Abstract
Cecal samples from laying chickens from 25 farms with a history of decreased egg production, diarrhea, and/or increased feed conversion ratios were examined for anaerobic intestinal spirochetes of the genus Brachyspira. Seventy-three samples positive in an immunofluorescence assay for Brachyspira species were further examined using selective anaerobic culture, followed by phenotypic analysis, species-specific PCRs (for Brachyspira hyodysenteriae, B. intermedia, and B. pilosicoli), and a Brachyspira genus-specific PCR with sequencing of the partial 16S rRNA gene products. Brachyspira cultures were obtained from all samples. Less than half of the isolates could be identified to the species level on the basis of their biochemical phenotypes, while all but four isolates (5.2%) were speciated by using PCR and sequencing of DNA extracted from the bacteria. Different Brachyspira spp. were found within a single flock and also in cultures from single chickens, emphasizing the need to obtain multiple samples when investigating outbreaks of avian intestinal spirochetosis. The most commonly detected spirochetes were the pathogenic species B. intermedia and B. pilosicoli. The presumed nonpathogenic species B. innocens, B. murdochii, and the proposed “B. pulli” also were identified. Pathogenic B. alvinipulli was present in two flocks, and this is the first confirmed report of B. alvinipulli in chickens outside the United States. Brachyspira hyodysenteriae, the agent of swine dysentery, also was identified in samples from three flocks. This is the first confirmed report of natural infection of chickens with B. hyodysenteriae. Experimental infection studies are required to assess the pathogenic potential of these B. hyodysenteriae isolates.
Avian intestinal spirochetosis (AIS) is a disease complex that affects commercial laying and meat breeder chickens, characterized by the colonization of the ceca by members of one or more species of anaerobic intestinal spirochetes of the genus Brachyspira (formerly Serpulina) (11, 14, 36). The condition is associated with delayed or reduced egg production and a chronic diarrhea in adult birds that can result in wet litter and fecal staining of eggshells. Moreover, reduced performance has been reported for broiler chicks hatched from eggs of breeder chickens infected with Brachyspira spp. (7, 33).
AIS was first clearly described in the 1980s, in both The Netherlands (5, 7, 8) and the United Kingdom (10), and subsequently has been reported in the United States (39, 43), Australia (24, 35), and a number of other European countries (2, 3). The nonspecific nature of the disease and the lack of availability of simple and specific diagnostic tests for these anaerobic spirochetes from chickens may have resulted in the condition being greatly underreported (14).
Studies using multilocus enzyme electrophoresis showed that intestinal spirochetes from chickens could be divided into at least six species groupings (25). These included the three species currently considered to be pathogenic to chickens, Brachyspira pilosicoli, B. intermedia, and B. alvinipulli (13, 37, 34, 40), as well as B. innocens, B. murdochii, and the proposed species “B. pulli” (35) that are generally considered to be nonpathogenic in chickens (35, 36). In several studies, other unidentified Brachyspira isolates of unknown pathogenic significance also have been identified in chickens (25, 27, 38).
In both Europe and Australia, the two most frequently reported pathogenic Brachyspira species in cases of AIS have been B. intermedia and B. pilosicoli (2, 3, 7, 25, 35, 38), while in the United States only pathogenic B. pilosicoli and B. alvinipulli have been recorded to date (39, 40, 43). In recent years, there has been only one brief conference report (45) regarding the occurrence of Brachyspira spp. infections in commercial poultry in The Netherlands; hence, the purpose of the current study was to determine which species are most commonly encountered in this region in poultry flocks with symptoms consistent with AIS. After selective anaerobic culture, isolates were identified using phenotypic traits and PCR-based methods. Significant new findings included the identification of the porcine pathogen B. hyodysenteriae (the agent of swine dysentery) in chickens from three flocks as well as the first identification of B. alvinipulli in chickens in Europe.
MATERIALS AND METHODS
Source of samples.
The cecal samples that were examined originated from laying chickens from 25 flocks with symptoms associated with AIS and were submitted to the Animal Health Service (GD), Deventer, The Netherlands, for diagnostic purposes. Live chickens were stunned using CO2 plus O2 and exsanguinated, and a general routine postmortem examination was performed. The cecal contents initially were subjected to an indirect immunofluorescence antibody test (IFAT) for the detection of Brachyspira spp., and then 73 samples positive by IFAT were selected for further examination. Forty-three of the 73 samples were derived from laying chickens on 23 farms located throughout The Netherlands, including farms with different breeds and housing systems. The housing systems included cage housing (n = 2), aviary housing (n = 2), floor housing without free-range access (n = 12), and free-range housing (n = 7). The age and flock size varied from 24 to 80 weeks of age and 5,800 to 60,000 chickens, respectively. All farms had a history of decreased egg production, diarrhea, and/or increased feed conversion. Single samples were collected from three to six chickens from the first six flocks, and then single samples were taken from the next 17 flocks. The submitted chickens were between 24 and 80 weeks of age.
Another 30 samples were collected from live birds submitted from two other farms with epidemiological links (farms 24 and 25). The samples from farm 24 were derived from 1, 4, and 17 chickens collected at 24, 48, and 56 weeks of age, respectively, while the samples from farm 25 were from 8 chickens of 50 weeks of age. On farm 24, there was reduced egg production and eggshell quality and slightly increased mortality (0.15%). Additionally, there was an above-average number of eggs with fecal staining of the eggshells due to wet feces. Egg production and eggshell quality on farm 25 also were decreased. Both farms were situated in Germany just across the Dutch borders (1 to 2 km) but were owned by a Dutch poultry farmer. They had the same caretaker and well for drinking water and were located 500 m from each other. On both farms, brown-layer hens of the same breed were housed in an aviary system with a free-range area. The free-range areas of the two farms were adjacent to each other. The farms housed 25,452 and 20,971 chickens, respectively. The flocks were reared at different farms and had different origins. Fattening pigs were housed within 300 m of both farms, while a swamp with waterfowl was present within 800 m of both farms.
IFAT.
A routine diagnostic IFAT for Brachyspira spp. was performed on a smear of the contents of one cecum from each of the sampled chickens. The other cecum was kept in sterile water and was used for Brachyspira culture within 24 h if the IFAT was positive. The general method for IFAT was carried out as described previously (20). Briefly, smears on glass slides were air dried and fixed in acetone for 5 min and then were incubated at 37°C for 30 min with a unabsorbed Brachyspira-specific antiserum raised in a rabbit by repeated intramuscular injection with a formalinized bacterin produced from a Dutch B. hyodysenteriae strain (CIDC, Lelystad, The Netherlands). The serum had been shown to cross-react with other Brachyspira spp. Subsequently, the smears were subjected to three 2-min washes in phosphate-buffered saline (PBS), incubated for 30 to 45 min with goat anti-rabbit fluorescein isothiocyanate-labeled conjugate (Nordic, Tilburg, The Netherlands), and again subjected to three 2-min washes in PBS. The slides were dried and examined for positive apple-green fluorescent spirochetes using a UV fluorescence microscope (DM2000; Leica Microsystems) at ×100 to ×400 magnification. Negative and positive control samples were examined with each batch of slides.
Spirochete culture.
Cecal contents were stirred with a sterile cotton-tipped swab that was used to inoculate two selective Trypticase soy agar (TSA) plates (BBL Microbiology Systems, Cockeysville, MD). One plate was supplemented with spectinomycin (200 μg/ml), spiromycin (25 μg/ml), rifampin (12.5 μg/ml), vancomycin (6.25 μg/ml), colistin (6.25 μg/ml), 0.1% yeast extract (Oxoid, Basingstoke, United Kingdom), and 5% defibrinated ovine blood (22), and the other plate was supplemented with spectinomycin (400 μg/ml), vancomycin (25 μg/ml), colistin (25 μg/ml), and 5% defibrinated ovine blood (18). The reference strains B. hyodysenteriae ATCC 27146 and B. pilosicoli ATCC 51139 each were used as positive controls on duplicate plates. The plates were incubated in an anaerobic jar under H2 and CO2 generated using an anaerogen gaspak (Oxoid) for 4 to 7 days at 42°C. If spirochetal growth was suspected, a piece of agar was removed and used to inoculate an agar plate supplemented with 5% defibrinated sheep blood (SBA) (Biotrading, Meidrecht, The Netherlands), which subsequently was incubated anaerobically at 42°C for 2 to 3 days. The purity and the presence of spirochetes were examined by Gram staining. In the case of pure cultures, an agar fragment was used to subculture the isolate onto four SBA plates and one TSA plate (without antibiotics). One SBA plate was used to harvest the spirochetes in a peptone-glycerol medium that was stored at −80°C. Only pure cultures were used for biochemical typing. In cases for which a pure culture could not be obtained (after repeating the procedure twice), the culture was harvested directly from the SBA plate. A total of 73 culture-positive samples were stored frozen at −80°C.
Phenotypic characteristics.
Selected phenotypic characteristics of pure primary isolates from the original samples, and of three secondary cultures from frozen samples from farm 24, were determined using the methods described by Hommez and others (15). The presence and extent of β-hemolysis was observed on the original SBA plate after it was further incubated for 2 days. Spirochetes from the TSA plates were tested for indole by the spot test, while those from three of the SBA plates were harvested and pooled for the determination of α-galactosidase, α-glucosidase, β-glucosidase, and hippurate activity. The biochemical identification of pure cultures was performed as described for porcine isolates by Hommez and others (15), but with the addition of data for “B. pulli” and B. alvinipulli (34, 40). The phenotypic definitions used to identify the species are shown in Table 1.
TABLE 1.
Phenotypic properties used for identification of Brachyspira species
| Species | Groupa | Hemolysis | Biochemical reactivity with:
|
||||
|---|---|---|---|---|---|---|---|
| Indole | α-Galactosidase | α-Glucosidase | β-Glucosidase | Hippurate | |||
| B. hyodysenteriae | I | Strong | +/− | − | +/− | + | − |
| B. intermedia | II | Weak | + | − | + | + | − |
| B. murdochii | III | Weak | − | − | +/− | + | − |
| B. innocens | IIIb | Weak | − | + | − | + | − |
| IIIc | Weak | − | + | + | + | − | |
| B. pilosicoli | IV | Weak | − | +/− | − | − | + |
| “B. pulli” | Weak | − | + | +/− | + | ? | |
| B. alvinipulli | Weak | − | +/− | − | + | + | |
Biochemical definitions of the first five species are as listed by Hommez and others (15), with the definition for “B. pulli” derived from McLaren and others (25) and the definition for B. alvinipulli derived from Swayne and others (40) and Stanton and others (34). The group definitions are from Fellström and others (9).
PCR tests and 16S rRNA gene sequencing.
DNA was extracted from the Brachyspira cells present on SBA plates using the QIAamp DNA stool Midi kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Initially, the extracted DNA was subjected to three previously described species-specific PCR tests for B. pilosicoli, B. intermedia, and B. hyodysenteriae (23, 28). The PCR primers for B. pilosicoli were designed to amplify an 823-bp region of the 16S rRNA gene, while the primers for both B. hyodysenteriae and B. intermedia were based on the amplification of species-specific NADH oxidase (nox) gene sequences of 354 and 567 bp, respectively. All of the DNA samples also were subjected to a Brachyspira genus-specific PCR based on the amplification of a 1,309-bp portion of the 16S rRNA gene (27). Twenty samples that were negative by the species-specific PCRs for B. pilosicoli and B. intermedia, and the five samples that were positive by the B. hyodysenteriae PCR, also were used for partial 16S rRNA gene sequencing, as previously described (27). Three samples were not sequenced. The nucleotide sequences obtained were compared to known sequences available in GenBank. An approximately 1,240-bp gene sequence for each isolate was incorporated individually into a Brachyspira phylogenetic tree, and based on their clustering in the tree, the sequences were assigned to known Brachyspira species. The tree was constructed using partial 16S rRNA gene sequences obtained from GenBank that represented the known Brachyspira species (Fig. 1). These sequences were from five strains of B. pilosicoli; four strains each of B. hyodysenteriae, B. intermedia, B. innocens, B. murdochii, B. aalborgi, “B. suanatina” (porposed name), “B. canis” (proposed name), and “B. pulli”; and three strains of B. alvinipulli. The phylogenetic tree was constructed using ClustalX (41) and Mega 3.1 (21).
FIG. 1.
Partial 16S rRNA gene phylogenetic tree representing the Brachyspira genus that was used to identify Brachyspira isolates that were negative in species-specific PCRs. The tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates. Accession numbers for each sequence used for the construction are shown. Species are distinct, except for B. innocens and B. murdochii, which tend to cluster together, and B. hyodysenteriae and B. intermedia, which also cluster together.
Six samples that were positive in the B. hyodysenteriae nox PCR and/or that showed a 16S rRNA gene sequence consistent with that of B. hyodysenteriae then were tested in another confirmatory PCR used for the specific amplification of B. hyodysenteriae tlyA genomic sequences (10).
Histopathology and immunohistochemistry.
For chickens from flocks 24 and 25, besides using cecal samples for culture and PCR, cecal tissue was obtained within 10 to 15 min of death for histopathology and immunohistochemistry. Tissue from the cecal wall was fixed in 4% formalin, paraffin embedded, and processed for histology. Sections were stained with hematoxylin and eosin. Sections were pretreated by incubation in a solution of methanol-H2O2 to neutralize endogenous peroxidase activity prior to immunohistochemistry. Samples then were incubated in the same rabbit polyclonal antiserum against Brachyspira that was used in the IFAT, rinsed in PBS, incubated with peroxidase-labeled goat anti-rabbit antibodies as described by the manufacturer (Nordic, Tilburg, The Netherlands), rinsed, and then incubated with 3,3′-diaminobenzidine tetrahydrochloride-H2O2 (Sigma-Aldrich). All sections were viewed at ×100 to ×400 magnification.
RESULTS
Spirochete cultures.
Anaerobic spirochetes presumed to be Brachyspira spp. were isolated from all 73 samples that were positive by IFAT.
Phenotypic identification of 43 isolates from 23 farms.
Only 16 (37%) of the 43 Brachyspira isolates from the first 23 farms could be identified to the species level on the basis of their phenotypic properties (Table 2). These comprised isolates of the nonpathogenic B. innocens (n = 7), B. murdochii (n = 6), and “B. pulli” (n = 2) and a single isolate of the potentially pathogenic B. pilosicoli.
TABLE 2.
Results of phenotype, species-specific PCRs, and partial 16S rRNA gene sequence analysis for identifying 43 Brachyspira spp. isolates from 23 chicken farms
| Farm no. | Chicken age (wk) | Flock size | Housing type | Sample no. | Species identification by:
|
||
|---|---|---|---|---|---|---|---|
| Phenotypea | Species-specific PCRb | 16S rRNA gene sequencingc | |||||
| 1 | 24 | 5,800 | Floor housing | 1 | NId | B. pilosicoli | NDe |
| 2 | NI | NI | B. pilosicoli | ||||
| 3 | NI | B. intermedia and B. pilosicoli | ND | ||||
| 4 | NI | B. pilosicoli | ND | ||||
| 2 | 40 | 17,000 | Floor housing | 5 and 6 | NI | B. intermedia | ND |
| 7 | NI | B. pilosicoli | ND | ||||
| 8 | B. murdochii | NI | “B. pulli” | ||||
| 3 | 28 | 25,500 | Floor housing+i | 9 | NI | NI | “B. pulli” |
| 10 | B. innocens | NI | NDg | ||||
| 11 | B. innocens | NI | “B. pulli” | ||||
| 12 | NI | NI | B. innocens/B. murdochii cluster | ||||
| 13 | B. innocens | NI | B. innocens/B. murdochii cluster | ||||
| 14 | NI | B. pilosicoli | ND | ||||
| 4 | 44 | 9,000 | Floor housing+ | 15 | “B. pulli” | B. intermedia and B. pilosicoli | ND |
| 16 | NI | B. intermedia and B. pilosicoli | ND | ||||
| 17 | NI | B. intermedia | ND | ||||
| 18 | NI | B. pilosicoli | ND | ||||
| 19 | NI | B. pilosicoli and B. hyodysenteriae | B. pilosicolih | ||||
| 5 | NAf | 18,000 | Floor housing | 20 | NI | NI | B. hyodysenteriae/B. intermedia cluster |
| 21 | NI | B. intermedia and B. pilosicoli | ND | ||||
| 22 | NI | B. intermedia | ND | ||||
| 6 | 36 | 9,000 | Floor housing | 23 | NI | B. intermedia | ND |
| 24 | B. innocens | B. intermedia | ND | ||||
| 25 | B. murdochii | B. intermedia | ND | ||||
| 26 | B. pilosicoli | B. pilosicoli | ND | ||||
| 7 | 52 | 33,500 | Floor housing | 27 | NI | NI | “B. pulli” |
| 8 | 35 | 15,000 | Cage | 28 | NI | B. intermedia | NI |
| 9 | 28 | 23,000 | Floor housing | 29 | “B. pulli” | B. pilosicoli | ND |
| 10 | 60 | 19,000 | Floor housing+ | 30 | NI | B. pilosicoli | ND |
| 11 | 68 | 8,000 | Floor housing+ | 31 | NI | B. intermedia | ND |
| 12 | 36 | 12,550 | Aviary | 32 | NI | NI | NDg |
| 13 | 64 | 60,000 | Aviary | 33 | B. murdochii | B. intermedia | ND |
| 14 | 64 | 15,000 | Floor housing | 34 | NI | B. intermedia | ND |
| 15 | NA | NA | Floor housing | 35 | B. murdochii | B. intermedia | ND |
| 16 | 32 | 29,000 | Cage | 36 | B. innocens | NI | B. innocens/B. murdochii cluster |
| 17 | NA | 20,000 | Floor housing | 37 | NI | NI | B. innocens/B. murdochii cluster |
| 18 | NA | 24,000 | Floor housing | 38 | B. innocens | B. intermedia | ND |
| 19 | 80 | 19,000 | Floor housing | 39 | B. innocens | B. intermedia | ND |
| 20 | NA | NA | Floor housing+ | 40 | murdochii | B. pilosicoli | ND |
| 21 | 44 | 12,000 | Floor housing+ | 41 | NI | NI | “B. pulli” |
| 22 | 28 | 30,000 | Floor housing | 42 | murdochii | NI | “B. pulli” |
| 23 | 29 | 21,000 | Floor housing+ | 43 | NI | B. pilosicoli | ND |
Phenotype identifications are based on hemolysis and biochemical analysis (Table 1).
Individual species-specific PCRs for B. pilosicoli, B. intermedia, and B. hyodysenteriae.
Only samples negative in the species-specific PCRs or positive in the B. hyodysenteriae PCR were subjected to partial 16S rRNA gene sequencing.
NI, not identified biochemically and/or by PCR, or no pure culture was available for biochemical identification.
ND, not done.
NA, not available.
Samples 10 and 32 were not sequenced, because they were negative in the genus-specific PCR.
Also positive in the supplementary B. hyodysenteriae tlyA PCR.
The plus indicates floor housing with free-range access.
Genus- and species-specific PCRs on 43 isolates from 23 farms.
Forty-one of the 43 DNA samples prepared from Brachyspira isolates from the first 23 farms were positive in the general Brachyspira 16S rRNA gene PCR. Two samples (samples 10 and 32) were negative, both in the general PCR and in the species-specific PCRs, possibly due to a too-low DNA concentration and poor DNA quality. In total, 29 of the samples could be identified by at least one of the species-specific Brachyspira PCRs (Table 2). These included 14 cultures identified as B. intermedia, 10 as B. pilosicoli, four as a mixture of B. intermedia and B. pilosicoli, and one as a mixture of B. pilosicoli and B. hyodysenteriae. The tlyA PCR for B. hyodysenteriae also was positive on the latter sample (sample 19). For five of the first six farms for which multiple isolates were examined, both B. intermedia and B. pilosicoli were present in each farm, while B. pilosicoli alone was identified in the samples from farm 3.
Partial 16S rRNA gene sequencing of isolates from 23 farms.
Twelve samples that were positive in the general PCR and negative in the species-specific PCR and one sample that was positive for both B. hyodysenteriae and B. pilosicoli (sample 19) were subjected to partial 16S rRNA gene sequencing. For sample 19, a sequence consistent with B. pilosicoli was obtained. Of the others, four were identified as belonging to the B. innocens/B. murdochii cluster, six as “B. pulli,” and one as B. pilosicoli, and one was consistent with the B. hyodysenteriae/B. intermedia cluster (Table 2).
Phenotypic identification of isolates from farms 24 and 25.
Of 33 isolates (including three secondary cultures) from farms 24 and 25 (Table 3), six isolates, all from farm 24, had the phenotype of B. hyodysenteriae (including two of the three secondary cultures). Nine isolates, including one subculture, were identified as B. murdochii, three as B. innocens, and two each as B. intermedia and B. pilosicoli; 11 isolates could not be identified.
TABLE 3.
Results of identification based on phenotype, species-specific PCRs, and partial 16S rRNA gene sequence analysis for identifying 33 Brachyspira isolates (including three secondary cultures) from farms 24 and 25
| Farma | Sample no. | Species identification by:
|
||
|---|---|---|---|---|
| Phenotypeb | Species-specific PCRc | 16S rRNA gene sequenced | ||
| 24 | 44 | B. hyodysenteriae | B. hyodysenteriae | NDf |
| 44 (secondary culture) | B. hyodysenteriae | B. hyodysenteriae | B. hyodysenteriae/B. intermedia clusterh | |
| 45 | NIe | B. intermedia | ND | |
| 46 | NI | NI | B. pilosicoli | |
| 47 | NI | B. pilosicoli | ND | |
| 48 | B. hyodysenteriae | B. hyodysenteriae | B. hyodysenteriae/B. intermedia clusterh | |
| 48 (secondary culture) | B. hyodysenteriae | B. hyodysenteriae | B. alvinipullih | |
| 49-50 | B. intermedia | B. intermedia | ND | |
| 51 | NI | NI | B. alvinipulli | |
| 52-57 | NI | B. pilosicoli | ND | |
| 58 | NI | B. intermedia + B. pilosicoli | ND | |
| 59 | B. murdochii | NI | B. alvinipulli | |
| 60 | B. innocens | B. pilosicoli | ND | |
| 61-62 | B. pilosicoli | B. pilosicoli | ND | |
| 63 | B. murdochii | NI | B. alvinipulli | |
| 64 | B. hyodysenteriae | NI | B. hyodysenteriae/B. intermedia clusteri | |
| 65 | B. hyodysenteriae | NI | B. alvinipulli | |
| 65 (secondary culture) | B. murdochii | NI | B. alvinipulli | |
| 25 | 66-69 | B. murdochii | NI | B. alvinipulli |
| 70 | B. murdochii | NI | B. murdochii/B. innocens cluster | |
| 71 | B. murdochii | NI | NDg | |
| 72 | B. innocens | B. hyodysenteriae | B. alvinipullih | |
| 73 | B. innocens | NI | Brachyspira species | |
For farm 24, sample 1 was the first submission, samples 45 to 48 were the second submission, and samples 49 to 65 were the third submission.
Phenotype identifications are based on hemolysis and biochemical analysis (Table 1).
Individual species-specific PCRs for B. pilosicoli, B. intermedia, and B. hyodysenteriae.
Only samples that were negative in the species-specific PCRs or that were B. hyodysenteriae positive were subjected to partial 16S rRNA gene sequencing.
NI, not identified biochemically and/or by PCR, or no pure culture was available for biochemical identification.
ND, not done.
Sample 71 was not sequenced, because it was negative in the genus-specific PCR.
Tested positive in the additional B. hyodysenteriae tlyA PCR.
Tested negative in the additional B. hyodysenteriae tlyA PCR.
Genus- and species-specific PCRs on isolates from farms 24 and 25.
Only one DNA sample was negative in the Brachyspira genus-specific PCR (sample 71). Five of the 33 isolates (including two secondary cultures) were positive for B. hyodysenteriae in the species-specific nox PCR, including one culture from farm 25 (Table 3). Ten cultures were identified as B. pilosicoli, three as B. intermedia, and one as a mixture of B. intermedia and B. pilosicoli; the other 11 samples were PCR negative. B. intermedia and B. pilosicoli were found only on farm 24. Of the five isolates from these two farms tested in the B. hyodysenteriae tlyA PCR (Table 3), only sample 64 was negative.
Partial 16S rRNA gene sequencing on isolates from farms 24 and 25.
Sequencing was undertaken on six isolates identified as B. hyodysenteriae either by phenotypic analysis or PCR testing, as well as on 11 isolates that did not amplify in the species-specific PCRs but were positive in the general PCR. Three of the six isolates identified as B. hyodysenteriae clustered in the B. hyodysenteriae/B. intermedia group on the 16S rRNA gene tree, while the other three clustered with B. alvinipulli. Of the other 12 isolates, 8 were identified as B. alvinipulli, 1 as B. pilosicoli, and 1 as B. murdochii/B. innocens, and one (sample 73) did not cluster with any species in the tree.
Postmortem findings in chickens from farms 24 and 25.
All but one chicken had productive ovaries at postmortem. A macroscopic examination showed the presence of localized enteritis with necrotic lesions in the duodenum in 12 of the chickens. A bacteriological examination suggested that the local enteritis in the duodenum was caused by a Clostridium perfringens infection. The presence of coccidia and other enteric parasites was ruled out by microscopic examination. Five chickens had unusual foamy cecal contents, but only one had obvious typhlitis.
Histopathology and immunohistochemistry.
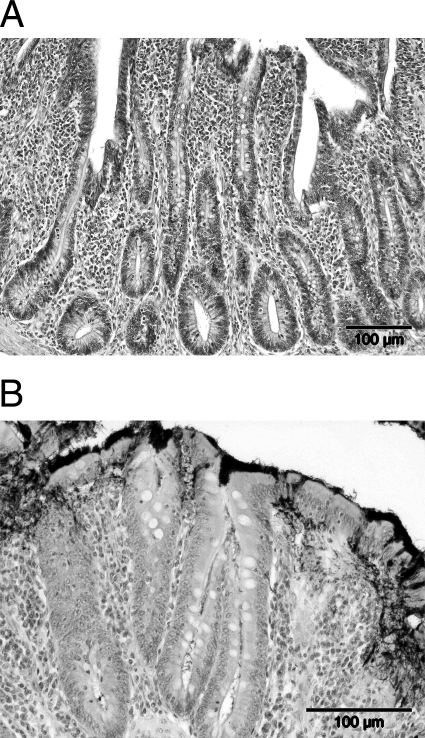
Cecal tissue from the chickens showed reactive and mild inflammatory changes morphologically consistent with Brachyspira infections. The changes were characterized by crypt hyperplasia, epithelial erosion, and increased numbers of goblet cells (Fig. 2A). Furthermore, mild to moderate infiltrates of macrophages and heterophils were observed in the lamina propria. In one sample from farm 24, which contained B. alvinipulli and showed typhlitis at postmortem, focal necrosis and necrotic material containing spirochetes was present in the lumen. Also, oocysts were found within the necrotic core.
FIG. 2.
Histopathological changes in a chicken colonized with Brachyspira pilosicoli. (A) Section of the cecal wall showing crypt hyperplasia, increased numbers of goblet cells, and mild to moderate infiltrates of macrophages and heterophils in the lamina propria. (B) Immunoperoxidase staining showing a dense fringe of spirochetes on the surface of the cecal enterocytes and penetration of spirochetes between cells.
Spirochetes were observed in three different locations in the samples: (i) forming a false border and frequently penetrating between enterocytes, sometimes to the level of the lamina propria (Fig. 2B), (ii) in the crypts in low numbers, and (iii) in the crypts in large numbers and penetrating into the lamina propria.
DISCUSSION
This study provides a description of the occurrence of different Brachyspira spp. in flocks of commercial laying chickens in the region serviced by the Animal Health Service (GD) in The Netherlands. Although it cannot be regarded as a detailed survey, particularly as only single isolates were examined from 17 of the 25 flocks, it does point to the fact that Brachyspira spp. are frequently present in chickens from laying flocks with production problems and diarrhea in this region.
In relation to the methodology used for identifying Brachyspira spp., it was evident that biochemical identification was problematic due to the frequent presence of mixed spirochete species (and possibly strains) and/or contamination of cultures with other bacteria. Only a minority of spirochete isolates could be identified by their biochemical profiles, and in many cases the phenotypic identity did not correspond to the species identified by PCR or by the clustering observed in the 16S rRNA gene phylogenetic tree. In part, this can be explained by the fact that the phenotypic identification systems originally were set up for the Brachyspira spp. isolated from pigs (9). Chickens are colonized by a greater range of Brachyspira spp. than are pigs, including a number of unidentified or new species groupings (25, 30, 38). Furthermore, typical biochemical profiles for B. alvinipulli and “B. pulli” have not yet been defined using large panels of isolates from these species. In this study, it was clear from the PCR results that quite a few of the individual cultures contained mixed species. Such mixtures would make determining biochemical reactivities problematic. In contrast, mixed infections of individuals with different Brachyspira spp. appear to occur less commonly in pigs (29). To help address the problems of mixed cultures and the presence of bacterial contamination, in future studies it will be necessary to be more careful in subculturing different colonies to purity, including, if necessary, making a series of limiting dilutions of the primary inocula before plating them out. Brachyspira spp. tend to form low flat confluent areas of growth on most agar media; however, it is possible to obtain individual colonies for subculturing by using fastidious anaerobe agar with 10% horse blood (29). The use of this sort of medium for subculturing is recommended for future studies.
The PCR methods that were used greatly improved the identification of species compared to the results of the examination of phenotypic properties. There were two discrepancies between the results of the species-specific PCRs and the 16S rRNA gene sequencing (for samples 19 and 72), but these could be explained by the likely presence of mixed species in the cultures tested. The sequence from the more common strain in a mixed culture likely would be amplified for sequencing at the expense of the less common strain. Despite the relatively small number of samples examined overall, seven different named or proposed Brachyspira spp. and many unidentified isolates were detected. Besides the presumed nonpathogenic species B. murdochii, B. innocens, and “B. pulli,” the potentially pathogenic species B. pilosicoli, B. intermedia, and B. alvinipulli were identified. B. alvinipulli was identified only by using partial 16S rRNA gene sequencing, as a diagnostic PCR for this species has not been developed to date. The seventh species identified was B. hyodysenteriae, which was confirmed by a minimum of two independent species-specific PCR tests (for nox and tlyA) in three samples from flock 24, one from flock 25, and one from flock 4. Two of the confirmed positive isolates from flock 24 (44 and 48) also had the typical phenotypic properties of B. hyodysenteriae.
The results show that B. intermedia and B. pilosicoli either alone or in combination are the two most common pathogenic species in Dutch laying flocks with clinical symptoms consistent with AIS. These observations agree with the results of several studies from other countries (2, 3, 35, 38, 42). In addition, in the examination of larger numbers of chickens from flocks 24 and 25, B. alvinipulli was identified. This species was first identified and described as a pathogen in laying chickens in the United States (34, 39, 40). It subsequently has been reported in Hungary in two large goose flocks that had severe fibrinonecrotic typhlitis (26), and there are recent brief reports of this species occurring in laying chickens in the United Kingdom (42) and Sweden (16). Our results confirm that B. alvinipulli is present as a potential pathogen in laying chickens in countries outside the United States and emphasize the importance of considering this species in cases of AIS. The findings also point to the need for the development of an improved species-specific PCR for the identification of B. alvinipulli in diagnostic samples from chickens.
Brachyspira hyodysenteriae is the etiological agent of swine dysentery, a severe mucohemorrhagic colitis of growing and fattening pigs that is still common and important in many swine-rearing countries (12). This spirochete also naturally infects and causes typhlocolitis in rheas (Rhea americana) (19, 31). More recently, strongly β-hemolytic B. hyodysenteriae was isolated from mallards in Sweden (17). There has been a brief unconfirmed report mentioning the isolation of B. hyodysenteriae from Dutch commercial poultry (45) and a very recent brief report of it being isolated from laying chickens in the United Kingdom (42). The current study clearly shows that B. hyodysenteriae can naturally infect laying chickens.
The experimental infection of day-old chicks with B. hyodysenteriae isolates from pigs has caused a necrotic typhlitis (1, 44). Although it appears likely that B. hyodysenteriae also has pathogenic potential in adult chickens, currently there is no experimental evidence available to support this. Clearly, it will be important to use our B. hyodysenteriae isolates for experimental infections of adult chickens to determine the extent of any pathological changes they may induce.
The original source of the B. hyodysenteriae isolates infecting the chickens was not established, but in the case of the chickens on farms 24 and 25, infection may have come from pigs on the nearby swine farm or from waterfowl in the local swampland. As the survival time of B. hyodysenteriae in pig manure can be up to 48 days (4), hypothetically pig (and duck) manure cannot be excluded as a potential epidemiological factor.
Knowledge of the origin and transmission routes of the spirochete are important, as this may help in devising means to reduce its spread to chicken farms. Unfortunately, it was not possible to sample the neighboring pigs or the waterfowl to determine if they were infected. It seems likely that transmission of B. hyodysenteriae and/or B. alvinipulli between the two neighboring farms (farms 24 and 25) also may have occurred. Movement between the two farms by the caretaker, contact between chickens across the adjacent free-range areas, or a contaminated common water supply could have enabled the spread of these spirochetes between the farms.
All the farms investigated had clinical and production problems consistent with AIS. Flocks with AIS have been reported to show reduced egg production, diarrhea, increased fecal fat content, fecal staining of eggshells, increased feed consumption, and/or maldigestion (5, 8, 11, 35, 39, 43). Nevertheless, in the current study it was difficult to attribute the presence of any one species or set of spirochete species to the particular problems in the farms, especially as many of the farms were shown to have, or may have had, mixed Brachyspira species infections. Moreover, other potential pathogens affecting the gastrointestinal tract were detected in chickens from these farms (e.g., coccidia, nematodes, infectious bronchitis virus, and Clostridium perfringens). Only the chickens from farms 24 and 25 were subjected to histologic examination of their ceca, and the chickens from both farms showed the presence of different pathogenic Brachyspira spp. in combination with enteric disease. Histology and immunohistochemistry of cecal tissue from both farms in general showed reactive and mild inflammatory changes that were morphologically consistent with Brachyspira spp. in chickens and in other animal species (5, 6, 32). Focal necrosis was seen in one chicken, and in some ceca spirochetes were attached to cecal enterocytes and/or penetrating the epithelium as far as the lamina propria. The occurrence of clinical signs and histologic lesions in chickens from those two farms suggests a causality with the pathogenic Brachyspira strains detected. Nevertheless, potentially pathogenic Brachyspira spp. have been found previously in flocks without clinical signs, suggesting that predisposing factors can be necessary for the full clinical expression of AIS (35, 36). On the other hand, even the presumed nonpathogenic species B. murdochii has been associated with clinical problems on certain farms (35, 38). Arguably, preexisting enteritis (for example, associated with coccidiosis) might be a predisposing factor that enhances the pathogenicity of some of the Brachyspira species found in these chickens.
In conclusion, a number of different pathogenic Brachyspira spp. were widely distributed within and across different Dutch commercial laying chicken farms with clinical symptoms consistent with AIS. Besides potentially pathogenic B. intermedia, B. pilosicoli, and B. alvinipulli, laying chickens were naturally infected with B. hyodysenteriae. Further work is required to study the epidemiology of AIS in general and to assess the pathogenic potential of B. hyodysenteriae isolates in laying chickens.
Acknowledgments
This research was supported by a grant from the National Board for Poultry and Eggs (PPE) of The Netherlands.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Adachi, Y., M. Sueyoshi, E. Miyagawa, H. Minato, and S. Shoya. 1985. Experimental infection of young broiler chicks with Treponema hyodysenteriae. Microbiol. Immunol. 296834-6838. [DOI] [PubMed] [Google Scholar]
- 2.Bano, L., G. Merialdi, P. Bonilauri, G. Dall'Anese, K. Capello, D. Comin, V. Cattoli, V. Sanguinetti, and F. Agnoletti. 2005. Prevalence of intestinal spirochaetes in layer flocks in Treviso province, Northern Italy, p. 56-57. In Proceedings of the 3rd International Conference on Colonic Spirochaetal Infections in Animals and Humans, University of Parma, Parma, Italy.
- 3.Burch, D. G. S., C. Harding, R. Alvarez, and M. Valks. 2006. Treatment of a field case of avian intestinal spirochaetosis caused by Brachyspira pilosicoli with tiamulin. Avian Pathol. 35211-216. [DOI] [PubMed] [Google Scholar]
- 4.Chia, S. P., and D. J. Taylor. 1978. Factors affecting the survival of Treponema hyodysenteriae in dysenteric pig faeces. Vet. Rec. 10368-78. [DOI] [PubMed] [Google Scholar]
- 5.Davelaar, F. G., H. F. Smit, K. Hovind-Hougen, R. M. Dwars, and P. C. van der Valk. 1986. Infectious typhlitis in chickens caused by spirochaetes. Avian Pathol. 15247-258. [DOI] [PubMed] [Google Scholar]
- 6.Duhamel, G. E. 2001. Comparative pathology and pathogenesis of naturally acquired and experimentally induced colonic spirochaetosis. Anim. Health Res. Rev. 23-17. [PubMed] [Google Scholar]
- 7.Dwars, R. M., F. G. Davelaar, and H. F. Smit. 1993. Infection of broiler parent hens (Gallus domesticus) with avian intestinal spirochaetes: effects on egg production and chick quality. Avian Pathol. 22693-701. [DOI] [PubMed] [Google Scholar]
- 8.Dwars, R. M., H. F. Smit, F. G. Davelaar, and W. T. van Veer. 1989. Incidence of spirochaetal infections in cases of intestinal disorder in chickens. Avian Pathol. 18591-595. [DOI] [PubMed] [Google Scholar]
- 9.Fellström, C., B. Petterson, M. Ulen, A. Gunnarsson, and K. E. Johansson. 1995. Phylogeny of Serpulina based on sequence analysis of the 16S rRNA gene and comparison with a scheme based on biochemical classification. Res. Vet. Sci. 595-9. [DOI] [PubMed] [Google Scholar]
- 10.Fellström, C., U. Zimmerman, A. Aspan, and A. Gunnarsson. 2001. The use of culture, pooled samples and PCR for identification of herds infected with Brachyspira hyodysenteriae. Anim. Health Res. Rev. 237-43. [PubMed] [Google Scholar]
- 11.Griffiths, I. B., B. W. Hunt, S. A. Lister, and M. H. Lamont. 1987. Retarded growth rate and delayed onset of egg production associated with spirochaete infection in pullets. Vet. Rec. 12135-37. [DOI] [PubMed] [Google Scholar]
- 12.Hampson, D. J., C. Fellström, and J. R. Thomson. 2006. Swine dysentery, p. 785-805. In B. E. Straw, J. J. Zimmerman, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, 9th ed. Blackwell Publishing, Oxford, United Kingdom.
- 13.Hampson, D. J., and A. J. McLaren. 1999. Experimental infection of laying hens with Serpulina intermedia causes reduced egg production and increased faecal water content. Avian Pathol. 28113-117. [DOI] [PubMed] [Google Scholar]
- 14.Hampson, D. J., and D. E. Swayne. 2006. Avian intestinal spirochetosis. In Y. M. Saif (ed.), Diseases of poultry, 12th ed. Blackwell Publishing, Oxford, United Kingdom.
- 15.Hommez, J., F. Castryck, F. Haesebrouck, and L. A. Devriese. 1998. Identification of porcine Serpulina strains in routine diagnostic bacteriology. Vet. Microbiol. 2163-169. [DOI] [PubMed] [Google Scholar]
- 16.Jansson, D. S., C. Fellström, T. Råsbäck, A. Gunnarsson, and K.-E. Johansson. 2007. Characterization of Brachyspira spp. from Swedish laying hens, abstr. 37. In Proceedings of the 4th International Conference on Colonic Spirochaetal Infections in Animals and Humans, Prague, the Czech Republic.
- 17.Jansson, D. S., K. E. Johansson, T. Olofsson, T. Råsbäck, I. Vågsholm, B. Pettersson, A. Gunnarsson, and C. Fellström. 2004. Brachyspira hyodysenteria and other strongly β-haemolytic and indole-positive spirochaetes isolated from mallards (Anas platyrhynchos). J. Med. Microbiol. 53293-300. [DOI] [PubMed] [Google Scholar]
- 18.Jenkinson, S. R., and C. R. Wingar. 1981. Selective medium for the isolation of Treponema hyodysenteriae. Vet. Rec. 109384-385. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, N. S., T. B. Stanton, and D. E. Swayne. 1996. Identification of the swine pathogen Serpulina hyodysenteriae in rheas (Rhea americana). Vet. Microbiol. 52259-269. [DOI] [PubMed] [Google Scholar]
- 20.Kiernan, J. A. 1990. Histological and histochemical methods: theory and practice, p. 337-339, 2nd ed. Pergamon Press, Oxford, United Kingdom.
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 22.Kunkle, R. A., and J. M. Kinyon. 1988. Improved selective medium for the isolation of Treponema hyodysenteriae. J. Clin. Microbiol. 262357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La, T., N. D. Phillips, and D. J. Hampson. 2003. Development of a duplex PCR assay for detection of Brachyspira hyodysenteriae and Brachyspira pilosicoli in pig feces. J. Clin. Microbiol. 413372-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren, A. J., D. J. Hampson, and S. J. Plant. 1996. The prevalence of intestinal spirochaetes in commercial poultry flocks in Western Australia. Aust. Vet. J. 7431-33. [DOI] [PubMed] [Google Scholar]
- 25.McLaren, A. J., D. J. Trott, D. E. Swayne, S. L. Oxberry, and D. J. Hampson. 1997. Genetic and phenotypic characterization on intestinal spirochetes colonizing chickens, and allocation of known pathogenic isolates to three distinct genetic groups. J. Clin. Microbiol. 35412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemes, C. S., R. Glávits, M. Dobos-Kovács, E. Ivanics, E. Kaszanyyitzky, A. Beregszászi, L. Szeredi, and L. Dencsō. 2006. Typhlocolitis associated with spirochaetes in goose flocks. Avian Pathol. 354-11. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, N. D., T. La, and D. J. Hampson. 2005. A cross-sectional study to investigate the occurrence and distribution of intestinal spirochaetes (Brachyspira spp.) in three flocks of laying hens. Vet. Microbiol. 105189-198. [DOI] [PubMed] [Google Scholar]
- 28.Phillips, N. D., T. La, and D. J. Hampson. 2006. Development of a two-step nested duplex PCR assay for the rapid detection of Brachyspira pilosicoli and Brachyspira intermedia in chicken faeces. Vet. Microbiol. 116239-245. [DOI] [PubMed] [Google Scholar]
- 29.Råsbäck, T., C. Fellström, A. Gunnarsson, and A. Aspen. 2006. Comparison of culture and biochemical tests with PCR for detection of Brachyspira hyodysenteriae and Brachyspira pilosicoli. J. Microbiol. Methods 66347-353. [DOI] [PubMed] [Google Scholar]
- 30.Råsbäck, T., D. S. Jansson, K. E. Johansson, and C. Fellström. 2007. A novel enteropathogenic, strongly haemolytic spirochaete isolated from pig and mallard, provisionally designated “Brachyspira suanatina” sp. nov. Environ. Microbiol. 9983-991. [DOI] [PubMed] [Google Scholar]
- 31.Sagartz, J. E., D. E. Swayne, K. E. Eaton, J. R. Hayes, K. D. Amess, R. Wack, and L. Kramer. 1992. Necrotizing typhlocolitis associated with a spirochete in rheas (Rhea americana). Avian Dis. 36282-289. [PubMed] [Google Scholar]
- 32.Shibahara, T., A. Kuwano, T. Ueno, Y. Katayama, T. Ohya, T. Taharaguchi, S. Yamamoto, T. Umemura, Y. Ishikawa, and K. Kadota. 2005. Immunohistochemical and ultrastructural detection of intestinal spirochetes in thoroughbred horses. J. Vet. Diagn. Investig. 17145-150. [DOI] [PubMed] [Google Scholar]
- 33.Smit, H. F., R. M. Dwars, F. G. Davelaar, and G. A. W. Wijtten. 1998. Observations on the influence of intestinal spirochaetosis in broiler breeders on the performance of their progeny and egg production. Avian Pathol. 27133-141. [DOI] [PubMed] [Google Scholar]
- 34.Stanton, T. B., D. Postic, and N. S. Jensen. 1997. Serpulina alvinipulli sp. nov., a new Serpulina species enteropathogenic to chickens. Int. J. Syst. Bacteriol. 471007-1012. [DOI] [PubMed] [Google Scholar]
- 35.Stephens, C. P., and D. J. Hampson. 1999. Prevalence and disease association of intestinal spirochaetes in chickens in eastern Australia. Avian Pathol. 28447-454. [DOI] [PubMed] [Google Scholar]
- 36.Stephens, C. P., and D. J. Hampson. 2001. Intestinal spirochaete infections of chickens: a review of disease associations, epidemiology and control. Animal Health Res. Rev. 283-91. [PubMed] [Google Scholar]
- 37.Stephens, C. P., and D. J. Hampson. 2002. Experimental infection of broiler breeder hens with the intestinal spirochaete Brachyspira (Serpulina) pilosicoli causes reduced egg production. Avian Pathol. 31169-175. [DOI] [PubMed] [Google Scholar]
- 38.Stephens, C. P., S. L. Oxberry, N. D. Phillips, T. La, and D. J. Hampson. 2005. The use of multilocus enzyme electrophoresis to characterise intestinal spirochaetes (Brachyspira spp.) colonising hens in commercial flocks. Vet. Microbiol. 107149-157. [DOI] [PubMed] [Google Scholar]
- 39.Swayne, D. E., A. J. Bermudez, J. A. Sagartz, K. A. Eaton, J. D. Montfort, J. W. Stoutenberg, and J. R. Hayes. 1992. Association of cecal spirochetes with pasty vents and dirty eggshells in layers. Avian Dis. 36776-781. [PubMed] [Google Scholar]
- 40.Swayne, D. E., K. A. Eaton, J. Stoutenburg, D. J. Trott, D. J. Hampson, and N. S. Jenssen. 1995. Identification of a new intestinal spirochaete with pathogenicity to chickens. Infect. Immun. 63430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomson, J. R., B. P. Murray, L. E. Henderson, J. Thacker, and D. G. S. Burch. 2007. Brachyspira species isolated from UK poultry samples, abstr. 39. In Proceedings of the 4th International Conference on Colonic Spirochaetal Infections in Animals and Humans, Prague, the Czech Republic.
- 43.Trampel, D. W., N. S. Jensen, and L. J. Hoffman. 1994. Cecal spirochetosis in commercial laying hens. Avian Dis. 38895-898. [PubMed] [Google Scholar]
- 44.Trott, D. J., and D. J. Hampson. 1998. Evaluation of day-old specific pathogen-free chicks as an experimental model for pathogenicity testing of intestinal spirochaete species. J. Comp. Pathol. 118265-381. [DOI] [PubMed] [Google Scholar]
- 45.Wagenaar, J. A., M. A. P. Van Bergen, I. Van der Graaf, and W. J. M. Landman. 2003. Free-range chickens show a higher incidence of Brachyspira infections in The Netherlands, abstr. 16. In Proceedings of the 2nd International Conference on Colonic Spirochaetal Infections in Animals and Humans, Barony Castle, Eddleston, Scotland, United Kingdom.