Abstract
The advent of T-cell assay methodologies that are amenable to high throughput coupled with the availability of large libraries of overlapping peptides have revolutionized the fields of vaccine efficacy testing and cellular immune response assessment. Since T-cell assay performance is critically dependent upon the quality and specificity of the stimulating peptides, assurance of high-quality and reliable input peptides is an important aspect of assay validation. Herein, we demonstrate that individual peptides from large human immunodeficiency virus (HIV)-based peptide library sets obtained directly from two independent custom peptide suppliers contained contaminating peptides capable of giving false-positive results, which were consistent with nominal antigen-specific CD8+ T-cell responses. In-depth investigation of the cellular response in terms of responding CD8+ T-cell frequency and human leukocyte antigen (HLA) restriction led to the conclusion that one set of HIV type 1 (HIV-1)-derived peptides was contaminated with a peptide from human cytomegalovirus (HCMV), which is commonly used in cellular immunology research applications. Analytical characterization of the original stock of the suspect HIV-1 peptide confirmed the presence of ∼1% by weight of the HCMV peptide. These observations have critical implications for quality assurance (QA) and quality control (QC) of peptides used in clinical trials where cellular immune-based assays are important end-point determinants. We propose a simple schema of biological QA/QC protocols to augment the standard biochemical QA/QC analyses as a means to circumvent this and other problems that can affect cellular immune-based assay outcome and interpretation.
The identification and characterization of immunogenic T-cell epitope-containing regions of the proteomes of infectious agents and candidate cancer- and tumor-specific proteins are an essential phase in the rational development of prophylactic vaccines and immunotherapeutic strategies. Recent methodological advances, particularly enzyme-linked immunospot (ELISPOT) assays and cytokine-based flow cytometry (CFC) assays coupled with overlapping pooled peptide technology, give the opportunity for detailed and precise analyses of specific cellular immune responses (1, 3, 11, 19, 22, 29). As cellular immune response assays proceed from being used primarily as research tools to be being used as tools for clinical evaluation and assessment of end points in different phases of vaccine development, the required standards of quality assurance (QA)/quality control (QC) for these assays rise dramatically (4, 10, 16, 17, 21, 27, 28). While assay techniques and equipment can be standardized by employing standard operating procedures and assay reagents and can be standardized by use of manufacturer-certified assay kits, a critical component of the assay that is not typically subject to standardized QA/QC is the synthetic peptides used for stimulating the T cells. Synthetic peptides are usually specific to the particular application of the assay, defined by the infectious agent or tumor antigen being studied, and are often ordered in bulk quantities from a variety of manufacturers specializing in, or expanding into, custom peptide synthesis (15). Most researchers depend upon the biochemical QA/QC performed postsynthesis by the peptide manufacturer because of the exorbitant monetary and time-consuming costs of performing such analyses. A drawback to this approach is that there is currently no consistent biological assay for synthetic peptide QA/QC, nor is there a single set of standards for defining peptide quality for cellular immune response assays. Hence, the validity of conclusions reached in assays using synthetic peptides depends critically upon the quality of the input peptide(s) obtained directly from the manufacturer.
In this report, we describe two case studies of problematic contamination of individual peptides from human immunodeficiency virus (HIV-1)-based peptide sets with a commonly used peptide from human cytomegalovirus (HCMV). In the first case, the contaminated peptide was proven to have come directly from the manufacturer, while in the second case, three different peptides from a single HIV-1-based peptide set were shown to have been most likely contaminated in the hands of the manufacturer. While the level of contaminating peptide was shown to be relatively low (∼1% or less of the total peptide weight), the extraordinary sensitivity of T cells for their cognate antigen resulted in detection of peptide-specific responses in both ELISPOT and CFC assays. Such contamination, if not detected, could lead to false-positive interpretations of important research and clinical trial data. Both research and clinical assessment laboratories should be aware of the possibility of peptide cross-contamination and other potential impurities and perform appropriate biological as well as biochemical QA/QC on all peptides. Peptide manufacturers and suppliers should also be made aware of this problem and implement the appropriate changes to their current QA/QC procedures to further ensure against this problem.
MATERIALS AND METHODS
Study subjects.
HIV-1-positive blood units were collected from Kericho District Hospital (Kericho), Rift Valley Provincial Hospital (Nakuru), and Kenyatta National Hospital (Nairobi), all in central/southern Kenya, between 1999 and 2000 under a study approved by both Kenyan- and U.S.-based institutional review boards. HIV-1 positivity was assessed by Serostrip (Saliva Diagnostic Systems, Medford, NY) and confirmed by enzyme-linked immunosorbent assay (Organon Teknika/BioMerieux, Inc., Marcy l'Etiole, France). HIV-1-exposed but -uninfected subjects were women selected from the HIV-1 Superinfection Study cohort based in Mbeya, Tanzania. The HIV-1 Superinfection Study is an institutional review board-approved study and is a collaborative effort between the University of Munich, the Mbeya Research Program, the U.S. Military HIV Research Program, and the National Institute of Communicable Diseases (Johannesburg, South Africa). The exposed yet uninfected status of the subjects was determined on the basis of reported engagement in unprotected sex work for more than 3 years prior to enrollment in the study and having remained HIV seronegative over the course of 2 years during the study. Leukapheresis samples from HIV-1-seronegative subjects were obtained from two different sources: BRT Laboratories (Baltimore, MD) and the NIH Blood Donor Center (Bethesda, MD). Peripheral blood mononuclear cells (PBMC) were collected by centrifugation over a Ficoll gradient and cryopreserved for subsequent batched analysis.
Synthetic peptides.
The sequence, origin, and location of single peptides used in this study are detailed in Table 1. Briefly, the Gag 114 peptide was derived from the HIV-1 Gag p6 protein (positions 453 to 467) of CRF01_AE isolate 90CF402 and was originally part of an overlapping-peptide set (15 amino acids in length with an 11-amino-acid overlap) designed for screening for subtype A Gag-specific T-cell responses from eastern African HIV-1-infected cohorts (6, 13). This peptide was obtained from “manufacturer A” in 20 individual vials of 1 mg each; each vial was hermetically sealed. The Gag (CM240 isolate), Pol (CM240 isolate), and Env (CM235 isolate) overlapping peptide sets (15 amino acids in length with an 11-amino-acid overlap) were obtained from “manufacturer B.” The HCMV pp65 NV9, Gag (90CF402) 114, Env (CM235) 97, Env (CM235) 137, and Pol (CM240) 31 peptides were resynthesized in-house (Henry M. Jackson Foundation) with free amino termini using 9-fluorenylmethoxy carbonyl (Fmoc) chemistry and standard solid-phase techniques (Excel automated synthesizer; Waters, Milford, MA). The CEF peptide set was also synthesized in-house (Henry M. Jackson Foundation). All peptides were >80% pure as determined by high-performance liquid chromatography (HPLC) and verified for correct sequence by mass spectroscopy (MS). In some cases, amino acid analysis and N-terminal sequencing were also performed. The HPLC-MS analysis was performed by JPT Peptide Technologies (Berlin, Germany) using an LC-MSD Trap LC device 1100 series (Agilent, Waldbronn, Germany).
TABLE 1.
Source proteins and sequences of peptides used in the study
| Peptide | Protein derivationa | Sequence |
|---|---|---|
| HCMV-NV9 | HCMV (AD169) pp65495-503 | NLVPMVATV |
| Gag 114 | HIV-1 (90CF402) Gag455-469 | PTAPPMESLGMGEEI |
| Pol 31 | HIV-1 (CM240) Pol116-130 | DQILIEICGKKAIGT |
| Env 97 | HIV-1 (CM235) Env229-241 | NNTCIENGTMGGCNG |
| Env 137 | HIV-1 (CM235) Env558-572 | AIEAQQHLLQLTVWG |
Numbering according to the HXB2 reference is positions 453 to 467.
ELISPOT assay.
All assays were performed using RPMI 1640 medium containing 10% normal human serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (complete medium [CM]). An ELISPOT assay for detecting gamma interferon (IFN-γ) was employed to measure the frequency of peptide-specific CD8 T cells in cryopreserved PBMC from HIV-1-infected and uninfected subjects directly. The assay was performed as described in detail elsewhere previously (5, 6). Briefly, cryopreserved PBMC were thawed, washed, and rested overnight in CM for 14 to 16 h. After overnight resting, the PBMC were enumerated prior to addition to prewetted, primary-antibody-coated (1-D1K; Mabtech AB, Sweden) ELISPOT assay plates (Multiscreen-IP plates, MAIP-type plates; Millipore, MA). Approximately 1 × 105 to 2 × 105 viable PBMC were plated per well for all assays. Plates were incubated for 18 to 20 h prior to washing and the addition of the secondary biotinylated mouse anti-human IFN-γ monoclonal antibody (7B6-1-biotin; Mabtech AB, Sweden). Assay spot development was performed using the avidin horseradish peroxidase complex system (Vectastain ABC kit; Vector Laboratories, CA). Peptides were used at a final concentration of 1 to 2 μg/ml and were prefiltered for some assays as noted. As a positive control for functional integrity of the PBMC, staphylococcal entertoxin B (SEB) was added to wells at a final concentration of 5 μg/ml, and cells were incubated with CM only as a negative control. The plates were evaluated with an automated ELISPOT reader system and KS 4.3 software (Carl Zeiss, Thornwood, NY). Plates were read and spots were counted by an independent scientist (Henry M. Jackson Foundation, Rockville, MD). Positive IFN-γ spot-forming cells (SFC) representing single cells were counted and expressed as SFC per 1 million input PBMC. Assays were considered to be valid if responses of greater than 500 SFC per 1 million input PBMC were detected with SEB.
Cytotoxic T-lymphocyte effector cell generation.
Effector cells were prepared by in vitro stimulation (IVS) of thawed, cryopreserved PBMC. Peptide-pulsed (10 μM overnight), irradiated (10,000-rad), autologous BLCL (Epstein-Barr virus-transformed B-cell line) or allogeneic (HLA-A0201-positive) BLCL (2 × 106 cells) were washed three times with CM and then cocultured with 10 × 106 to 20 × 106 PBMC in 10 ml CM supplemented with 5 ng/ml recombinant human interleukin-7 (rhIL-7) (R&D Systems, Minneapolis, MN) for 4 days. A total of 5 ng/ml rhIL-2 (R&D Systems, Minneapolis, MN) was then added to the cocultures, and the cultures were maintained and split with fresh CM and rhIL-2 for up to 24 days. Effector cell lines were then maintained by restimulation every 7 to 14 days with irradiated, peptide-pulsed BLCL (HLA-A0201-positive) and 5 ng/ml rhIL-2 in CM.
CFC assay.
Cryopreserved PBMC were thawed rapidly, washed twice with CM, and either used immediately or incubated at 2 ×106 to 5 ×106 cells/ml in CM for 14 to 16 h. PBMC rested overnight were recounted prior to assay. Effector cells (prepared above) or PBMC were added at 0.5 × 106 to 1.0 × 106 cells per well into 96-well polypropylene tissue culture trays (catalog no. 3790; Costar) and stimulated either directly with peptide or with autologous or allogeneic peptide-pulsed BLCL. BLCL were pulsed overnight with the relevant peptide (10 μg/ml), washed five times after overnight incubation, and distributed at 1 ×105 cells per well (ratio of PBMC or effector cells to BLCL of 5:1 to 10:1). As a positive control for functional integrity of the cryopreserved cells, SEB was added to a single well at a final concentration of 5 μg/ml. The assay mixtures were incubated for 6 h at 37°C (5% CO2) in the presence of the protein transport inhibitor brefeldin A (10 μg/ml; Sigma, St. Louis, MO) and were then interrupted by transfer to a reduced temperature (either 4°C or 18°C). Cells were stained for surface markers and intracellular IFN-γ expression (CFC assay) on the following day. Cocultured PBMC or effector cells (prepared as described above) were washed once with flow buffer (DPBS-0.1% bovine serum albumin-0.1% sodium azide) and incubated in 96-well tissue culture trays for 10 min in 200 μl flow buffer at room temperature (the same volume, temperature, and base buffer were used for all subsequent washings and incubations) containing 1 mM EDTA. Cells were washed once, fixed in 2% formaldehyde for 30 min, and washed again. Fixed cells were permeabilized with 0.5% saponin (Sigma, St. Louis, MO) for 30 min, washed, and resuspended in 0.5% saponin containing the following monoclonal antibodies: fluorescein isothiocyanate (FITC)-conjugated anti-IFN-γ (clone 25723.11), phycoerythrin (PE)-conjugated anti-CD69 (clone L78); PerCP-Cy5.5-conjugated anti-CD8 (clone SK1), and allophycocyanin (APC)-conjugated anti-CD3 (clone SK7) (BD Biosciences, San Jose, CA). After 30 min of incubation, cells were washed three times with flow buffer and finally resuspended in 200 μl flow buffer. Stained cells were stored at 4°C and analyzed by flow cytometry within 24 h.
Flow cytometry and analysis.
Phenotypic analysis of PBMC and cytotoxic T-lymphocyte effector cells for the expression of T-cell receptors specific for the HCMV-NV9 epitope was performed using the iTAg major histocompatibility complex class I human tetramer PE system (Beckman Coulter Immunomics, San Diego, CA). Cells were stained for 30 min at room temperature in flow buffer containing tetramer and either FITC- or APC-conjugated anti-CD8 (clone SK1) and then fixed with 2% formaldehyde prior to flow cytometry acquisition and analysis. Data acquisition was performed using a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA). Initial gating was performed using a total lymphocyte gate based on forward- and side-scatter characteristics and the acquisition of 50,000 to 200,000 cells within this gate. Color compensation was performed using similarly prepared cells from an HIV-1-seronegative donor and staining singly labeled cells with anti-CD3 labeled with FITC, PE, PerCP-Cy5, and APC fluorochromes (BD Biosciences). Data sets were analyzed using FlowJo software (version 4; TreeStar, Cupertino, CA).
Statistical analysis.
In the ELISPOT assays, the cutoff for positivity was a count of >55 SFC per 1 million input PBMC and more than four times the background (mean of the negative-control wells) (12). Nonparametric tests were used for comparing the nonpaired peptide responses between the HIV-1-seronegative and -seropositive groups (Mann-Whitney rank test) and for comparing the paired peptide responses within the groups (Wilcoxon signed-rank test).
RESULTS
HCMV pp65495-503 NV9 peptide and HIV-1 (isolate 90CF402) Gag455-469 peptide 114 stimulate equivalent numbers IFN-γ-producing CD8 cells from PBMC of HLA-A0201 individuals independent of their HIV serostatuses.
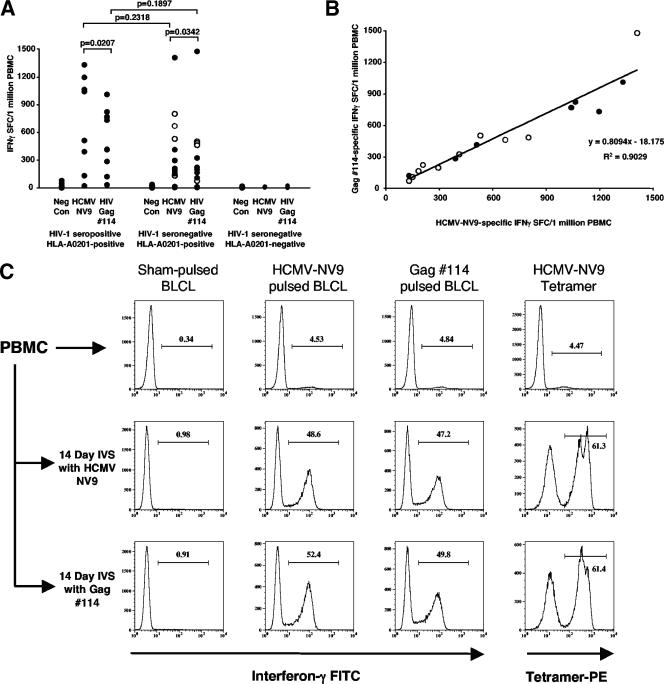
An immunodominant response directed against HIV-1 Gag peptide 114 (provided by manufacturer A) was observed while screening for HIV-specific CD8+ T-cell responses in a cohort of HIV-1 subtype A-infected individuals and in a cohort of HIV-1-exposed yet -uninfected subjects. In addition, a similar frequency of IFN-γ SFCs were detected in response to either Gag 114 or a pool of positive-control peptides (CEF peptide pool) (5), representing optimal broadly recognized CD8 epitopes, in four HEPS subjects. High-resolution HLA typing revealed that all subjects responding to Gag 114 possessed the HLA-A0201 allele. Since preliminary data had shown that the majority of the response against the CEF peptide pool in HLA-A0201-positive subjects was directed toward the HCMV-NV9 peptide, we tested the hypothesis that there was an association between the responses to the HCMV-NV9 peptide and the Gag 114 peptide. Figure 1A shows that both the HCMV-NV9 and Gag 114 peptides induced positive IFN-γ ELISPOT responses in PBMC from 7/8 HIV-1-seropositive, HLA-A0201-positive subjects; 10/12 HIV-1-seronegative, HLA-A0201-positive subjects; and 0/9 HIV-1-seronegative, HLA-A0201-negative subjects. The ability to respond to either peptide was independent of HIV-1 serostatus and was dependent upon the possession of the HLA-A0201 allele, since only HLA-A0201-positive subjects respond to both peptides, and HLA-A0201-negative subjects respond to neither peptide. Importantly, four of the HIV-1-seronegative, HLA-A0201-positive subjects were exposed to HIV-1 but were uninfected, and each subject demonstrated strong and equivalent HCMV-NV9 and Gag 114 responses (Fig. 1A, open circles). There was no significant difference in the magnitudes of the responses to HCMV-NV9 or Gag 114 between HIV-1-seropositive and -seronegative subjects (Mann-Whitney rank test) (Fig. 1A). There was, however, a significant difference in HCMV-NV9 and Gag 114 responses within both seropositive and seronegative subject groups (Wilcoxon signed-rank test) (Fig. 1A). A strong positive correlation (r2 = 0.90) between the magnitudes of responses to each peptide in all responders, irrespective of HIV-1 serostatus, was demonstrated (Fig. 1B). The slope of the linear regression equation for HCMV-NV9 versus Gag 114 responses shows that the magnitude of the response to Gag 114 was approximately 80% of the response to HCMV-NV9. Importantly, three HLA-A0201-positive subjects (one HIV-1-seropositive subject and two HIV-1-seronegative subjects) who did not respond to the HCMV-NV9 peptide also did not respond to the Gag 114 peptide. These data strongly imply that either a large proportion of the T cells responding to the HCMV-NV9 peptide were cross-reactive with the HIV-1 Gag 114 peptide or the Gag 114 peptide was contaminated with the HCMV-NV9 peptide.
FIG. 1.
Cumulative ELISPOT responses to the HCMV-NV9 and Gag 114 peptides from 8 HIV-1-seropositive, HLA-A0201-positive subjects; 12 HIV-1-seronegative, HLA-A0201-positive subjects, and 9 HIV-1-seronegative, HLA-A0201-negative subjects. (A) Responses are plotted on a linear scale of IFN-γ SFC/106 PBMC, with subjects stratified according to HIV-1 serostatus and possession of the HLA-A0201 allele. P values for comparing the HCMV-NV9 and Gag 114 responses within each group (Wilcoxon signed-rank test) and for comparing HCMV-NV9 or Gag 114 responses between groups (Mann-Whitney rank test) are shown. HIV-1-exposed yet -uninfected subjects within the HIV-1-seronegative groups are indicated with open circles. Neg Con, negative control. (B) Scatter plot of all HCMV-NV9 and Gag 114 responder subjects (n = 17) showing the positive correlation of the HCMV-NV9 response with the Gag 14 response. HIV-1-seropositive (filled circles) (n = 7) and -seronegative (open circles) (n = 10) subjects are indicated. (C) Cryopreserved PBMC from an HIV-1-seronegative subject and PBMC subjected to a 14-day IVS with either HCMV-NV9 or Gag 114 peptide were tested for their abilities to respond to sham-pulsed, HCMV-NV9-pulsed, or Gag 114-pulsed autologous BLCL in a CFC assay for IFN-γ production. After IVS with either peptide, an approximately 10-fold expansion of CD8+ T cells specific for both peptides was observed (middle row, HCMV-NV9; bottom row, Gag 114). The percentage of HCMV-NV9 tetramer-positive cells among the total CD8+ T-cell population in the IVS is indicated above the gate marker in each histogram. All panels were derived from the gate containing the CD3+ CD8+ T-cell population.
The Gag 114 peptide and HCMV-NV9 peptide stimulate the same CD8+ T cells despite little primary sequence homology.
Cryopreserved PBMC from an HIV-1-seronegative HCMV-NV9 peptide responder (BC238) were tested for the direct recognition of HCMV-NV9 and Gag 114 peptides presented by an autologous BLCL before and after a 14-day IVS with either HCMV-NV9 or Gag 114 peptide. Direct analysis of the PBMC from subject BC238 using the CFC assay for IFN-γ demonstrated a robust and equivalent frequency of CD3+ CD8+ T cells responding to either HCMV-NV9 or Gag 114 peptide (4.53% and 4.84%, respectively) (Fig. 1C, top row). An HLA-A201-NV9 tetramer bound directly at a frequency of CD8+ T cells (4.47%) similar to that detected by the IFN-γ CFC assay. After the IVS with either HCMV-NV9 or Gag 114 peptides, there was a striking increase (∼10-fold) in the number of HCMV-NV9- and Gag 114-responsive CD8+ T cells in both culture systems. Confirmation of the cross-stimulatory capacity between HCMV-NV9 and Gag 114 peptides came from the tetramer staining of both 14-day IVS cultures (Fig. 1C, right panels). Virtually identical numbers of HLA-A0201-NV9 tetramer-positive cells were expanded in each culture (61.3% versus 61.4%). An alignment of the two peptides (Table 1) shows that they share very little primary sequence identity. Hence, direct cross-reactivity between the two peptides seemed unlikely, and contamination of Gag 114 with HCMV-NV9 was the most likely reason for these unusual observations.
Confirmation that the manufacturer-provided Gag 114 peptide was contaminated with the HCMV-NV9 peptide.
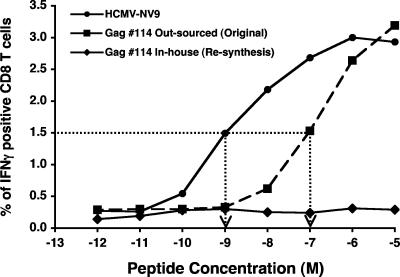
To test whether the Gag 114 peptide was indeed contaminated with the HCMV-NV9 peptide, the Gag 114 peptide was resynthesized in-house (Henry M. Jackson Foundation) and tested for its ability to stimulate T cells from an HCMV-NV9 responder HIV-1-seronegative subject. A CFC assay for IFN-γ was set up to directly compare the original Gag 114, the in-house-synthesized Gag 114, and the HCMV-NV9 peptides. Autologous BLCL were pulsed with serially 10-fold-titrated concentrations of each peptide and then used to stimulate PBMC from responder subject BC238. Figure 2 shows clearly that peptide HCMV-NV9 stimulates CD8 T cells at much lower concentrations than does the original Gag 114 peptide. Half-maximal stimulation for the HCMV-NV9 peptide (∼1 nM) was approximately 100-fold greater than that for the original Gag 114 peptide (∼100 nM). The in-house-synthesized Gag 114 had no such stimulatory capacity throughout the same concentration range. This result is consistent with a small amount of HCMV-NV9 contaminating the original Gag 114 peptide.
FIG. 2.
Titration of the CD8+ T-cell response to the original Gag 114 peptide, the resynthesized Gag 114 peptide, and the HCMV-NV9 peptide. Autologous BLCL were pulsed with serial 10-fold dilutions of each peptide and then used as antigen-presenting cells in a CFC assay for IFN-γ up-regulation in cryopreserved PBMC from a known HIV-1-seronegative, HCMV-NV9 responder subject. Results are expressed as the percentages of IFN-γ-positive cells in the CD3+ CD8+ T-cell population. The dotted line shows the interpolated peptide concentration at which the 50% maximal response to the HCMV-NV9 peptide occurred.
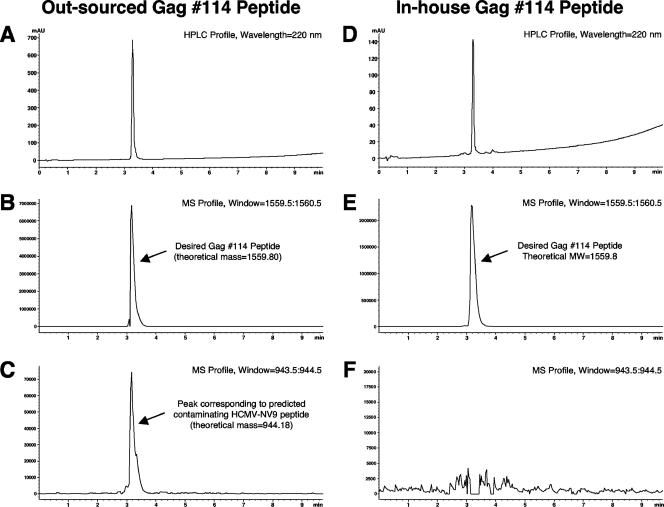
To confirm the nature of the contaminating product and to rule out the possibility that the outsourced Gag 114 peptide was contaminated in our laboratory, an unopened vial of the original stock peptide from the manufacturer and the in-house Gag 114 peptide were sent to a third party for HPLC-MS analysis. Evaluation of the UV trace of the HPLC spectrum for the outsourced Gag 114 peptide showed a single peak, consistent with a high-purity peptide (Fig. 3A). The corresponding MS analysis of the main peak revealed the overwhelming presence of ions corresponding to the theoretical Gag 114 peptide (calculated, 1,559.80 [M + H]+ and 780.4 [M + 2H]2+; found, 1,560.1 [M + H]+ and 780.2 [M + 2H]2+) (Fig. 3B). However, a significant number of ions matching the mass of the HCMV-NV9 peptide could be detected when a mass filter corresponding to the mass of that peptide was applied (calculated, 944.18 [M + H]+) at exactly the same retention time as that of the Gag 114 peptide (Fig. 3C). The relative signal intensity detected by MS showed a ratio of approximately 1:100 for the suspected HCMV-NV9 contaminant peptide and the Gag 114 peptide. This was consistent with the titration experiment for the HCMV-NV9 and Gag 114 peptides, which indicated that the Gag 114 peptide was about 100 times less efficient than the HCMV-NV9 peptide for stimulating CD8 T cells from an HCMV-NV9 responder. As a control experiment, Gag 114 peptide resynthesized in-house (Henry M. Jackson Foundation) was tested by LC-MS using the same conditions (Fig. 3D). A strong MS signal revealed the presence of ions related to the target peptide, whereas no MS signal relating to the HCMV-NV9 peptide was encountered when a corresponding mass filter was applied (Fig. 3E and F).
FIG. 3.
LC-MS analysis of the suspect contaminated Gag 114 peptide from manufacturer A (A to C) and the in-house-resynthesized Gag 114 peptide (D to F). (A and D) UV traces of HPLC. High peptide purity is evident. The selected ion mode with mass filter adjusted to the calculated theoretical mass of the Gag 114 peptide (1,559.80 [M + H]+) is shown for each peptide (B and E). The desired product is clearly present in both profiles. The selected ion mode with mass filter adjusted to the calculated theoretical mass of the HCMV-HCMV-NV9 peptide (944.18 [M + H]+) is shown (C and F). A signal corresponding the exact theoretical molecular weight (MW) of HCMV-NV9 is present in the outsourced Gag 114 peptide. Note the different scales on the y axes.
Multiple peptide contamination in two HIV-1-derived overlapping peptide sets.
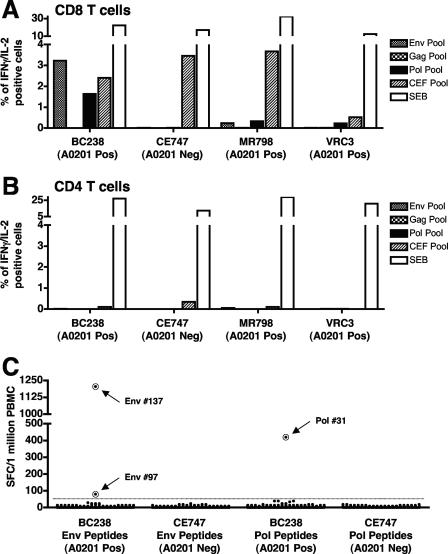
Initially, three overlapping peptide sets representing the Gag (CM240 isolate), Pol (CM240 isolate), and Env (CM235 isolate) proteins of HIV-1 were synthesized (by manufacturer B) for use in assays that assess cellular immune responses to candidate HIV-1 vaccines. Prior to use, the individual peptides were dissolved in dimethyl sulfoxide (DMSO) (Sigma Chemical Co., St. Louis, MO), pooled together to mimic the parent proteins, and then screened for their intrinsic capacity to nonspecifically inhibit, or to activate, T-cell function in a selection of individuals who were either seronegative or seropositive for HIV-1. Paradoxically, a subset of the seronegative subjects responded to the Env- and Pol-derived peptide sets with detectable CD8 T-cell-mediated production of IL-2 and/or IFN-γ that was comparable to their response to the CEF control peptide pool. No inhibitory activity was detected in any of the subjects tested (n = 23) (data not shown). Figure 4 shows the responses of four HIV-1-seronegative subjects to the Gag, Env, and Pol peptide pools. Three subjects responded to the Pol pool, two responded to the Env pool, and none responded to the Gag pool. All four subjects responded well to the positive-control CEF peptide pool, and all responses were CD8 mediated. Of note was that all three HIV-1-derived peptide pool-responding subjects possess the HLA-A0201 allele. Testing of all the individual peptides that make up each pool was carried out to determine which peptide(s) was responsible for the spurious CD8 T-cell activation. An IFN-γ ELISPOT assay of 326 individual peptides (179 from Env and 147 from Pol) in two subjects (one HLA-A0201 positive and one HLA-A0201 negative) revealed that three peptides could stimulate PBMC from the HLA-A0201-positive subject (Fig. 4C). These peptides, Env 97, Env 137, and Pol 31, were then confirmed to be HLA-A0201 restricted in a CFC assay for IFN-γ. As shown in Fig. 5, the response to all three peptides was HLA-A0201 restricted. Filtering of the peptide pools to remove possible insoluble peptide complexes, which have been shown to give false-positive responses in ELISPOT assays (18), did not result in any diminution of the response in an ELISPOT assay for IFN-γ (data not shown). Furthermore, the Env and Pol peptide pools were remade without the identified contaminating peptides, and IFN-γ ELISPOT responses were screened using 23 subjects. Only one subject exhibited a marginally positive response by our stringent positive-response cutoff criteria (63 SFC/106 PBMC; cutoff, 55 SFC/106 PBMC), a response which we consider to be “noise” in the assay system. Resynthesis of the Env 97, Env 137, and Pol 31 peptides was carried out, and each of the new peptides was retested in an ELISPOT assay for IFN-γ using PBMC from the donors for whom responses were detected previously. None of the three resynthesized peptides stimulated a response in our assay system. Taken together, these data provide evidence that three peptides from manufacturer B were capable of stimulating immune responses consistent with classical HLA restriction.
FIG. 4.
Evidence of peptide contamination within two pools of overlapping peptide sets obtained from manufacturer B. Cryopreserved PBMC from four HIV-1-seronegative subjects were tested for their ability to respond to three different HIV-1-derived peptide pools (Gag, Pol, and Env) in a CFC assay for IFN-γ production. Note that only the subjects possessing the HLA-A0201 allele respond to the Env and Pol peptide sets, and the response is observed within the CD3+ CD8+ T-cell population (A) but not the CD3+ CD4+ T-cell population (B). (C) IFN-γ ELISPOT response from an HIV-1-seronegative subject to all individual peptides (326 peptides) from the Env (179 peptides) and Pol (147 peptides) peptide sets. Three peptides, Env 97, Env 137, and Pol 31 (indicated with circles), were identified clearly as positive stimulators of IFN-γ production in the HLA-A0201-positive subject (subject BC238). Pos, positive; Neg, negative.
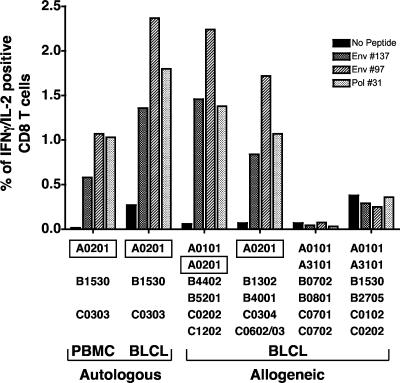
FIG. 5.
The three stimulatory peptides, Env 97, Env 137, and Pol 31, mediate their effect in an HLA-A0201-dependent manner. Cryopreserved PBMC from three HIV-1-seronegative subjects were tested for their abilities to respond to Env 97, Env 137, and Pol 31 peptides alone or pulsed onto autologous and partially HLA-matched BLCL in a CFC assay for IFN-γ production. The percentage of IFN-γ-positive cells within the gated CD3+ CD8+ T-cell population is shown. The HLA type of each subject and cocultured BLCL is shown below the bar graphs. The common allele shared by the responding subject and stimulating peptide-pulsed BLCL is shown.
DISCUSSION
The advent of T-cell assay methodologies that are amenable to high throughput coupled with the availability of large libraries of overlapping peptides have revolutionized the fields of vaccine efficacy and cellular immune response assessment (1, 3, 11, 19, 22, 29). The widespread availability of large peptide libraries has in part been driven by and in part contributed to the application of high-throughput assays for the assessment of cellular immune responses. Custom peptide manufacturers and suppliers have responded to this demand by increasing their supply capacity to meet the growing demand in both research and therapeutics markets (15). Since peptide libraries used for screening cellular immune responses may contain hundreds or even thousands of individual peptides, and each peptide is synthesized and purified independently, QA/QC of peptide libraries used in both basic research and clinical research protocols has become a significant and daunting task (4, 16, 24, 27, 28). Stringent QA/QC is an absolute requirement for clinical trials where biological assay outcome may be the end-point determinant for advancement or nonadvancement of a vaccine or therapeutic product (10, 25). Herein, we have clearly demonstrated that biological as well as biochemical QA/QC procedures are necessary requirements for synthetic peptides that are to be utilized in vaccine trial assessment assays and thus provide a clear experimental schema that can be implemented to achieve this goal.
As described above, during screening of HIV-1-seropositive and -seronegative subjects with an HIV-1 Gag protein-based peptide set (derived from isolate 90CF402), we found that one particular peptide (Gag 114) gave consistently high CD8+ T-cell-mediated IFN-γ responses regardless of the HIV-1 serostatus of the subject. The subsequent demonstration that this was the result of a false-positive effect caused by the contamination of the HIV-1 Gag peptide with a commonly recognized peptide from HCMV was both surprising and alarming. Clear evidence that a peptide obtained directly from a supplier contained a significant amount of another peptide (∼1% of the total weight) should be disconcerting to all investigators currently studying cellular immune responses using synthetic peptides. The identified contaminating peptide (HCMV pp65495-503) is used extensively as a positive control for ELISPOT and CFC assays in clinical trials and as a reagent for the study of cellular immune responses to chronic viral infection (5, 14, 26, 30). Furthermore, responses detected in subsequent ELISPOT and CFC assays would ostensibly have all the hallmarks of a classical T-cell-mediated response: a high frequency of IFN-γ-producing cells, HLA restriction, and immunodominance. Hence, peptide cross-contamination carries with it the likelihood that it would be easily mistaken as a T-cell response against the test peptide and therefore be misinterpreted as a false-positive result. Further cause for concern was raised following routine quality control testing of an additional three HIV-1-derived overlapping peptide sets obtained from a different supplier, two of which demonstrated convincing evidence of contamination of three more individual HIV-1 peptides. While conventional biochemical analysis could not be used to verify the identity of the contaminating peptide(s) involved in this case (all of the peptides were dissolved in DMSO), circumstantial evidence indicated that it was another commonly recognized HLA-A0201-restricted peptide, possibly the same HCMV-derived peptide responsible for the first case of contamination. Such potential contamination has also been described in a recent report, where false-positive responses to a synthetic peptide pool were detected in subjects with a particular HLA type (7). Resynthesis of the suspect individual peptides resulted in the removal of the false-positive response and presumably the contaminant. Therefore, it seems unlikely that the effects that we have observed are spurious events or are specific to our laboratory.
Certainly, the vast majority of peptides obtained from custom peptide suppliers are of the highest quality and are not cross-contaminated; however, the consequence of even a single contaminated peptide making its way into a clinical trial is considerable. This leads to an important question: how did the peptides become contaminated? Since the vast majority of the lyophilized peptide received from the supplier was indeed the intended product, the possibility of simple mislabeling or mix-up of the product vials during or after synthesis can be ruled out. Therefore, contamination at some point during the synthesis, purification, or vialing of the product was the likely source of the contamination. Since contamination may occur simply as a result of improperly cleaned glassware or other apparatuses, this implies that synthetic peptide suppliers need to implement more stringent QA/QC practices for synthetic peptide manufacture.
Both the nature of the contamination—peptides commonly recognized by CD8+ T cells—and the fact that it has been documented in four different peptides obtained from two different manufacturers raise important questions for future QA/QC assessment of peptides slated for future use in cellular immune function assays. While nonspecific cellular immune response-inhibitory and -stimulatory functions of peptide sets can be, and usually are, easily screened for prior to use, potential cross-contamination of peptides with other peptides (often used for positive controls) is not screened for routinely and adds an extra level of complexity to the QA/QC process. It is important that the currently employed methods for QA/QC of synthetic peptides, various combinations of HPLC, mass spectroscopy, amino acid content analysis, and amino acid sequencing, cannot be used routinely to detect potential cross-contaminating peptides. In fact, it is the extraordinary sensitivity of T cells for their cognate antigens that facilitated the detection of the contaminating peptides. T cells can recognize their specific cognate peptides at subnanomolar concentrations, levels at which it is impossible for conventional analytical chemical and biochemical methods to discern a possible contaminant in the presence of overwhelming quantities of another peptide.
The most sobering prospect that emerges from these findings is that we have managed to detect the particular contaminating peptide only because it represents an epitope from HCMV that is recognized frequently in the human populations that we were studying. It is therefore not unreasonable to assume that peptide cross-contamination occurs more frequently and that the general research community has not yet screened for it appropriately. A recent study has shown that Fmoc-modified peptides, a common minor contaminant in synthetic peptides, can directly stimulate human CD4+ T-cell clones (23). As with the cross-contamination that we have described here, the contaminating Fmoc-modified peptides were present at very low levels within the desired product (<0.5%). Therefore, cross-contaminating peptides and peptide synthesis side-reaction adducts could both contribute to spurious false-positive responses in assays for cellular immune responses even when present in sparing quantities.
We recommend that a robust biochemical and biological QA/QC protocol (Table 2) be followed prior to the use of custom synthetic peptides in clinical trial protocols involving measurements of cell-mediated immunity. While important QA procedures such as careful peptide set design (2, 8, 9, 20), biochemical analysis, and selection of appropriate peptide solvent (18) are standard practice for most laboratories, real-world biological assay screening of synthetic peptide sets is not routinely conducted. The selection of an appropriate sample size for the biological screening of the peptides will be dependent upon the HLA diversity in the population studied. Obviously, suppliers and manufacturers must be alerted to the potential problem of peptide cross-contamination that can occur during synthesis and fine-tune their synthesis protocols to account for this problem. Current standard biochemical QA/QC analyses such as HPLC, mass spectrometry, and amino acid sequencing are still recommended, as they ensure that the vast majority of the synthesis product is indeed the correct peptide. Biological QA/QC would complement the biochemical QA/QC procedures (Table 2) and provide an explicit validation of both the peptide and any unavoidable impurities in the assay system to be used for a clinical trial or research protocol. In light of the potential peptide cross-contamination issues that we have described here, researchers should be advised to perform their own regular and robust QA/QC of synthetic peptides used for all research and clinical assay protocols.
TABLE 2.
Critical parameters and checkpoints in the biochemical and biological QA/QC of synthetic peptides prior to use in assays of cell-mediated immunity
| Stage of QC/QA | Common problem(s) | Proposed solution |
|---|---|---|
| Peptide set design | Difficult to synthesize peptides | Avoid problematic C- and N-terminal residues where possible |
| Length and overlap of peptides | 15-mer-18-mer-length peptides with an overlap of 11-12 amino acids are optimal for CD4 and CD8 responses | |
| Biochemical characterization | HPLC and MS reveal undesired impurities and side reaction products | Resynthesize peptides using a realistic, cost-effective cutoff for desired purity; may need to redesign or even omit particular peptides |
| Dissolution and storage | Peptide insolubility in aqueous buffers | 100% DMSO is the most universally applicable buffer for generic peptide solubility and is also compatible with downstream assay applications |
| Storage | Keep all resolved peptides at −80°C and minimize freeze-thaw cycles | |
| Biological characterization | Potential for inhibitory or stimulatory activity of individual or pooled peptides in downstream assay applications; this activity may not be predicted from the primary peptide sequence or from the biochemical analysis and requires empirical pretesting in the assay system of choice | Screen peptide pools in biological QA/QC assays using PBMC from as many as 50 subjects representing the HLA background in which the trial is to take place; screen for both inhibitory and stimulatory activity using the same assay to be implemented in the trial; deconvolute peptide pools to identify immunostimulatory or immunoinhibitory individual peptides; resynthesize and retest any problematic peptides |
Acknowledgments
We thank Kelly Smith, James Graziano, and Marvin Walker for expert technical assistance in performing the cellular immunology experiments, in peptide dissolution and pooling, and for synthetic peptide synthesis and analysis.
Financial support was provided by Department of Defense Collaborative agreement DAMD17-98-2-8007.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Departments of the Army and Defense.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beattie, T., R. Kaul, T. Rostron, T. Dong, P. Easterbrook, W. Jaoko, J. Kimani, F. Plummer, A. McMichael, and S. Rowland-Jones. 2004. Screening for HIV-specific T-cell responses using overlapping 15-mer peptide pools or optimized epitopes. AIDS 18:1595-1598. [DOI] [PubMed] [Google Scholar]
- 3.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, J. H., G. Ferrari, S. A. Kalams, W. Lopaczynski, N. Oden, and P. M. D'Souza. 2005. Results of an ELISPOT proficiency panel conducted in 11 laboratories participating in international human immunodeficiency virus type 1 vaccine trials. AIDS Res. Hum. Retrovir. 21:68-81. [DOI] [PubMed] [Google Scholar]
- 5.Currier, J. R., E. G. Kuta, E. Turk, L. B. Earhart, L. Loomis-Price, S. Janetzki, G. Ferrari, D. L. Birx, and J. H. Cox. 2002. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 260:157-172. [DOI] [PubMed] [Google Scholar]
- 6.Currier, J. R., U. Visawapoka, S. Tovanabutra, C. J. Mason, D. L. Birx, F. E. McCutchan, and J. H. Cox. 2006. CTL epitope distribution patterns in the Gag and Nef proteins of HIV-1 from subtype A infected subjects in Kenya: use of multiple peptide sets increases the detectable breadth of the CTL response. BMC Immunol. 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Beukelaar, J. W., J. W. Gratama, P. A. Smitt, G. M. Verjans, J. Kraan, T. M. Luider, and P. C. Burgers. 2007. The impact of impurities in synthetic peptides on the outcome of T-cell stimulation assays. Rapid Commun. Mass Spectrom. 21:1282-1288. [DOI] [PubMed] [Google Scholar]
- 8.Draenert, R., M. Altfeld, C. Brander, N. Basgoz, C. Corcoran, A. G. Wurcel, D. R. Stone, S. A. Kalams, A. Trocha, M. M. Addo, P. J. Goulder, and B. D. Walker. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19-29. [DOI] [PubMed] [Google Scholar]
- 9.Draenert, R., C. Brander, X. G. Yu, M. Altfeld, C. L. Verrill, M. E. Feeney, B. D. Walker, and P. J. Goulder. 2004. Impact of intrapeptide epitope location on CD8 T cell recognition: implications for design of overlapping peptide panels. AIDS 18:871-876. [DOI] [PubMed] [Google Scholar]
- 10.Findlay, J. W., W. C. Smith, J. W. Lee, G. D. Nordblom, I. Das, B. S. DeSilva, M. N. Khan, and R. R. Bowsher. 2000. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective. J. Pharm. Biomed. Anal. 21:1249-1273. [DOI] [PubMed] [Google Scholar]
- 11.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, T. M., S. A. Dubey, D. V. Mehrotra, D. C. Freed, W. L. Trigona, L. Adams-Muhler, J. H. Clair, T. G. Evans, R. Steigbigel, J. M. Jacobson, P. A. Goepfert, M. J. Mulligan, S. A. Kalams, C. Rinaldo, L. Zhu, K. S. Cox, L. Guan, R. Long, N. Persaud, M. J. Caulfield, J. C. Sadoff, E. A. Emini, S. Thaler, and J. W. Shiver. 2007. Evaluation of cellular immune responses in subjects chronically infected with HIV type 1. AIDS Res. Hum. Retrovir. 23:67-76. [DOI] [PubMed] [Google Scholar]
- 13.Geldmacher, C., J. R. Currier, M. Gerhardt, A. Haule, L. Maboko, D. Birx, C. Gray, A. Meyerhans, J. Cox, and M. Hoelscher. 2007. In a mixed subtype epidemic, the HIV-1 Gag-specific T-cell response is biased towards the infecting subtype. AIDS 21:135-143. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, L., G. Piccinini, D. Lilleri, M. G. Revello, Z. Wang, S. Markel, D. J. Diamond, and K. Luzuriaga. 2004. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8+ T cell responses in children with congenital or postnatal human cytomegalovirus infection. J. Immunol. 172:2256-2264. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, V. 2006. Market growing for custom-made peptides. GEN 26:38-40. [Google Scholar]
- 16.Hudgens, M. G., S. G. Self, Y. L. Chiu, N. D. Russell, H. Horton, and M. J. McElrath. 2004. Statistical considerations for the design and analysis of the ELISpot assay in HIV-1 vaccine trials. J. Immunol. Methods 288:19-34. [DOI] [PubMed] [Google Scholar]
- 17.Janetzki, S., J. H. Cox, N. Oden, and G. Ferrari. 2005. Standardization and validation issues of the ELISPOT assay. Methods Mol. Biol. 302:51-86. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson, R. K., W. Jennes, K. Page-Shafer, D. F. Nixon, and B. L. Shacklett. 2004. Poorly soluble peptides can mimic authentic ELISPOT responses. J. Immunol. Methods 285:89-92. [DOI] [PubMed] [Google Scholar]
- 19.Kern, F., I. P. Surel, C. Brock, B. Freistedt, H. Radtke, A. Scheffold, R. Blasczyk, P. Reinke, J. Schneider-Mergener, A. Radbruch, P. Walden, and H. D. Volk. 1998. T-cell epitope mapping by flow cytometry. Nat. Med. 4:975-978. [DOI] [PubMed] [Google Scholar]
- 20.Los Alamos National Laboratory. 2006. PeptGen: creates sets of overlapping peptides for proteins to aid in peptide design for mapping epitopes. HIV Molecular Immunology Database. http://www.hiv.lanl.gov/content/hiv-db/PEPTGEN/PeptGenSubmitForm.html.
- 21.Maecker, H., J. Moon, S. Bhatia, S. Ghanekar, V. Maino, J. Payne, K. Kuus-Reichel, J. Chang, A. Summers, T. Clay, M. Morse, H. K. Lyerly, C. DeLaRosa, D. Ankerst, and M. Disis. 2005. Impact of cryopreservation on tetramer, cytokine flow cytometry, and ELISPOT. BMC Immunol. 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maecker, H. T., H. S. Dunn, M. A. Suni, E. Khatamzas, C. J. Pitcher, T. Bunde, N. Persaud, W. Trigona, T. M. Fu, E. Sinclair, B. M. Bredt, J. M. McCune, V. C. Maino, F. Kern, and L. J. Picker. 2001. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J. Immunol. Methods 255:27-40. [DOI] [PubMed] [Google Scholar]
- 23.Mannering, S. I., A. W. Purcell, M. C. Honeyman, J. McCluskey, and L. C. Harrison. 2003. Human T-cells recognise N-terminally Fmoc-modified peptide. Vaccine 21:3638-3646. [DOI] [PubMed] [Google Scholar]
- 24.Moodie, Z., Y. Huang, L. Gu, J. Hural, and S. G. Self. 2006. Statistical positivity criteria for the analysis of ELISpot assay data in HIV-1 vaccine trials. J. Immunol. Methods 315:121-132. [DOI] [PubMed] [Google Scholar]
- 25.Najafian, N., A. D. Salama, E. V. Fedoseyeva, G. Benichou, and M. H. Sayegh. 2002. Enzyme-linked immunosorbent spot assay analysis of peripheral blood lymphocyte reactivity to donor HLA-DR peptides: potential novel assay for prediction of outcomes for renal transplant recipients. J. Am. Soc. Nephrol. 13:252-259. [DOI] [PubMed] [Google Scholar]
- 26.Ohnishi, M., T. Sakurai, Y. Heike, R. Yamazaki, Y. Kanda, Y. Takaue, H. Mizoguchi, and Y. Kawakami. 2005. Evaluation of cytomegalovirus-specific T-cell reconstitution in patients after various allogeneic haematopoietic stem cell transplantation using interferon-gamma-enzyme-linked immunospot and human leucocyte antigen tetramer assays with an immunodominant T-cell epitope. Br. J. Haematol. 131:472-479. [DOI] [PubMed] [Google Scholar]
- 27.Russell, N. D., M. G. Hudgens, R. Ha, C. Havenar-Daughton, and M. J. McElrath. 2003. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J. Infect. Dis. 187:226-242. [DOI] [PubMed] [Google Scholar]
- 28.Trigona, W. L., J. H. Clair, N. Persaud, K. Punt, M. Bachinsky, U. Sadasivan-Nair, S. Dubey, L. Tussey, T. M. Fu, and J. Shiver. 2003. Intracellular staining for HIV-specific IFN-gamma production: statistical analyses establish reproducibility and criteria for distinguishing positive responses. J. Interf. Cytok. Res. 23:369-377. [DOI] [PubMed] [Google Scholar]
- 29.Whiteside, T. L., Y. Zhao, T. Tsukishiro, E. M. Elder, W. Gooding, and J. Baar. 2003. Enzyme-linked immunospot, cytokine flow cytometry, and tetramers in the detection of T-cell responses to a dendritic cell-based multipeptide vaccine in patients with melanoma. Clin. Cancer Res. 9:641-649. [PubMed] [Google Scholar]
- 30.Wills, M. R., A. J. Carmichael, K. Mynard, X. Jin, M. P. Weekes, B. Plachter, and J. G. Sissons. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]