Abstract
Three strains of a novel Bartonella species (Bartonella tamiae) were isolated from human patients from Thailand. Sequence analysis of six chromosomal regions (16S rRNA, gltA, groEL, ftsZ, rpoB, and the intergenic spacer region) and phenotypical analysis supported the similarity of the three strains and placed them within the genus Bartonella separately from previously described species.
Several species of the genus Bartonella cause numerous disorders, and most are thought to be zoonoses (1, 3). These Bartonella-associated illnesses occur worldwide, including in Asia, and they encompass a broad clinical spectrum, including fever, skin lesions, lymphadenopathy, endocarditis, and abnormalities of the central nervous system, liver, eye, and bone tissues (5). There has been a report indicating that humans are being exposed to Bartonella species in Thailand, though the investigators did not isolate the agents (7).
We report the characterization of three Bartonella strains isolated from blood samples of patients from Khon Kaen Province, Thailand, that belong to a novel Bartonella species. These strains were identified during the screening of blood clot specimens from a prospective study performed to determine the etiology of febrile illnesses in Thailand. It is the first report of culture-confirmed Bartonella infection in humans in Thailand.
Blood clots were separated from sera, stored at −70°C, and shipped to the U.S. CDC (Fort Collins, CO) for testing. Two approaches were used for the isolation of Bartonella from human clots: (i) blood clots were cocultivated with Vero E6 cells at 35°C with 5% carbon dioxide for 7 days and then subcultured onto rabbit blood-enriched agar and (ii) blood clots were inoculated into a preenrichment liquid, the Bartonella-Alphaproteobacteria growth medium (BAPGM) developed by Maggi et al. (6), and after 7 days of incubation at 35°C with 5% carbon dioxide were plated onto rabbit blood agar. The agar plates were incubated at 35°C with an aerobic atmosphere of 5% carbon dioxide for up to 30 days. The cultured bacteria were visualized with Gram stain by using standard light microscopy with an oil immersion objective at a magnification of 1,000×. For negative staining, a drop of suspension was placed on a copper grid coated with Formvar-carbon film and allowed to adhere for 10 min. The grids with adherent bacteria were stained by placing them on a drop of 2% potassium-phosphotungstic acid and air dried. The grids were examined with a Phillips 201 electron microscope. For ultrathin sectioning, the bacterial suspension was fixed in a mixture of 2.5% formaldehyde, 0.1% glutaraldehyde, 0.03% trinitrophenol, and 0.03% CaCl2 in 0.05 M cacodylate buffer (pH 7.2). Ultrathin sections were cut with a Leica-Reichert Ultracut S ultramicrotome, stained with 2% aqueous uranyl acetate and lead citrate, and examined with a Phillips 201 electron microscope.
The MicroScan rapid anaerobe identification panel (Dade Behring, Inc., West Sacramento, CA) was used to test the activities of preformed bacterial enzymes in accordance with the manufacturer's instructions on the preparation, incubation, and interpretation of the test results. For the study of antibiotic susceptibility, microbial suspensions were prepared according to a 0.5 McFarland standard from 5-day-old agar cultures and diluted 10-fold. Eight antibiotics (penicillin, cefotaxime, gentamicin, erythromycin, clindamycin, doxycycline, ciprofloxacin, and rifampin) diluted in Columbian agar supplemented with 5% sheep blood were tested. Results were read at the third and fifth days postinoculation.
Bacterial DNA was heat extracted at 95°C for 10 min from whole bacterial cells. Oligonucleotide primers were used for the amplification of single regions of the Bartonella citrate synthase (gltA), the cell division protein (ftsZ), the RNA polymerase beta-subunit (rpoB), the heat shock protein (groEL), and 16S rRNA genes, as well as the 16S-to-23S rRNA intergenic spacer (ITS) region. Positive and negative controls were included in each PCR to evaluate the presence of appropriately sized amplicons and possible contamination. Each PCR was conducted with a PTC-200 Peltier automated thermal cycler (MJ Research, Waltham, MA). PCR products were analyzed for the presence of amplicons of the correct size by electrophoresis of 5 μl of the product in 1.5% agarose gels containing ethidium bromide. Amplicons of the expected size were identified by size comparison to the positive control, and the resultant PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germantown, MD) and sequenced in both directions using the same primers that were used for the PCR assay. Sequencing reactions were carried out with a PTC-200 Peltier thermal cycler, using the Quick Start dye terminator cycle sequencing kit (Beckman Coulter, Fullerton, CA).
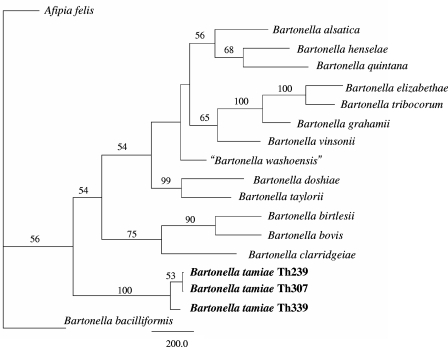
Sequences were analyzed using Lasergene sequence analysis software (DNAStar, Madison, WI). The SeqMan program (DNAStar) was used to determine consensus sequences for the amplified region of the target genes, and the Clustal V program within Megalign (DNAStar) was used to align and compare homologous sequences. The sequences were then analyzed using PAUP 4.0 (Center for Biodiversity, Illinois Natural History Survey, Champaign, IL), and phylogenies were constructed using the maximum parsimony algorithm. To improve the statistical support of the phylogeny of the Bartonella genus, partial nucleotide sequences of six chromosomal regions (16S rRNA, 1,216 bp [final sequence length]; ftsZ, 788 bp; the ITS region, 1,076 bp; gltA, 323 bp; groEL, 825 bp; and rpoB, 1,209 bp) from 17 Bartonella strains were aligned and trimmed individually using MEGA version 3.1 and then concatenated in a multilocus sequence typing approach (Fig. 1). Multiple-sequence alignments were analyzed using the PAUP 4.0 software program, and phylogenies were constructed by using a distance-based likelihood method with the GTR+I+G DNA substitution model, as chosen by Modeltest v3.1. Trees were rooted using Afipia felis as the outgroup; because it was intended to be used solely for rooting the tree and not for phylogenetic comparison, only the 16S rRNA gene of A. felis was used. Missing sequences were treated as missing data.
FIG. 1.
Phylogenetic tree showing the positions of strains Th239, Th307, and Th339 among members of the genus Bartonella based on comparisons of concatenated sequences of the following six genes: the 16S rRNA gene, the citrate synthase gene gltA, the RNA polymerase beta-subunit gene rpoB, the cell division gene ftsZ, the heat shock protein genes groEL, and the 16S-to-23S rRNA ITS region sequences. The 16S rRNA gene from Afipia felis was included for outgroup comparison. Bootstrap values strongly support the position of these strains in a novel clade within the genus Bartonella; however, the values do not support separating the three strains into distinct species. Trees were constructed using a maximum likelihood-based distance algorithm and a GTR+I+G DNA substitution model using PAUP software. Numbers on branches indicate the bootstrap values derived from 500 replications. The bar indicates the number of nucleotide changes.
Strain Th239 was isolated from a 38-year-old male patient who was admitted to the hospital with fatigue, myalgia, a headache, a maculopapular rash that had lasted for 22 days, and a fever that had lasted for 6 days. Strain Th239 was obtained by the cocultivation of blood with Vero E6 cells. After 7 days, the suspension was found to be positive by PCR amplification of a Bartonella-specific fragment of the gltA gene. Subsequently, the inoculated Vero E6 cells were subcultured on rabbit blood agar, and very small colonies appeared on the agar 17 days postinoculation. The growth rate increased after several sequential subculturing passages of the collected bacterial suspension. After each passage, the suspensions of the Vero E6 cells were PCR positive for amplification of the targeted piece of the gltA gene.
Strain Th307 was obtained from a 41-year-old female admitted to the hospital with an initial diagnosis of a pterygium in each eye. The strain was isolated after the inoculation of the patient's blood clot into preenrichment BAPGM. After a 7-day incubation, the suspension was found to be positive for Bartonella organisms by PCR using the gltA gene as a target. The inoculated medium was then placed on rabbit blood agar and incubated at 35°C with 5% CO2. Bartonella-like colonies were observed after 5 days of cultivation and confirmed to be positive by PCR.
Strain Th339 was obtained from a 12-year-old male patient admitted to the hospital with fever, fatigue, myalgia, a headache, and a petechial rash on his arms and legs that had lasted for 2 days. The strain was isolated after the inoculation of the patient's blood clot into preenrichment BAPGM and a 10-day incubation. Inoculated medium was then placed on rabbit blood agar and incubated at 35°C with 5% CO2. Bartonella-like colonies were observed on the agar plate after an additional 12 days.
BLAST searches indicated that all sequences of the 16S rRNA, gltA, groEL, ftsZ, rpoB, and the ITS region of strains Th239, Th307, and Th339 are closely related to the homologous sequences of various Bartonella species and unnamed Bartonella strains. Phylogenetic analyses based on the parsimony method (heuristic search), neighbor joining, and MegAlign alignments of all sequences (16S rRNA, gltA, groEL, ftsZ, rpoB, and the ITS region) supported the novelty of the new isolate and suggested a distant phylogenetic lineage in the genus Bartonella. Phylogenetic analysis of a concatenated, multiple-sequence alignment of the 16S rRNA, gltA, ftsZ, groEL, rpoB, and ITS region nucleotide sequences using a distance-based maximum likelihood algorithm supported the placement of the new isolates within a distinct phylogenetic lineage of the Bartonella genus (Fig. 1).
The structures of phylogenetic trees and the percentages of divergence between the sequences of the 16S ribosomal genes of strains Th239, Th307, and Th339 and sequences in the GenBank nucleic acid database demonstrated that these strains were close to representatives of the genus Bartonella. The genetic distances between the isolates and representatives of the genus Brucella, which is the taxonomic group closest to Bartonella, were evidently greater than the distances between these isolates and other Bartonella species. The 16S rRNA gene sequence analysis indicated that these three strains represent a distant phylogenetic lineage in the genus Bartonella. The 16S rRNA sequences of strains Th239, Th307, and Th339 exhibited the closest phylogenetic relationship with three sequences of three uncultured Bartonella species clones (pAJ203 [AY370185], pAJ208 [AY370186], and pAJ210 [AY370187]) obtained from the honeybee Apis mellifera (4).
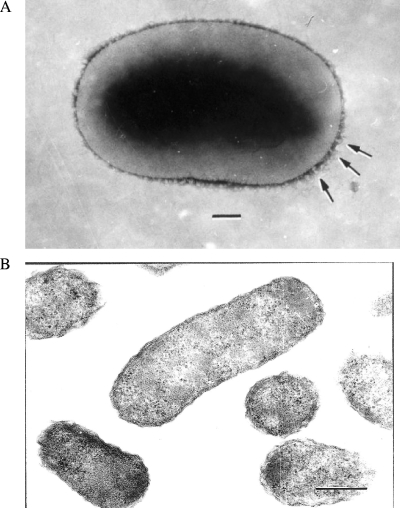
Because of the genetic similarity among the strains, only one strain, Th239, was used for further phenotypical characterization. Gram staining of isolate Th239 revealed rod-shaped, gram-negative bacilli that were small, straight, and slightly curved. Transmission electron microscopy of negatively stained bacteria showed small, rounded rods ranging from 0.7 to 1.2 μm in length and 0.5 to 0.6 μm in width. Some bacteria had bundles of fibrils approximately 15 nm thick and 30 nm in length with a periodicity ranging from 30 to 120 nm and with polar distribution (Fig. 2A). Examination of ultrathin sections revealed typical gram-negative morphology with a wavy cell wall. The bacteria appeared to be approximately 0.3 μm by 0.8 μm (Fig. 2B). Therefore, the sizes of the organisms, taking into account measurements from both ultrathin sections and negative staining, appear to range from 0.3 to 0.6 μm in width and from 0.7 to 1.2 μm in length.
FIG. 2.
Ultrastructure of Bartonella tamiae (strain Th239). (A) Strain Th239 after gram-negative staining showing bundles of fibrils closer to its pole (arrows). Bar = 250 nm. (B) Ultrathin section of bacterial suspension showing typical gram-negative cell walls of Bartonella tamiae organisms. Bar = 250 nm.
Most of the biochemical properties of strain Th239 were typical for bacteria of the Bartonella genus. Specifically, enzymatic hydrolysis results were negative for p-nitrophenyl-beta-d-galactopyranoside, p-nitrophenyl-alpha-d-galactopyranoside, p-nitrophenyl- N-acetyl-beta-d-glucosaminide, p-nitrophenyl-alpha-d-glucopyranoside, o-nitrophenyl-beta-d-glucopyranoside, p-nitrophenyl-alpha-l-fucopyranoside, p-nitrophenyl-alpha-d-mannopyranoside, l-proline-beta-naphthylamide, l-pyrrolidonyl-beta-naphthylamide, trehalose, urea, indole, and nitrate. Positive reactions were observed in the enzymatic hydrolysis of bis-p-nitrophenyl-phosphate, p-nitrophenyl-phosphate, l-leucine-beta-naphthylamide, l-lysine-beta-naphthylamide (acid and alkaline), dl-methionine-beta-naphthylamide, glycylglycine-beta-naphthylamide, glycine-beta-naphthylamide, l-arginine-beta-naphthylamide, l-tryptophane-beta-naphthylamide, and 3-indoxyl-phosphate. The unique property of strain Th239 in contrast to other Bartonella strains was its 3-indoxylphosphate activity.
The following MICs of antibiotics were observed with strain Th239: penicillin, 4 μg/ml; cefotaxime, >4 μg/ml; gentamicin, 0.5 μg/ml; erythromycin, >4 μg/ml; clindamycin, 32 μg/ml; doxycycline, 4 μg/ml; ciprofloxacin, 8 μg/ml; and rifampin, 4 μg/ml.
Our findings support the possibility of Bartonella strains being causes of human disease in Thailand. Two of the three patients were febrile, and all three had clinical and laboratory findings similar to those found for patients infected with other forms of bartonellosis (5). Mild anemia observed in all patients is presumed to have resulted from the infection of red blood cells, and headache, myalgia, and abnormalities in liver function, consistent features in these three patients, are commonly identified in other Bartonella infections.
All three patients reported trapping or killing rats in their houses, and two of them reported recent rat exposures within the 2 weeks prior to the onset of illness. Further investigation is needed to determine the animal reservoir and any possible vectors for B. tamiae. Previous investigations of rodents of northern Thailand demonstrated that 9% of the tested animals were Bartonella culture positive (2), and phylogenetic analysis indicated a high diversity of the Bartonella strains obtained from Thai rodents. Also, Bartonella species have been identified in cats and fleas from Thailand (8, 9). However, to date, no homologous mammalian or flea Bartonella sequences phylogenetically similar to B. tamiae have been identified.
Based on a combination of genetic and phenotypic characteristics, we consider these described strains to be representatives of a novel Bartonella species. Bartonella tamiae (tam.i′ae. N.L. fem. gen. n. tamiae, of Tami) is the name proposed to honor the late Tamara (Tami) Fisk, who organized the febrile illness study in Thailand from which this bacterial species originated. The prototype strain (Th239) isolated from the blood of a Thai patient has been deposited in the American Type Culture Collection (ATCC BAA-1343), the National Collection of Type Cultures (United Kingdom, NCTC 13398), and the Japan Collection of Microorganisms (JCM 14580).
Nucleotide sequence accession numbers.
Sixteen unique nucleotide sequences were identified among the obtained three isolates. The three gltA sequences were assigned the GenBank accession numbers DQ395177 (strain TH239), EF605279 (strain TH307), and EF605280 (strain TH339). The ftsZ sequences were assigned the accession numbers DQ395178 (TH239), EF605281 (TH307), and EF605282 (TH339). The 16S-to-23S ribosomal ITS region sequences were assigned the accession numbers DQ395180 (TH239), EF605283 (TH307), and EF605284 (TH339). The groEL sequence was assigned the GenBank accession number DQ395179 (TH239). The rpoB sequences were assigned the accession numbers EF091855 (TH239), EF605285 (TH307), and EF672730 (TH339). The novel 16S ribosomal gene sequences were assigned the accession numbers DQ395176 (TH239), EF672728 (TH307), and EF672729 (TH339).
Acknowledgments
This project was supported by the International Emerging Infections Program and the Global Disease Detection Network of the U.S. Centers for Disease Control and Prevention.
For their support and collaboration, we are indebted to Khanchit Limpakarnjanarat of the IEIP; Akradeth Pensiri, Chawalit Nilvarangkul, Kamol Srilom, and Somjit Dechasatien of the Kohn Kaen Provincial Health Office; Kasem Pataralitikun and the staff at Nong Song Hong Hospital; and Aumporn Ratanaparinya and the staff at Kranuan Crown Prince Hospital.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Anderson, B. E., and M. A. Neuman. 1997. Bartonella spp. as emerging human pathogens. Clin. Microbiol. Rev. 10203-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castle, K. T., M. Kosoy, K. Lerdthusnee, L. Phelan, Y. Bai, K. L. Gage, W. Leepitakrat, T. Monkakka, N. Khlaimanee, K. Chandranoi, J. W. Jones, and R. E. Coleman. 2004. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am. J. Trop. Med. Hyg. 70429-433. [PubMed] [Google Scholar]
- 3.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 98-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeyaprakash, A., M. A. Hoy, and M. H. Allsopp. 2003. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J. Invertebr. Pathol. 8496-103. [DOI] [PubMed] [Google Scholar]
- 5.Koehler, J. E. 1996. Bartonella infections. Adv. Pediatr. Infect. Dis. 111-27. [PubMed] [Google Scholar]
- 6.Maggi, R. G., A. W. Duncan, and E. B. Breitschwerdt. 2005. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J. Clin. Microbiol. 432651-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruyama, S., S. Boonmar, Y. Morita, T. Sakai, S. Tanaka, F. F. Yamaguchi, H. Kabeya, and Y. Katsube. 2000. Seroprevalence of Bartonella henselae and Toxoplasma gondii among healthy individuals in Thailand. J. Vet. Med. Sci. 62635-637. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama, S., T. Sakai, Y. Morita, S. Tanaka, H. Kabeya, S. Boonmar, A. Poapolathep, T. Chalarmchaikit, C. Chang, R. Kasten, B. Chomel, and Y. Katsube. 2001. Prevalence of Bartonella species and 16S rRNA gene types of Bartonella henselae from domestic cats in Thailand. Am. J. Trop. Med. Hyg. 65783-787. [DOI] [PubMed] [Google Scholar]
- 9.Parola, P., O. Y. Sonogo, K. Lerdthusnee, Z. Zeaiter, G. Chauvancy, J. P. Gonzalez, R. S. Miller, S. R. Telford III, C. Wongsrichanalai, and D. Raoult. 2003. Identification of Rickettsia spp. and Bartonella spp. in fleas from Thai-Myanmar border. Ann. N. Y. Acad. Sci. 990173-181. [DOI] [PubMed] [Google Scholar]