisense miR-21. Using this proteomics approach, these investigators demonstrated that the tumor suppressor, tropomyosin, is a target of miR-21 [36].
miRNA Effects on Target Biological Function
Once a given miRNA has been experimentally confirmed to be involved in the regulation of a target gene (i.e. repress protein expression levels), it then becomes necessary to demonstrate that this regulation equates to changes in biological function (Criterion 4). Depending upon the protein target of interest, biological assays could include signaling pathways, cell proliferation, cell differentiation, cell death, cell migration, etc. When a biological pathway is being studied, phenotypic changes may be assayed as an indirect measure of miRNA effect on protein levels as long as the phenotypic assay is accompanied by a direct protein assay (i.e. Criterion 3). To determine if the miR-155/hAT1R target gene pair meets Criterion 4, we investigated whether the anti-mir-155-mediated increase in hAT1R density also resulted in enhanced Ang II-induced signal transduction. Importantly, transfection of anti-miR-155 resulted in increased phosphoERK1/2 levels suggesting that the efficacy of Ang II signaling can be modulated by regulating hAT1R expression (Fig. 6A-B).
Figure 6.
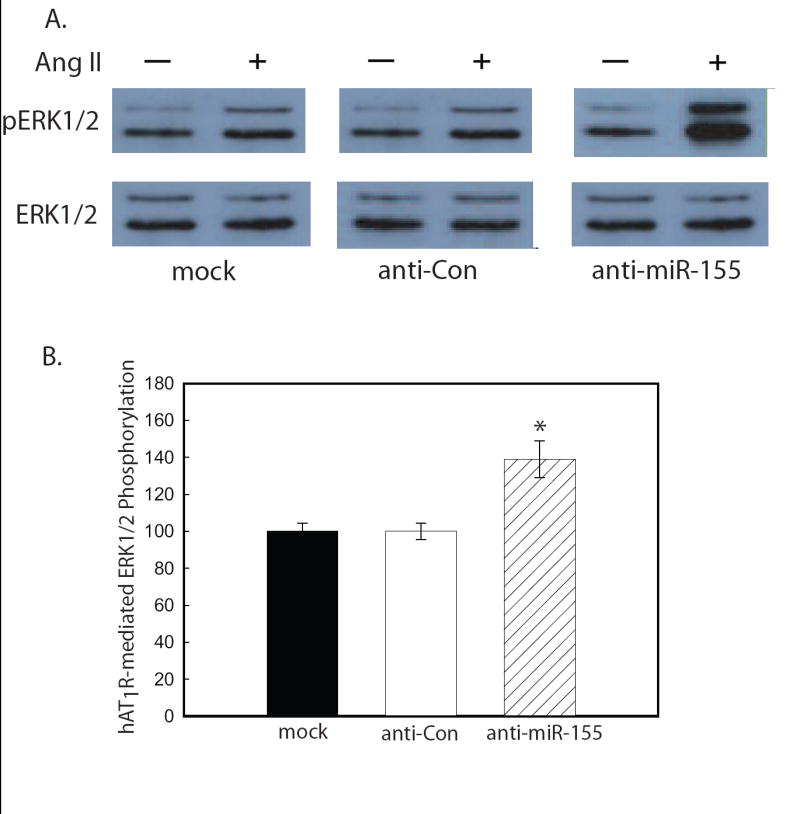
Anti-miR-155 enhances Ang II-induced signaling in VSMCs. VSMCs were transfected with the anti-miRNA oligonucleotides as indicated. A. Ang II-induced phosphoERK1/2 experiments were performed utilizing serum-starved, transiently-transfected cells as described [21,22]. A representative immunoblot is shown. Results are representative of four independent experiments. B. The quantitation of Ang II-(1 μM for 5 min) induced ERK1/2 phosphorylation was determined by densitometry. Values are expressed as a percent of the maximal phosphorylation of ERK1/2 in response to Ang II in mock transfected cells and represents the mean ± S.E. from four independent transfection experiments (*p<0.01).
Concluding Remarks
A growing body of evidence suggests that miRNAs are important regulators of cell growth [39], differentiation [40], and apoptosis [41]. In support of the significance of miRNAs in normal development and physiology, recent mouse miRNA “knockout” studies demonstrated that depending upon which miRNA gene was deleted, mice were left immuno-deficient [42,43] or with heart defects [24,44]. Implications drawn from these studies support the hypothesis that dysregulation of miRNA function may lead to human disease.
As disease-specific miRNAs are identified, the validation of novel targets within a disease pathway of interest may lead to novel therapeutic strategies. Therefore, it is critically important to be able to identify and validate miRNA/mRNA target pairs. Although not perfect, computational algorithms and ΔG analyses allow for the identification of putative miRNA/mRNA targets. Once identified, the authenticity of a functional miRNA/mRNA target pair can be validated by fulfilling four criteria. First, miRNA/mRNA target interaction must be verified. Second, the predicted miRNA and mRNA target gene must be co-expressed. Third, a given miRNA must have a predictable effect on target protein expression. That is, if a gene is a true target of a given miRNA, its miRNA mimic will decrease the target gene expression level while a miRNA ASO inhibitor will increase the target gene expression level. Fourth and final, miRNA-mediated regulation of target gene expression should equate to altered biological function.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. Ann Review of Cell Develop Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Stricker HM, Gou O, Liu L. Frontiers in Bioscience. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzila A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 6.Berezikov E, van Tetoring G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, Wetering M, Guryev V, Takada S, Zonneveld AT, Mano H, Plasterk R, Cuppen E. Genome Res. 2006;16:1286–98. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Standart N, Jackson RJ. Genes Develop. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 8.Du T, Zamore PD. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- 9.Pillai RS, Bhattachcryya SN, Filipowicz W. Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Bentwich I. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 12.Rajewsky N. Nat Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 13.Maziere P, Enright AJ. Drug Discovery Today. 2007;12:452–458. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Doran J, Strauss WM. DNA and Cell Biology. 2007;26:353–360. doi: 10.1089/dna.2006.0546. [DOI] [PubMed] [Google Scholar]
- 15.Diadian D, Hobert O. Nature Struc Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 16.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih I, Jones-Rhoades JW, Bartel DP, Burge CB. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein DJ, MacMenamin P, daPiedade I, Gunsalus KC, Stoffel M, Rajewsky N. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 20.Chen K, Rajewsky N. Nature Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 21.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. J Biol Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 22.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Zhao Y, Samal E, Srivastava D. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Cell. 2007;129:1–15. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Zucker M. Nucleic Acid Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeGasparo M, Catt KJ, Inagami T, Wright JW, Unger T. Pharmacol Rev. 2000;52:415–472. [PubMed] [Google Scholar]
- 27.Saito Y, Berk BC. Curr Hypertens Rep. 2002;4:161–171. doi: 10.1007/s11906-002-0042-1. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler G, Valoczi A, Havelda Z, Dalmay T. DNA Cell Biol. 2007;26:251–255. doi: 10.1089/dna.2006.0538. [DOI] [PubMed] [Google Scholar]
- 29.Nuovo GJ. PCR in situ hybridization: Protocols and Applications. 3. Lippincott, Williams and Wilkins Press; Baltimore, MD: 1997. [Google Scholar]
- 30.Sethupathy P, Corda B, Hatzigeorgiou AG. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin MM, Buckenberger JA, Jiang J, Malana GE, Knoell DL, Feldman DS, Elton TS. Am J Physiol Lung Cell Mol Physiol. 2007;293:L790–799. doi: 10.1152/ajplung.00099.2007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 34.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 35.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang M-L, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MVG, Hoydal M, Autore C, Russo MA, Dorn GW, II, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 36.Zhu S, Si M-L, Wu H, Mo Y-Y. J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 37.Easow G, Teleman AA, Cohen SM. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. RNA Biology. 2007;4:e1–e9. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 39.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 40.Hwang HW, Mendell JT. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wienholds E, Plasterk RH. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 42.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]


